Usher Syndrome 1D and Nonsyndromic Autosomal Recessive Deafness DFNB12 Are Caused by Allelic Mutations of the Novel Cadherin-Like Gene CDH23 (original) (raw)
Abstract
Genes causing nonsyndromic autosomal recessive deafness (DFNB12) and deafness associated with retinitis pigmentosa and vestibular dysfunction (USH1D) were previously mapped to overlapping regions of chromosome 10q21-q22. Seven highly consanguineous families segregating nonsyndromic autosomal recessive deafness were analyzed to refine the DFNB12 locus. In a single family, a critical region was defined between D10S1694 and D10S1737, ∼0.55 cM apart. Eighteen candidate genes in the region were sequenced. Mutations in a novel cadherin-like gene, CDH23, were found both in families with DFNB12 and in families with USH1D. Six missense mutations were found in five families with DFNB12, and two nonsense and two frameshift mutations were found in four families with USH1D. A northern blot analysis of CDH23 showed a 9.5-kb transcript expressed primarily in the retina. CDH23 is also expressed in the cochlea, as is demonstrated by polymerase chain reaction amplification from cochlear cDNA.
Introduction
Usher syndrome (USH) is an autosomal recessive disorder characterized by sensorineural hearing loss and retinitis pigmentosa. There are three clinical subtypes that can be differentiated on the basis of the severity of auditory and vestibular dysfunction and the age at onset of retinitis pigmentosa: USH1, characterized by congenital profound hearing loss, vestibular areflexia, and prepubertal onset of retinitis pigmentosa; USH2, associated with moderate-to-severe hearing loss, normal vestibular function, and onset of retinitis pigmentosa during the 2d decade of life; and USH3, characterized by progressive hearing loss, variable vestibular function, and adult-onset retinitis pigmentosa (Smith et al. 1994). At least six loci for USH1 (A–F) have been mapped (Kaplan et al. 1992; Kimberling et al. 1992; Smith et al. 1992; Wayne et al. 1996, 1997; Chaib et al. 1997), and two of the USH1 genes have been identified. Mutations in MYO7A cause USH1B (MIM 276903 [Weil et al. 1995]), and mutations in USH1C have recently been shown to underlie USH1C (MIM 605242 [Bitner-Glindzicz et al. 2000; Verpy et al. 2000]).
Nonsyndromic deafness is also genetically heterogeneous. At present, 38 autosomal dominant and 30 recessive nonsyndromic-deafness loci have been mapped, and 10 genes underlying autosomal dominant hearing loss and 7 genes underlying autosomal recessive hearing loss have been identified (Morell 1999; Willems 2000; E. R. Wilcox, Q. L. Burton, S. Naz, S. Riazuddin, T. N. Smith, I. Belyantseva, T. Ben-Yosef, N. A. Liburd, R. J. Morell, B. Kachar, D. K. Wu, A. J. Griffith, S. Riazuddin, and T. B. Friedman, unpublished data). One of the unidentified loci for autosomal recessive nonsyndromic deafness, DFNB12, had been mapped to 10q21-q22 (MIM 601386 [Chaib et al. 1996]), where its critical region was overlapped by the 15-cM interval that subsequently defined the USH1D region (MIM 601067 [Wayne et al. 1996]).
We have refined the genetic interval of the DFNB12 locus and have screened 18 candidate genes in the linked region for mutations, in families with DFNB12 and USH1D. We demonstrate that DFNB12 and USH1D are allelic disorders caused by mutations in a novel cadherin-like gene, CDH23.
Subjects and Methods
Family Enrollment and Diagnosis
Numerous consanguineous families segregating autosomal recessive, profound, congenital deafness were ascertained, including nine Pakistani families (304, PKSR13a, PKSR3, PKSR46a, PKSR5b, PKSR7a, PKZA3, PKZA46, and PKZA56), two Indian families (INJ5 and 5020A-AN), and two families of Pakistani and Turkish descent, respectively, that were living in England (USH05 and USH08). These 13 families demonstrated linkage to 10q21-q22. In addition to audiometry, participating family members underwent medical history interviews, physical examination (including fundoscopy), and, in a subset of patients, electroretinography. Family 304, the original family with USH1D, cosegregated symptoms and signs of retinitis pigmentosa with hearing loss (Wayne et al. 1996). Appropriate institutional-review board approval and informed consent were obtained for this study.
Linkage Analysis
Genomic DNA was extracted from blood samples by either a phenol-extraction protocol (Grimberg et al. 1989) or a Puregene DNA-isolation kit (Gentra Systems); in some cases, buccal epithelial DNA was prepared (Epicenter Technologies). Samples were genotyped for markers flanking known DFNB loci, by use of marker information provided by the Hereditary Hearing Loss Homepage. Short tandem-repeat polymorphisms (STRPs) were amplified with fluorescent primer pairs synthesized on an Applied Biosystems 394 DNA/RNA synthesizer (PE Biosystems) and were analyzed using standard procedures (Lalwani et al. 1999). For family 5020A-AN, pools of genomic DNA from affected and from nonaffected persons were screened with 169 STRPs evenly spaced across the human autosomal genome (Screening set 8A; Research Genetics).
LOD scores were calculated by either the FASTLINK version of the LINKAGE program package (Lathrop and Lalouel 1984; Schaffer 1996; Lab of Statistical Genetics, Rockefeller University) or MAPMAKER/HOMOZ (Kruglyak et al. 1995; Whitehead Insitute for Biomedical Research, Center for Genome Research FTP). The disease-allele frequency was set at .00075, on the basis of estimates in the Indian population (Zbar et al. 1998), and the disease was coded as fully penetrant and autosomal recessive. An allele frequency of .1 for each allele and a 1/1,000 phenocopy rate were assumed. Linkage to markers at 10q21-q22 was observed in each of these families. Linkage analysis for family 304 has been reported elsewhere (Wayne et al. 1996).
Candidate Gene Screening
Candidate genes in the DFNB12 interval were screened by direct sequencing of PCR products generated from genomic DNA of one affected individual from each family. Primers were designed to amplify all exons of each candidate gene, including the intron-exon boundaries. Primer pairs used to sequence exons of CDH23 from genomic DNA are listed in the Appendix. Genomic DNA was amplified by PCR in a 20-μl reaction volume. Reactions contained 20–50 ng of genomic DNA, 5 pmol each of forward and reverse primer, 200 mM each dNTP, 1× PCR buffer (PE Biosystems), 1.5–2.5 mM MgCl2 (PE Biosystems), and 0.5 U of thermostable polymerase. The thermal cycling conditions were as follows: 95°C for 2 min; 35 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 45 s; and a final extension at 72°C for 15 min. Five microliters of the PCR reaction products were analyzed by 2.0% agarose gel electrophoresis. The remaining 15 μl were treated with 0.3 U of shrimp alkaline phosphatase (Amersham) and 3 U of exonuclease I (USB) at 37°C for 1 h, followed by 80°C for 15 min, then were diluted with 1 vol of dH2O. Six microliters were then used in 10-μl sequencing reactions containing 3.2 pmol of primer, 2 μl of Big Dye Terminator Ready Reaction mix (PE Biosystems), and 1 μl of a dilution buffer (400 mM Tris-HCl pH 9.0 and 10 mM MgCl2). Cycling conditions were 96°C for 2 min and 45 cycles of 96°C for 10 s, 55°C for 10 s, and 60°C for 4 min. DNA-sequencing reactions were ethanol precipitated with NaOAc and Pellet Paint NF Co-Precipitant (Novagen) and were resuspended in 2–5 μl of formamide loading dye. Products were separated on 4.25% DNA-sequencing PAGE gels, were visualized, and were analyzed using a PE Biosystems ABI Prism 377 DNA Sequencer and ABI Prism software. The resulting chromatograms were analyzed using the PHRED/PHRAP/CONSED software suite (Ewing et al. 1998; Gordon et al. 1998).
Mutations were confirmed by sequence analysis of DNA from all affected individuals of a given family. Random normal control DNA samples from either Pakistan or India were also analyzed, depending on the ethnic origin of the family (table 1), as were panethnic control samples from The Human Diversity Panel (Coriell Cell Repositories).
Table 1.
Family Data, Mutations of CDH23 (cadherin-23), and Allele Frequencies
| Allele Frequency in | ||||||
|---|---|---|---|---|---|---|
| Family | Phenotype(s)a | LOD Scoreb | Mutation(s) | Protein Domain | Control Population | HD Panelc |
| PKZA56 | A, B, C | 1.62 | Q492X | EC5 | 0/96d | 0/182 |
| 304e | A, B, C | >3.0f | IVS23+1G→A | EC12 | 0/96d | 0/64 |
| PKSR46a | A | 3.96 | D1243N | 0/158d | 0/160 | |
| USH05 | A, B, C | 4.11f | R1305X | EC13 | 0/78d | 0/62 |
| PKSR5b | A | 2.56 | D1400N | EC14 | 0/78d | 0/62 |
| PKZA3 | A | 2.31 | D188N | EC2 | 0/126d | 0/64 |
| INJ5 | A | 3.28 | I2148N, R2154C | MPED | 0/96,g 0/48d | 0/96 |
| 5020A-AN | A | 2.53f | P2257T | Prox to TM | 0/96g | 0/64 |
| PKSR7a | A, D | 4.80 | IVS44+1G→A | Cytoplasmic | 0/96d | 0/64 |
| PKSR13a | A, B, C | 4.4 | ||||
| PKSR3 | A | 3.29 | ||||
| PKZA46 | A | 1.71 | ||||
| USH08 | A, B, C | 1.32f |
Gene-Prediction Programs
Unfinished genomic sequence from bacterial artificial chromosomes (BACs) spanning the DFNB12 region was obtained from GenBank. Known genes were detected by BLASTN comparison of the BAC sequence against the nucleotide databases, and novel genes were detected by sequence similarity to proteins in the public database, by BLASTX, and/or were predicted by GeneMachine (GENSCAN and FGENES) software.
cDNA Analysis
PCR primers were designed to hybridize to selected exons predicted by GENSCAN analysis of genomic sequence of BAC 472K8. Overlapping PCR fragments were generated from aliquots of RACE-ready retinal cDNA (Clontech) or from human cochlear cDNA prepared using mRNA kindly provided by Dr. C. C. Morton (Harvard University). PCR reactions were performed in 40 μl as described above, except that 2.5 U of Takara LA-Taq polymerase (PanVera) were used. Cycling conditions were 95°C for 1 min and 45 cycles of 96°C for 10 s and 68°C for 8 min. Overlapping PCR products were gel purified and were cloned into PCR-XL-TOPO vector (Invitrogen). Primers used to generate clones were clone p132A (5′-CCACAAGGCTTCCCTGTCTA-3′ and 5′-CACCGTGAATTACCTGGACTACGAG-3′), clone p132B (5′-CATGGACTTCCTCATCAACAGCAG-3′ and 5′-CAGAGTGATGTAGACGCTGCAGAAG-3′), and clone p116A (5′-TTGGTTGAGGTTGTCGATGG-3′ and 5′-ACGTGGGTGGAGGTACTGCT-3′). In addition, the predicted CDH23 sequence was used as a BLAST query against a database of expressed-sequence tags generated from cDNA libraries of various ocular tissues (G. Wistow and S. L. Bernstein, unpublished data). Two clones from a human retinal pigmented epithelium library were identified, one of which (cs53A06) was used in the determination of the cDNA sequence. cDNA fragments were sequenced as described above (622 bp for clone p116A, 2,083 bp for clone cs53A06, 1,104 bp for clone p132B, and 6,177 bp for clone p132A) and were assembled using Seqman software (DNAStar).
Northern Blot Analysis
Human donor eyes (ages 54 and 60 years) and human occipital cortex (age 74 years) were obtained from the National Disease Research Interchange. Total RNA was isolated from dissected ocular tissues and brain, by use of RNAzolB (Tel-Test). Ten micrograms each of total RNA from retina, ciliary body, retinal pigmented epithelium-choroid, lens, iris, and human brain, along with a 9.5–0.24-kb RNA standard ladder (Life Technologies), were electrophoresed on an agarose-formaldehyde denaturing gel and were transferred to GeneScreen Plus membrane (Dupont-NEN). RNA loading per lane was monitored using the 18S rRNA band (Correa-Rotter et al. 1992). This northern blot and a commercially obtained multiple-tissue northern blot (MTN Human Blot; Clontech) were hybridized with a random primed, triple-labeled ([α32P]-dATP, -dCTP, and -dGTP) 488-bp cDNA probe generated from the cytoplasmic domain of CDH23, by use of primers rm102 (5′-GCGGAGCATGAGGATGACCT-3′) and rm103 (5′-AGGCCCTTCTGGTCTTCCTC-3′) to amplify cochlear cDNA. Northern blots were hybridized at 63°C in Hybrisol II (Intergen) for 18 h and were washed at 63°C in 0.2× SSC and 0.1% SDS.
Results
Clinical Data
Nonsyndromic autosomal recessive sensorineural hearing loss was diagnosed in affected individuals in families 5020A-AN (age ⩽12 years), INJ5 (age ⩽55 years), PKSR46a (age ⩽25 years), PKSR5b (age ⩽20 years), PKZA3 (age ⩽16 years), and PKZA46 (age ⩽9 years). Both the absence of retinitis pigmentosa or other cosegregating extra-auditory features and normal developmental motor milestones were confirmed by the medical histories and physical examinations. Ocular fundoscopy was performed on selected (usually the two oldest) affected individuals of each family, to determine the absence of retinitis pigmentosa.
Family PKSR3 segregates profound sensorineural deafness and demonstrates evidence for linkage to the DFNB12 region (table 1). No obvious fundoscopic abnormalities were observed in affected family members; however, they were not examined by an ophthalmologist. The two oldest affected individuals (ages 35 and 29 years) had delayed onset of independent ambulation (age 1.5–2 years), and only the oldest had decreased night vision and abnormal vestibular function. In light of the available clinical data, it is not clear whether the deafness segregating in this family is nonsyndromic or syndromic.
Affected individuals of families 304 (Wayne et al. 1996), USH05, USH08, PKSR13a, and PKZA56 have typical USH1 phenotypes, according to the criteria recommended by the Usher Syndrome Consortium (Smith et al. 1994). All affected individuals have profound deafness, evidence of vestibular dysfunction, and retinitis pigmentosa that, in almost all affected individuals, began during the 1st decade of life. For example, the affected individuals in family PKZA56 are ages 5 and 11 years, and both manifest signs of peripheral vestibulopathy, including delayed onset of independent ambulation (age 2 years) and persistent imbalance in walking and bicycle riding. Both children have congenital profound sensorineural hearing loss and pigmentary retinopathy. A diagnosis of retinitis pigmentosa was established by fundoscopic examination of both children and was confirmed by electroretinographic evaluation in the 11-year-old child.
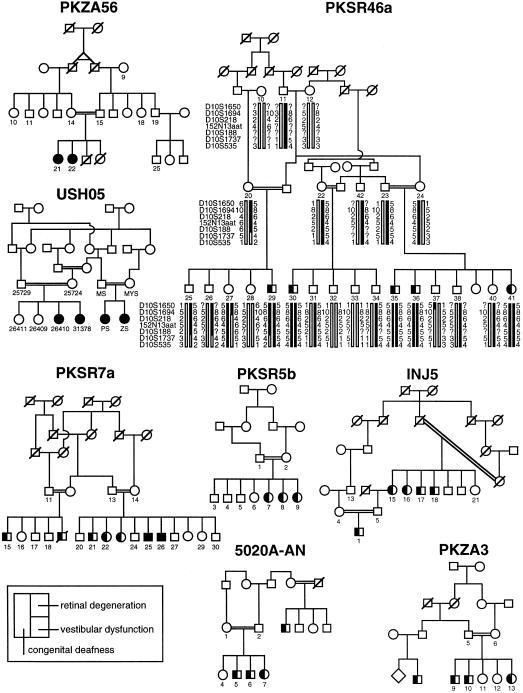
In contrast, family PKSR7a segregates an atypical USH1 phenotype. All five affected individuals have congenital profound sensorineural hearing loss. Although the two oldest affected individuals exhibit borderline abnormal vestibular function in the tandem-gait and Romberg tests, they had normal developmental motor milestones, similar to their siblings. With the exception of the oldest affected individual (age 28 years), who has slightly decreased night vision, the affected individuals of PKSR7a denied decreased night or peripheral vision. The two oldest affected subjects (ages 27 and 28 years) demonstrated mild pigmentary retinopathy by fundoscopic examination, consistent with early pathological changes of retinitis pigmentosa (fig. 1). The younger siblings had no obvious fundoscopic evidence of retinitis pigmentosa, although they were not examined by an ophthalmologist and did not undergo electroretinography. Pedigrees for the consanguineous families segregating mutant alleles of CDH23 are shown in figure 1, except for family 304, which has been described elsewhere (Wayne et al. 1996).
Figure 1 .
Pedigrees of five consanguineous families with DFNB12 and of three families with USH1D, segregating mutant alleles of CDH23. The pedigree for family 304, who are affected by USH1D, has been published elsewhere (Wayne et al. 1996). Blackened symbols represent individuals with clinically documented congenital deafness, retinal degeneration, and vestibular dysfunction, as described in the key. Genotype and haplotype data for family PKSR46a are shown. For the DFNB12 locus, the haplotype of unaffected individual 27 demonstrates the proximal breakpoint, at D10S1694, and that of affected individual 29 demonstrates the distal breakpoint, at D10S1737. 152N13aat is a polymorphic trinucleotide repeat identified on BAC 152N13.
Genetic and Physical Map
Genotype analysis of markers in the DFNB12 region was performed on all participating members of each family. Genotype and haplotype data for PKSR46a define the smallest critical interval, shown in figure 1. Individual 27 of PKSR46a has normal hearing levels, as shown by pure-tone audiometry, and is homozygous for markers in the proximal portion of the linked haplotype, with a recombination, at D10S1694, defining the proximal breakpoint of the linked region. Individual 29 of PKSR46a is profoundly deaf and has a crossover, at D10S1737, defining the distal breakpoint of the linked region.
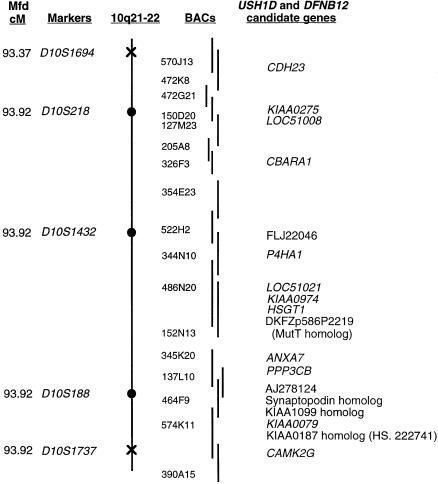
The genetic and physical maps of the linked region are depicted in figure 2. Also shown is a BAC contig that almost completely spans this region. These BACs are being sequenced by the Sanger Centre and Human Genome Therapeutics, as part of the Human Genome Project. Known or predicted genes that were sequenced in the families with DFNB12/USH1D are listed in the right margin of figure 2. Numerous homozygous sequence changes were found in these candidate genes during our mutation screen, but, in 16 of the 18 genes sequenced, all changes showed high allele-carrier rates among normal controls and were considered to be polymorphisms (data not shown). One exception was a homozygous C→G change in exon 7 of HSGT1 in family PKSR7a, which is predicted to result in the substitution of a glycine for arginine at amino acid position 281. This change was not found either in 70 chromosomes from ethnically matched controls or in 92 chromosomes from panethnic controls. However, no other nonpolymorphic changes in HSGT1 were found in our other families with DFNB12 and USH1D, and the significance of this amino acid substitution, if any, is not known. All other nonpolymorphic changes were found in the novel gene CDH23.
Figure 2 .
Genetic and physical map of the DFNB12 locus defined by proximal and distal breakpoints in family PKSR46a. The Center for Medical Genetics, Marshfield Medical Research Foundation map distance of polymorphic markers is shown on the left. The proximal breakpoint, marker D10S1694, and the distal breakpoint, marker D10S1737, are shown as crosses on the chromosome, and the linked markers are shown as blackened circles on the chromosome. Both the BACs spanning the linked region and the 18 candidate genes that were sequenced for mutations are shown. Locus designations are italicized. Nonitalicized entries represent either clones that have no associated locus designation or novel genes that have significant homology to other genes. There are two gaps in the BAC contig, and 150D20 is the only BAC in the region with a finished sequence.
Identification of USH1D and DFNB12
CDH23 was detected initially as three separate GENSCAN-predicted genes distributed among several sequence contigs from BACs 570J13 and 472K8, spanning ⩾120 kb. Several exons of one of the predicted genes are identical to a partial cDNA sequence of clone DKFZp434P2350 recovered from testis (GenBank accession number AL122081). The conceptual translations of all three predicted genes showed homology to cadherin-like genes, which encode integral membrane proteins involved in cell adhesion. Cadherins typically have large extracellular domains (characterized by cadherin repeats), a membrane-spanning region, and cytoplasmic domains that are highly divergent among family members (Nollet et al. 2000).
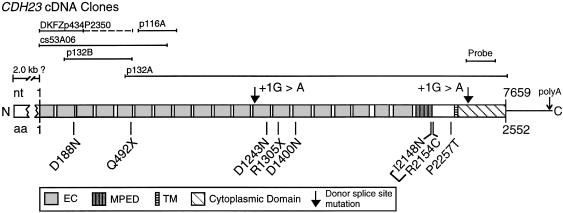
Two of the three cadherin-like predicted genes in the _DFNB12_-linked region encode only extracellular domains with cadherin repeats, whereas the third predicted gene encodes cadherin repeats and a predicted transmembrane-spanning domain, followed by ∼300 amino acids. It was possible to orient the sequence contigs relative to one another and to design primers to the predicted exons to amplify overlapping cDNA products that spanned the three predicted genes. By use of retinal and cochlear cDNAs as PCR templates, as well as a cDNA clone from a human retinal pigmented epithelium library identified by sequence similarity, it was possible to deduce 7.5 kb of continuous sequence of a single transcript with an open reading frame encoding 2,552 amino acids (fig. 3).
Figure 3 .
CDH23 cDNAs and the structure of the cadherin-23 protein (GenBank accession number AY010111). Mutations described in the text are indicated below the protein structure, and the +1G→A donor–splice-site mutations are indicated above the structure, by arrows. Intron 15 of CDH23 contains a nested gene (FLJ00041 [GenBank accession number AK024449]) encoded on the opposite strand. BLAST analysis reveals that the probable mouse orthologues of CDH23 and FLJ00041 are encoded on BAC RP23-161B11. EC denotes the extracellular cadherin repeat, and TM denotes a transmembrane spanning region, as predicted by TMpred.
The predicted protein shows 28% identity and 43% similarity to the Drosophila fat gene (GenBank accession number M80537) and 27% identity and 41% similarity to the human FAT gene (GenBank accession number X87241). This novel gene has been assigned the locus designation “_CDH23,_” for cadherin-related 23, by the HUGO nomenclature committee. The extracellular domain contains 20 cadherin repeat (EC) domains, as predicted by BLAST-CD, with expect scores ranging from 1-8 to 0.45. There is also a single transmembrane domain and a 268-amino-acid cytoplasmic domain that shows no similarity to either any other proteins or any protein domains in the _pfam_- or _smart_-motif databases.
Northern blots of human RNA probed with 488 bp of sequence from the unique cytoplasmic domain demonstrate a 9.5-kb mRNA, possibly a doublet, expressed specifically in the retina, indicating that another ∼2.0 kb of cDNA sequence remains to be identified (fig. 4). The 5′ end of our transcript does not contain a methionine start codon in Kozak (1987) consensus context. Therefore, it is likely that the 5′ end of the CDH23 transcript is incomplete. A polyadenylation site for CDH23 is predicted, by GENSCAN, to be 651 bp downstream of the TGA stop codon (fig. 3), and only 1.6 kb of genomic sequence separates the TGA stop codon of CDH23 from the 3′ UTR of the PSAP (prosaposin) gene.
Figure 4 .
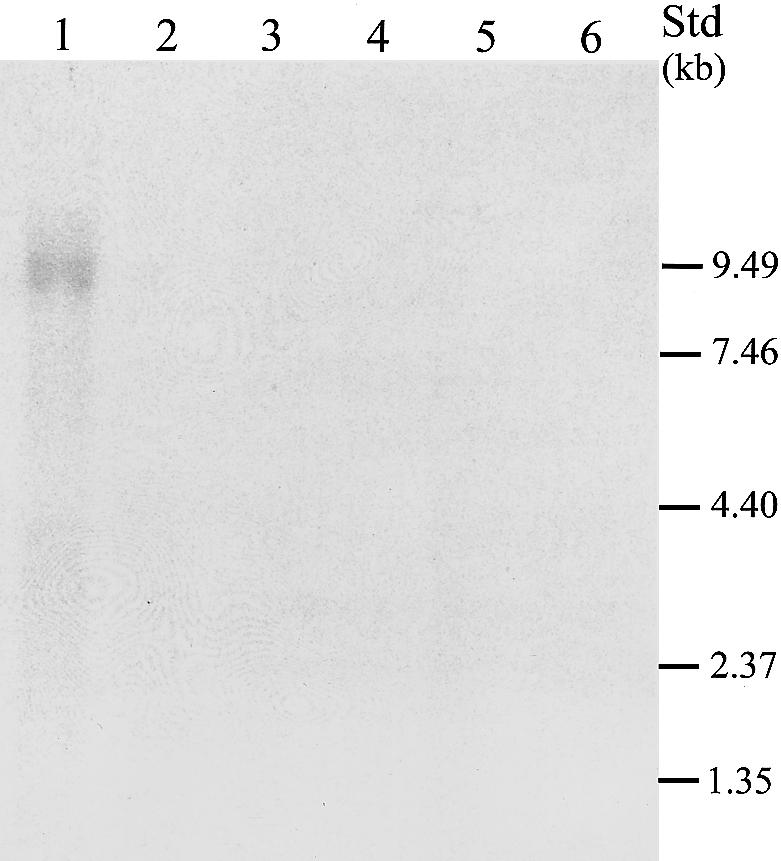
Northern blot analysis. Lanes 1–6 contain 10 μg each of total RNA from retina, ciliary body, retinal pigmented epithelium-choroid, lens, iris, and brain. Positions of bands from a 9.5–0.24-kb RNA ladder are indicated along the right margin. Shown is a 4-d exposure to autoradiograph film. The 9.5-kb band for human retina is possibly a doublet. A 1-wk exposure to a PhosphoImager screen shows identical results, with the presence of no additional bands (data not shown).
Hybridization to macula and peripheral retinal total RNA from monkey indicated that the 9.5-kb message is expressed in both retinal regions (data not shown). No signal was seen for human RNA from heart, brain, placenta, lung, liver, skeletal muscle, or kidney when a multiple-tissue northern blot was hybridized with the same probe, whereas a single 1.35-kb band was observed in pancreas (data not shown).
The existence of alternative splice forms of CDH23 is suggested by the northern blot analyses. During the generation of the northern blot probe, PCR primers were designed to amplify a 488-bp fragment from cochlear cDNA. Sequence analyses of several cloned fragments identified a 383-bp fragment in which exon 46 was omitted. The reading frame is preserved in these clones, with the resulting predicted peptide lacking 35 amino acids from the cytoplasmic domain. Moreover, the partial cDNA sequence of clone DKFZp434P2350 shows exact sequence similarity to 703 bp of CDH23, but the last 463 bp of DKFZp434P2350 apparently represents either alternate exons not recovered in our experiments or intronic sequence from a partially spliced message. Given the possibility of alternative splicing, we screened for mutations of CDH23 in an additional 14 exons predicted from genomic DNA, as well as in 42 of the 47 exons represented in our cDNAs. We found two nonsense and two splice-site mutations among four families with USH1D and six missense mutations among five families with DFNB12 (table 1). All of the mutations were in the 47 exons represented in the cDNA depicted in figure 3.
The six amino acid substitutions were found in all families with DFNB12: PKSR46a, PKSR5b, 5020A-AN, PKZA3, and INJ5 (table 1 and figs. 3 and 5). Of the six substitutions, five (D1243N, D1400N, D188N, I2148N, and R2154C) occur in EC domains. Two mutations cosegregate in the INJ5 family—I2148N and R2154C. Both substitutions occur at the distal end of the membrane proximal extracellular cadherin domain (MPED). It is not clear whether one or a combination of these substitutions causes the mutant phenotype, although neither was observed in 240 control chromosomes.
Figure 5 .
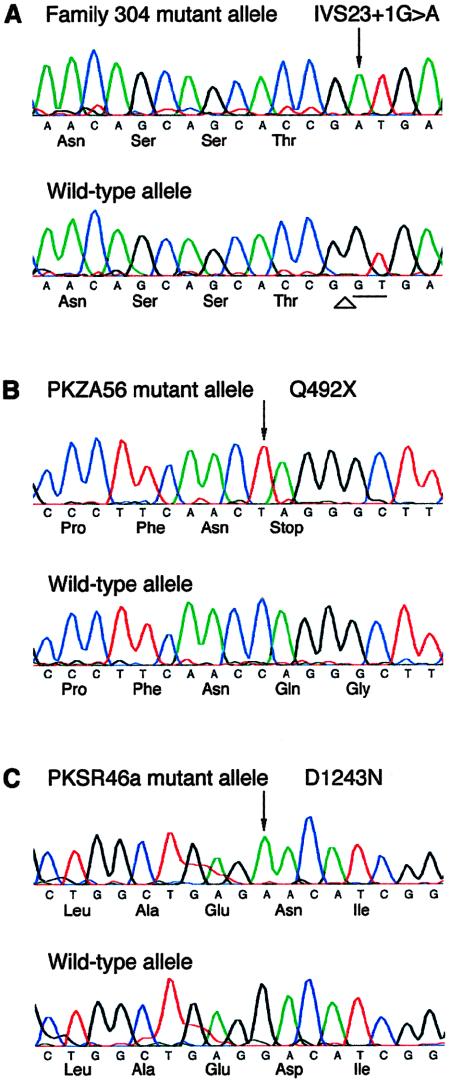
Nucleotide sequence of mutations in representative families with USH1D and DFNB12, with wild-type sequence shown below the mutant allele. The nucleotide and protein sequences of CDH23 and cadherin-23 are based on data for GenBank accession number AY010111. Arrows indicate the altered nucleotide of the mutant CDH23 allele. A, Mutation of conserved donor splice site in family 304, with USH1D (Wayne et al. 1996). The canonical GT donor dinucleotides are underlined, and the splice site is indicated by an arrowhead. B, Nonsense mutation in family PKZA56, with USH1D leading to premature termination of translation. C, Missense mutation in family PKSR46a, with DFNB12, with aspartic acid replaced by an asparagine.
The mutations segregating in the four families with USH1D (PKZA56, 304, UHS05, and PKSR7a) were either nonsense (Q492X and R1305X) or splice-site mutations (IVS23+1G→A and IVS44+1G→A) (table 1 and fig. 5). The nonsense mutations in families PKZA56 and USH05 both occur in EC domains. Splice-site mutations in families 304 and PKSR7a alter the invariant G of the donor–splice-site GT dinucleotide to an A. The IVS23+1G→A mutation segregating in family 304 is in the EC12 domain (figs. 3 and 5). The functional consequence of this mutation is difficult to predict. GENSCAN analysis predicts an alternate donor site 43 nucleotides downstream. Use of this alternate donor site would maintain the reading frame and would predict the use of an additional exon in intron 23 and the resumption of the EC domains downstream of exon 24. The mutation in the +1 donor site of intron 44 in family PKSR7a, which has USH1D, is in the region encoding the cytoplasmic domain. GENSCAN predicts an alternate donor site 37 nucleotides downstream and the use of an alternate acceptor site for exon 45, which is 20 nucleotides upstream. Use of these alternate splice sites is predicted to add a total of 19 in-frame amino acids and to preserve the open reading frame of the protein.
We did not identify mutant alleles in four of the families having homozygosity by descent in the DFNB12 region. Two of these families are too small to support significant evidence of linkage, and it is possible that they exhibit spurious homozygosity in the DFNB12 region. However, two of these families, PKSR13a and PKSR3, support evidence for linkage to CDH23 (table 1). Mutations causing USH1D and DFNB12 in these families may be found in the ∼2.0 kb of CDH23 cDNA that we have not yet recovered from our libraries.
Discussion
Cadherins comprise a large family of intercellular adhesion proteins. The classical cadherins encode repeated extracellular motifs that have been demonstrated to provide cell-to-cell adhesion via homophilic, calcium-dependent interactions, whereas cytoplasmic domains interact with the cytoskeleton via β-catenin (Nollet et al. 2000). An extensive family of atypical cadherins, or cadherin-like proteins, shares homology in the EC domains and presumably mediates similar functions involving cell-cell or cell–extracellular membrane interactions. One of these atypical cadherins, fat, has been characterized in Drosophila, where it was demonstrated that fat mutations resulted in imaginal disc overgrowth (Mahoney et al. 1991).
The novel cadherin-like gene described here has similarity to fat and to a human homologue, FAT. The similarity to fat proteins is entirely in the EC domains. CDH23 differs from fat and FAT in not having a laminin A-G domain—nor does it have any similarity in its cytoplasmic domain. Thus, CDH23 may represent a new class of cadherin-like proteins.
CDH23 is also remarkable in its limited tissue distribution. FAT transcripts are detected on northern blots of numerous epithelial tissues, including kidney, lung, breast, colon, arterial smooth muscle, and retina (Dunne et al. 1995). Northern blot analysis of CDH23 suggests that it is limited to the neuroretina. The pancreas may express a highly homologous or short alternate (∼1.4 kb) transcript (data not shown). We also have demonstrated cochlear expression of CDH23, by sequencing of PCR products from a human cochlear cDNA library.
Cochlear and retinal expression of CDH23 is consistent with the observed phenotypes of nonsyndromic deafness (DFNB12) and USH1D. CDH23 is the second known example of a gene associated with both nonsyndromic deafness and USH. Mutations in the unconventional myosin MYO7A can cause DFNA11, DFNB2, or USH1B (Weil et al. 1995, 1997; Liu et al. 1997_a_, 1997_b_), although there is no clear correlation between the type of MYO7A mutation and the phenotype (Keats and Corey 1999). In contrast, the six missense CDH23 mutations segregate in families with DFNB12, whereas the two nonsense mutations segregate in families with USH1D. The CDH23 splice mutations were associated with typical USH1D in one family and with a mild, atypical USH1D in a second family. In this latter family, affected persons had no obvious vestibular dysfunction, and the retinal degeneration observed in the two oldest individuals (ages 27 and 28 years) was mild and had late onset. It is possible that the young affected individuals of the families with DFNB12 may develop retinitis pigmentosa at an atypically late age; however, their normal developmental motor milestones and lack of vestibular abnormalities are most consistent, at this time, with DFNB12 deafness.
CDH23 is the second gene encoding a protein involved in adhesion in which mutations cause USH. The USH2A gene encodes a protein with homology to laminin, a constituent of the extracellular matrix (ECM; Eudy et al. 1998 [MIM 276901]). The presence of repeated laminin-like and fibronectin-like motifs in USH2A has led to the speculation that this protein is either a tissue-specific component of the ECM or an adhesion molecule that regulates cellular interactions with the ECM (Eudy et al. 1998). The genotype-phenotype correlation of USH2A parallels that of CDH23, in that frameshift mutations of USH2A cause deafness and retinitis pigmentosa, whereas homozygosity for a missense mutation (C759F) causes retinitis pigmentosa alone (Rivolta et al. 2000). Compound heterozygosity for a frameshift mutation and C759F leads to an intermediate phenotype, with retinitis pigmentosa and mild hearing loss (Rivolta et al. 2000).
The role of cadherin-23 in the cochlea and retina, the molecular partners interacting with the cytoplasmic domain, and the effects of the mutations described here may be addressed by direct experimentation with expression constructs and animal models. An animal model for CDH23 mutations may already exist, since the region surrounding CDH23 shows conserved synteny with mouse chromosome 10, where several mutations causing deafness have been mapped, including waltzer, jackson circler, ames waltzer, and age-related hearing loss (Ahl) (Bryda et al. 1997; Johnson et al. 1997; Alagramam et al. 2000). Possible homology to the Ahl gene is particularly intriguing, since age-related hearing loss occurs in almost 50% of the population by age 80 years (Morton 1991). Nonsyndromic autosomal recessive deafness occurs in 1/1,000 live births, with an unknown percentage attributable to mutations at the DFNB12 locus. Recently, it has been proposed that USH1D is the second most common type of USH1 (Astuto et al. 2000). Thus, the elucidation of the role of cadherin-23 in cochlear and retinal development and function may significantly influence the diagnosis and treatment of deafness and deafness-blindness worldwide.
Acknowledgments
We thank all of the families, whose participation made this project possible. We also thank Judith Willner, Lori Hampton, Barbara Ploplis, David Anderson, Tenesha Smith, Nikki Liburd, Jianhong Mo, Yan Guo, Breege MacArdle, Kaukab Rajput, Qasim Mehdi, and Cynthia C. Morton. We thank Dennis Drayna and James Battey for critical reading of the manuscript. S.L.B. was supported by the V. Kann Rasmussen Foundation (Denmark) and by a career-development award from Research to Prevent Blindness. C.R.S.S. was supported by the University Grants Commission Research Scientist Scheme, New Delhi. R.J.H.S. was supported in part by National Institutes of Health (NIH) grant R01-DC02842. M.B.-G. receives funding from Defeating Deafness. Research at both the Institute of Child Health and Great Ormond Street Hospital for Children National Health Service (NHS) Trust benefits from research and development funding received from the NHS Executive. X.-Z.L. and W.E.N. were supported by NIH grants R03-DC04530 and R01-DC02530, respectively, and by the Deafness Research Foundation. Research at the Center of Excellence in Molecular Biology was supported by University Grants Commission, Islamabad, Pakistan. Laboratory of Molecular Genetics staff were supported by National Institute on Deafness and Other Communication Disorders/NIH intramural funds 1Z01-DC00039-04 and 1Z01-DC00035-04 (to E.R.W. and T.B.F.).
Appendix
Table A1.
Genomic Primer Sequences from the Introns Adjacent to 42 Exons of CDH23
| Exon | Forward | Reverse | Product Length(bp) |
|---|---|---|---|
| 1a | ctcagtgaaggggtctgctc | gtaccccagaggcccagt | 299 |
| 2 | ctcaccacttgccttcttcc | cctatccttttcctgccaca | 396 |
| 3 | gaagcatccatcccagtgtc | cacactgagcacacagcaga | 316 |
| 4 | tgactcccttgggaattcat | cagaggctaaagcccaacag | 406 |
| 5 | ccactcctggactcaccatc | aggcaccctgtgtgaactct | 283 |
| 6 | gaagtgtgcccctctctcag | ctctggtgccactgagcat | 297 |
| 7 | ctgggtggcattcaagaagt | cctcatctcccagacctttg | 195 |
| 8 | aggaggggactggtgaactt | tgttctagctgtgggcttgg | 313 |
| 9 | ggaaagcagtgaccacacaa | tggggaggtttgctctga | 239 |
| 10 | tctatctgggactgcacagc | tgcacacagaaggagctcaa | 499 |
| 11 | acgtgacaggccttgtccta | aggacatgggattggaagtg | 579 |
| 12 | agggtttgctgatgttccag | ggatcctggctgtttcactc | 304 |
| 14 | ttagccctgactccagttgc | cccccgtatgtccagctat | 205 |
| 15 | tacaggagcaggtgccagac | ttgggcagatggactaaagg | 454 |
| 16 | tcgcagacataggagtggtg | ataggtttcgccccttgtct | 506 |
| 17 | cagtggttccccatcacag | aggacaggggtcttggattt | 365 |
| 18 | tggctaagcttttccaccat | ggtgttctcgccacatctct | 405 |
| 19 | ggcttgctagaggaagcaga | cccaaagagttgctggatgt | 400 |
| 20 | ttctccatgaccaactgcac | tagggcagggtcctcttttt | 398 |
| 21 | aatttagggaggccaagcag | aatggaggccaagaggagtt | 401 |
| 22 | ccaacctaccaaccctcctc | acccactggggtctaggttc | 301 |
| 23 | ctcccctctcatccatcgt | gctgtgaaaaatggggtctc | 361 |
| 24 | gcaccccttttctgtgtgtt | aactccgtgtccaacctgag | 338 |
| 25 | ctcccttttccctctccaac | gagaaagaagggggaagcag | 561 |
| 26 | cagccacaagtcccagattc | ttggctctgagtgaccaatg | 297 |
| 27 | ggtcacctagcccttcctct | cttggggactaagcaagcag | 502 |
| 28 | aatgaggagtggccaaaatg | gggcagggagagagagtagg | 413 |
| 29 | gggactgaccttggcctact | acttttggtggctgctcagt | 259 |
| 30 | tggccatagtaggtgctcaa | ccacttgctagaggctttgc | 268 |
| 31 | ctccttacctttggccttga | cacgcttccctctactcctg | 301 |
| 32 | tactctcctgctcccactgc | ccactcttctaggccacagc | 303 |
| 34 | actgtgctcttccgctccta | tgtttccgtgtctagccaaa | 251 |
| 35 | ctgaaacagggactggaagc | gggcatatgtgggtcatctc | 260 |
| 36 | ccatgatcccaccctcag | ggaagaggcaggtggtgtaa | 343 |
| 37 | ttacaccacctgcctcttcc | ccagatgacagtccatgcag | 256 |
| 38 | tgagtctctgagccgtaccc | ccttccttctccctccactg | 346 |
| 39 | gggtctatttgcagggaagg | cagacaggctgacagtccaa | 242 |
| 40 | gctgaggaggagagctgaga | aggaggatagccaggacgat | 263 |
| 44 | tcacaccccaagtcagtgaa | ccccttaaagaacccagctt | 350 |
| 45 | gcctctgctccagctaacat | aacctcagccaagggactg | 202 |
| 46 | tgtaccccttactcccagagg | ccggctactcagagatggag | 199 |
| 47 | ccacatagccagtgggtctc | ccacaaggcttccctgtcta | 435 |
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST-CD, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html
- GeneMachine, Division of Intramural Research, http://genome.nhgri.nih.gov/genemachine/
- Hereditary Hearing Loss Homepage, http://www.uia.ac.be/dnalab/hhh/ (for markers flanking known DFNB loci)
- Lab of Statistical Genetics, Rockefeller University, ftp://linkage.rockefeller.edu/ (for LINKAGE software)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MYO7A [MIM 276903], USH1C [MIM 605242], DFNB12 [MIM 601386], USH1D [MIM 601067], and USH2A [MIM 276901])
- Whitehead Institute for Biomedical Research, Center for Genome Research, FTP server, ftp://ftp-genome.wi.mit.edu/distribution/software/homoz (for MAPMAKER/HOMOZ software)
References
- Alagramam KN, Zahorsk-Reeves J, Wright CG, Pawlowski KS, Erway LC, Stubbs L, Woychik RP (2000) Neuroepithelial defects of the inner ear in a new allele of the mouse mutation Ames waltzer. Hear Res 148:181–191 [DOI] [PubMed] [Google Scholar]
- Astuto LM, Weston MD, Carney CA, Hoover DM, Cremers CWRJ, Wagenaar M, Moller C, Smith RJH, Pieke-Dahl S, Greenberg J, Ramesar R, Jacobson SG, Ayuso C, Heckenlively JR, Tamayo M, Gorin MB, Reardon W, Kimberling WJ (2000) Genetic heterogeneity of Usher syndrome: analysis of 151 families with Usher type I. Am J Hum Genet 67:1569–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, Barnes PD, O'Brien RE, Farndon PA, Sowden J, Liu X-Z, Scanlan MJ, Malcolm S, Dunne MJ, Aynsley-Green A, Glaser B (2000) A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet 26:56–60 [DOI] [PubMed] [Google Scholar]
- Bryda EC, Ling H, Flaherty L (1997) A high-resolution genetic map around waltzer on mouse chromosome 10 and identification of a new allele of waltzer. Mamm Genome 8:1–4 [DOI] [PubMed] [Google Scholar]
- Chaib H, Kaplan J, Gerber S, Vincent C, Ayadi H, Slim R, Munnich A, Weissenbach J, Petit C (1997) A newly identified locus for Usher syndrome type I, USH1E, maps to chromosome 21q21. Hum Mol Genet 6:27–31 [DOI] [PubMed] [Google Scholar]
- Chaib H, Place C, Salem N, Dode C, Chardenoux S, Weissenbach J, el Zir E, Loiselet J, Petit C (1996) Mapping of DFNB12, a gene for a non-syndromal autosomal recessive deafness, to chromosome 10q21-22. Hum Mol Genet 5:1061–1064 [DOI] [PubMed] [Google Scholar]
- Correa-Rotter R, Mariash CN, Rosenberg ME (1992) Loading and transfer control for Northern hybridization. Biotechniques 12:154–158 [PubMed] [Google Scholar]
- Dunne J, Hanby AM, Poulsom R, Jones TA, Sheer D, Chin WG, Da SM, Zhao Q, Beverley PC, Owen MJ (1995) Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics 30:207–223 [DOI] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao S, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, Cremers C, Davenport S, Moller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280:1753–1757 [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A (1989) A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res 17:8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Erway LC, Cook SA, Willott JF, Zheng QY (1997) A major gene affecting age-related hearing loss in C57BL/6J mice. Hear Res 114:83–92 [DOI] [PubMed] [Google Scholar]
- Kaplan J, Gerber S, Bonneau D, Rozet JM, Delrieu O, Briard ML, Dollfus H, Ghazi I, Dufier JL, Frezal J, Munnich A (1992) A gene for Usher syndrome type I (USH1A) maps to chromosome 14q. Genomics 14:979–987 [DOI] [PubMed] [Google Scholar]
- Keats BJ, Corey DP (1999) The Usher syndromes. Am J Med Genet 89:158-166 [PubMed] [Google Scholar]
- Kimberling WJ, Möller CG, Davenport S, Priluck IA, Beighton PH, Greenberg J, Reardon W, Weston MD, Kenyon JB, Grunkemeyer JA, Pieke-Dahl S, Overbeck LD, Blackwood DJ, Brower AM, Hoover DM, Rowland P, Smith RJH (1992) Linkage of Usher syndrome type I gene (USH1B) to the long arm of chromosome 11. Genomics 14:988–994 [DOI] [PubMed] [Google Scholar]
- Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15:8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Lander ES (1995) Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping. Am J Hum Genet 56:519–527 [PMC free article] [PubMed] [Google Scholar]
- Lalwani AK, Luxford WM, Mhatre AN, Attaie A, Wilcox ER, Castelein CM (1999) A new locus for nonsyndromic hereditary hearing impairment, DFNA17, maps to chromosome 22 and represents a gene for cochleosaccular degeneration. Am J Hum Genet 64:318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, Brown SD (1997_a_) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet 16:188–190 [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD (1997_b_) Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet 17:268–269 [DOI] [PubMed] [Google Scholar]
- Mahoney PA, Weber U, Onofrechuk P, Biessmann H, Bryant PJ, Goodman CS (1991) The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell 67:853–868 [DOI] [PubMed] [Google Scholar]
- Morell RJ (1999) Recent progress in hereditary hearing loss. Curr Opin Otolaryngol Head Neck Surg 7:259–265 [Google Scholar]
- Morton NE (1991) Genetic epidemiology of hearing impairment. Ann N Y Acad Sci 630:16–31 [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F (2000) Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol 299:551–572 [DOI] [PubMed] [Google Scholar]
- Rivolta C, Sweklo EA, Berson EL, Dryja TP (2000) Missense mutation in the USH2A gene: association with retinitis pigmentosa without hearing loss. Am J Hum Genet 66:1975–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA (1996) Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered 46:226–235 [DOI] [PubMed] [Google Scholar]
- Smith RJ, Berlin CI, Hejtmancik JF, Keats BJ, Kimberling WJ, Lewis RA, Moller CG, Pelias MZ, Tranebjaerg L (1994) Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am J Med Genet 50:32–38 [DOI] [PubMed] [Google Scholar]
- Smith RJH, Lee EC, Kimberling WJ, Daiger SP, Pelias MZ, Keats BJ, Jay M, Bird A, Reardon W, Guest M, Ayyagari R, Hejtmancik JF (1992) Localization of two genes for Usher syndrome type I to chromosome 11. Genomics 14:995–1002 [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu X-Z, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C (2000) A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet 26:51–55 [DOI] [PubMed] [Google Scholar]
- Wayne S, Der Kaloustian VM, Schloss M, Polomeno R, Scott DA, Hejtmancik JF, Sheffield VC, Smith RJ (1996) Localization of the Usher syndrome type ID gene (Ush1D) to chromosome 10. Hum Mol Genet 5:1689–1692 [DOI] [PubMed] [Google Scholar]
- Wayne S, Lowry RB, McLeod DR, Knaus R, Farr C, Smith RJ (1997) Localization of the Usher syndrome type 1F (Ush1F) to chromosome 10. Am J Hum Genet Suppl 61:A300 [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, Kelley PM, Kimberling WJ, Wagenaar M, Levi-Acobas F, Larget-Piet D, Munnich A, Steel KP, Brown SDM, Petit C (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374:60–61 [DOI] [PubMed] [Google Scholar]
- Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, Ayadi H, Petit C (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 16:191–193 [DOI] [PubMed] [Google Scholar]
- Willems PJ (2000) Genetic causes of hearing loss. N Engl J Med 342:1101–1109 [DOI] [PubMed] [Google Scholar]
- Zbar RI, Ramesh A, Srisailapathy CR, Fukushima K, Wayne S, Smith RJ (1998) Passage to India: the search for genes causing autosomal recessive nonsyndromic hearing loss. Otolaryngol Head Neck Surg 118:333–337 [DOI] [PubMed] [Google Scholar]