Diversity, topographic differentiation, and positional memory in human fibroblasts (original) (raw)
Abstract
A fundamental feature of the architecture and functional design of vertebrate animals is a stroma, composed of extracellular matrix and mesenchymal cells, which provides a structural scaffold and conduit for blood and lymphatic vessels, nerves, and leukocytes. Reciprocal interactions between mesenchymal and epithelial cells are known to play a critical role in orchestrating the development and morphogenesis of tissues and organs, but the roles played by specific stromal cells in controlling the design and function of tissues remain poorly understood. The principal cells of stromal tissue are called fibroblasts, a catch-all designation that belies their diversity. We characterized genome-wide patterns of gene expression in cultured fetal and adult human fibroblasts derived from skin at different anatomical sites. Fibroblasts from each site displayed distinct and characteristic transcriptional patterns, suggesting that fibroblasts at different locations in the body should be considered distinct differentiated cell types. Notable groups of differentially expressed genes included some implicated in extracellular matrix synthesis, lipid metabolism, and cell signaling pathways that control proliferation, cell migration, and fate determination. Several genes implicated in genetic diseases were found to be expressed in fibroblasts in an anatomic pattern that paralleled the phenotypic defects. Finally, adult fibroblasts maintained key features of HOX gene expression patterns established during embryogenesis, suggesting that HOX genes may direct topographic differentiation and underlie the detailed positional memory in fibroblasts.
Fibroblasts are mesenchymal cells with many vital functions during development and in adult organisms. They are responsible for much of the synthesis of extracellular matrix (ECM) in connective tissues and play major roles in wound healing. Many diseases are associated with fibroblasts, either because fibroblasts are implicated in their etiology or because of the fibrosis that accompanies damage to other cell types in tissues.
During development, reciprocal epithelial-mesenchymal interactions are required for the development of many organs, including the skin, eyes, lung, and other visceral organs. Heterotopic recombination experiments with tissue explants showed that mesenchymal identity determines the epidermal appendages that subsequently develop, indicating that the mesenchyme has an essential role in establishing positional identity (1). In some instances, purified human fibroblasts can substitute for mesenchymal tissue in epithelial induction (2). The molecular signals that underlie epithelial-mesenchymal interaction in many instances remain incompletely understood; the precision and extent of their roles in specifying the differentiation and architecture of epithelial tissues remain to be defined.
Although fibroblasts are among the most accessible normal mammalian cell types and still the most amenable to culture in vitro, they remain poorly defined in molecular terms. In practice, fibroblasts are usually identified by their spindle-shaped morphology, ability to adhere to plastic culture vessels, and the absence of markers for other cell lineages. Despite the evidence that the cells we call fibroblasts from different stromal sites may comprise a host of distinct differentiated cell types, neither the diversity of these cells nor the extent or nature of local specificity in their differentiation has been systematically examined.
Recent advances in microarray technology and bioinformatics have made it possible to appreciate previously unknown heterogeneity in cells and diseases based on the genome-wide gene expression profiles (3, 4). We hypothesized that fibroblasts, as they are traditionally defined, consist of several distinct, functional cell types separable based on their gene expression profiles. In this article, we assess the intrinsic diversity of fetal and adult fibroblasts derived from different sites by culturing them in standardized in vitro conditions. Comparison of the genome-wide mRNA expression programs provides a much richer portrait of fibroblast physiology than was previously possible and indicates that fibroblasts of distinct anatomical origin can be distinct differentiated cell types.
Materials and Methods
Reagents and Cells.
Human foreskin fibroblasts were gifts from J. Brooks (Stanford University). Other primary human fibroblasts were obtained from Coriell Cell Repositories, Camden, NJ or derived from autopsy skin samples after removal of keratinocytes and endothelial cells as described (5). The demographic information of fibroblasts is summarized in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.
In Vitro Propagation of Fibroblasts.
Fibroblasts were propagated in DMEM supplemented with 10% FBS (HyClone), glutamine, and 100 units penicillin-streptomycin (GIBCO). Cells were passaged for at least 10 population doublings in vitro before mRNA harvest. A total of 5 × 106 cells were harvested 48 h after the last passage for asynchronously growing cells or after 48 h in DMEM with 0.1% FBS for serum-starved samples as described (6).
Immunofluorescence.
Cells (104) were plated in 8-well chamber slides (Lab-Tek II, Nalge Nunc). Cells were fixed in 4% paraformaldehyde and stained with the indicated antibodies and 4′,6-diamidino-2-phenylindole as described (7).
Microarray Procedures.
Human cDNA microarray construction and hybridization were as described (3). mRNA was purified by using FastTrack according to the manufacturer's instructions (Invitrogen). A standard reference mixture of mRNAs derived from 11 cell lines was used in all experiments as an internal standard for quantitative measurement (4). Primary data and supplemental figures are available at http://genome-www.stanford.edu/fibroblast/.
Statistical Analysis.
Hierarchical clustering with array-weighted average linkage clustering (8) and Significance Analysis of Microarrays (SAM) (9) were performed as described. For SAM, 14 classes (fetal lung, fetal skin, abdomen, arm, foreskin, toe, and gum in either asynchronous or serum-starved condition) where replicate samples were available were used for multiclass analysis (9). The genes identified by SAM were then analyzed from all samples. The similarity score among clustering results is calculated as follows. The known sites of origin identify k classes. Fibroblast samples are clustered based on the expression levels of varying sets of genes by using the Partitioning Around Medoids algorithm (10), implemented in the R function pam from the cluster package. For n samples and k clusters, each application of the clustering algorithm produces a vector of n integer labels ranging from 1 through k. The similarity score for comparing two clusterings is defined as the maximum overlap of the two vectors of labels. More precisely, consider all possible permutations of the integers 1 through k for one of the vectors of cluster labels. For each such permutation, compute the number of entries at which the two vectors agree, and then take the maximum over permutations.
Results and Discussion
Fifty primary human fibroblast cultures obtained from 10 different sites in 16 donors were propagated in vitro. We chose fibroblasts from adult arm, abdomen, back, scalp, foreskin, thigh, gum, and toe, as well as fetal lung and skin to highlight potential differences among fetal vs. adult, dermal vs. visceral and mucosal, proximal vs. acral, and glabrous vs. hair bearing sites. The genome-wide gene expression program of each fibroblast culture was determined in two different culture conditions: asynchronously growing or serum-starved. Cultured fibroblasts from the diverse sites showed similar morphology, appearing as elongated, spindle-shaped cells; immunofluorescence microscopy confirmed that the fibroblast cultures were uniformly positive for vimentin, a mesenchymal marker, but negative for markers of epithelial, smooth muscle, endothelial, perineural, and histiocytic cells (Fig. 6, which is published as supporting information on the PNAS web site).
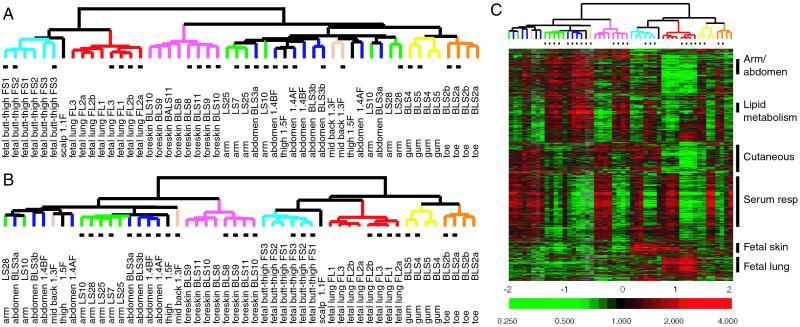
Gene expression patterns were obtained by using cDNA microarrays containing approximately 43,000 elements, corresponding to approximately 36,000 genes. The data set of 50 microarray experiments comprises ≈2.1 million gene expression measurements. Unsupervised hierarchical cluster analysis of the data revealed a striking grouping of the fibroblast gene expression patterns according to their sites of origin (Fig. 1A). For a majority of cases, fibroblasts were clustered into discrete groups with other fibroblasts derived from the same site irrespective of whether the cells were asynchronously growing or in serum starved conditions. In three pairs of fetal skin and lung fibroblasts derived from the same individuals (termed FS and FL1–3) and adult fibroblast cultures derived from four sites of the same donor (termed 1.1F to 1.5F), the fibroblasts were clustered next to other fibroblasts from the same site rather than cells from the same individual. Different passages of the same fibroblast culture (labeled with suffix –a or –b) clustered with each other, indicating that their in vitro phenotypes are stable through several passages in culture.
Figure 1.
Topographic differentiation of fibroblasts. (A) Unsupervised hierarchical clustering of cultured fibroblasts. The global gene expression patterns of 50 fibroblast cultures were sorted based on similarity by hierarchical clustering. Approximately 1,400 genes were selected from the total data set based on variance more than 3-fold in at least two arrays. The site of origin of each fibroblast culture is indicated and color-coded. Fibroblasts cultured in minimal-serum medium (0.1% FCS) are indicated by black dots below the dendrogram. (B) Supervised hierarchical clustering of cultured fibroblasts was performed by using approximately 1,600 genes identified by SAM (9) that varied according to fibroblast site of origin. Serum-starved samples are indicated by black dots below the dendrogram. (C) Topography transcriptome of fibroblasts. The variation in expression of approximately 1,600 genes described in B are shown in matrix format (8). The scale extends from 0.25- to 4-fold over mean (−2 to +2 in log2 space) as is indicated on the bottom. Gray represents missing data. Gene clusters are indicated on the right.
We used a permutation-based, statistical method termed SAM (9) to identify genes that vary in expression specifically in accordance to the fibroblast site of origin. Approximately 1,600 genes were identified by SAM with an estimated false discovery rate of 0.02%. The genes identified by SAM, which we collectively term the topography transcriptome, allowed improved clustering of the fibroblasts as demonstrated by the shorter branches of the dendrograms (Fig. 1B). In this analysis, all of the fibroblasts were clustered according to their site of origin with the singular exception that subsets of arm and abdomen fibroblasts were separated based on whether the cells were grown in the presence or absence of serum (Fig. 1B). The topography transcriptome of fibroblasts is shown in Fig. 1C. Gene expression signatures, comprised of large groups of genes that are coordinately regulated, are named by the predominant topographic sites or process that they represent. Together, these results demonstrate that fibroblasts from different anatomic sites have characteristic, distinct phenotypes, a phenomenon we term topographic differentiation. Moreover, topographic differentiation is maintained in vitro when fibroblasts are isolated from the influence of other cell types.
Topographic Transcriptome of Fibroblasts.
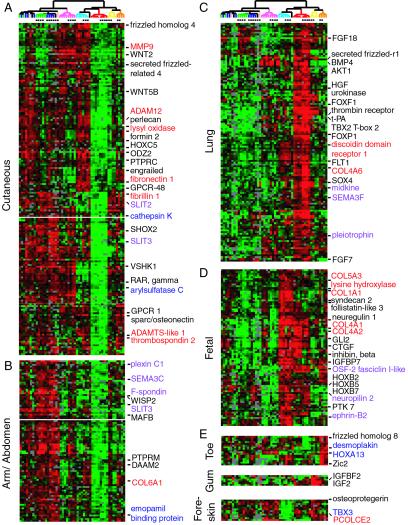
The patterns that distinguish fibroblast types contain few genes that are uniquely expressed in fibroblasts from any particular site. Rather, it is the combinatorial patterns of large groups of genes that define fibroblasts from different sites (Fig. 1C); the same was found in previous studies of tumor subtype classification (3, 4). For instance, the gene expression program of fetal lung fibroblasts differs from all other fibroblasts: the most prominent differences are two large groups of genes that segregate fetal lung fibroblasts from fetal and adult fibroblasts from cutaneous sites (Fig. 2). Still, there are genes whose expression is high in both types of fetal fibroblasts examined relative to the adult fibroblasts. The diversity of gene expression programs reveals several biological themes that can be related to known or inferred physiologic functions of fibroblasts in vivo.
Figure 2.
Features of the topography transcriptome. Select genes from the cutaneous cluster (A), arm/abdomen (B), fetal lung (C), fetal lung and skin (D), and other sites (E) are shown. The names of genes involved in ECM synthesis (red), cell signaling or fate determination (black), cell migration guidance (purple), and genes mutated in inherited human diseases (blue) are labeled by the indicated colors. The order of samples and scale are the same as Fig. 1 B and C.
Synthesis of ECM.
Fibroblasts have long been considered the main cell type that synthesizes ECM in connective tissues. Consistent with this idea, fibroblasts in vitro expressed appropriate ECM genes characteristic their sites of origin (Fig. 2, gene names shown in red). This distinction was most evident comparing cutaneous to fetal lung fibroblasts. For example, fibroblasts from fetal skin and fetal lung expressed relatively high levels of type IV collagen (Fig. 2D), a net-like collagen that forms the basement membrane of the skin and lines the lung alveoli (www.ncbi.nlm.nih.gov/omim). However, fetal skin but not lung fibroblasts strongly expressed type I and V collagen (Fig. 2D), which are essential for the tensile strength of dermis. Mutations in type I and type V collagen cause osteogenesis imperfecta and Ehler–Danlos disease (EDS) type I, two congenital diseases with abnormal skin texture (OMIM, Online Mendelian Inheritance in Man, www.ncbi.nlm.nih.gov/omim). Osteogenesis imperfecta is characterized by translucent and thin skin whereas EDS patients have hyperextensible skin that is easily scarred. Collagen maturation requires posttranslational hydroxylation and oxidative crosslinking of lysine residues; mutations in lysine oxidase and lysine hydroxylase lead to EDS type V and type VI, respectively (OMIM, www.ncbi.nlm.nih.gov/omim). Indeed, high expression of lysine hydroxylase and lysine oxidase was observed in the fetal skin and adult cutaneous signature, respectively (Fig. 2 A and D). Finally, high levels of fibronectin and fibrillin expression also characterized the adult cutaneous signature. Fibronectin and fibrillin are ECM proteins that are essential for normal skin elasticity; mutation of these genes cause EDS type X and Marfan syndrome, respectively (OMIM, www.ncbi.nlm.nih.gov/omim). Thus, one aspect of the topographic genomic program in fibroblasts is the coordinate regulation and synthesis of the ECM proteins in a site-specific manner.
Cell type-specific growth/differentiation factors.
One of the intriguing questions considered in this study is the role that fibroblasts play in patterning and regulating the associated epithelial structures. We found topographic variation in the expression of a rich array of genes known or suspected to be involved in cell fate determination and inductive interactions. For instance, in the developing lung, epithelial-mesenchymal interactions are required for branching morphogenesis and generation of distinct epithelial cell types. FOXF1 and FOXP1, two previously identified, lung-specific forkhead family transcription factors, were specifically expressed in fetal lung fibroblasts (Fig. 2C). Haploinsufficiency of FOXF1 in mice disrupts the branching morphogenesis of lungs and causes lobe fusion (11). Several genes previously shown to be important for lung development, and thought to regulate FOXF1 function, including HGF, BMP4, FLT1, and FGF7 (11, 12), were also identified in the lung signature. Thus, several essential components of this pathway appear to be coordinately expressed in lung fibroblasts, pointing to a role for these cells in establishing or maintaining the lung architecture.
Growth and differentiation signaling molecules that are topographically regulated in fibroblasts include components of transforming growth factor (TGF)-β signaling (BMP4, inhibin, follistatin), Wnt signaling (Wnt2, Wnt5, frizzled homologs 1, 4, and 8, WISP2, DAAM2), and G protein signaling (GPRC1, GPRC48) pathways (Fig. 2). TGF-β and Wnt families are well-established growth and differentiation factors in development and are known to play essential roles in epithelial appendage specification and homeostasis (13–15). Many ligands for receptor tyrosine kinases (FGF18, neuregulin, insulin-like growth factor 2, connective tissue growth factor), ligand binding proteins that antagonize signaling (IGFBP7 and IGFBF2), and receptor tyrosine phosphatases (PTPRC, PTPRM) were also identified. The topographic variation of a large number of cell signaling molecules hint at the richness and specificity of the fibroblasts' role in inductive signaling and reciprocal epithelial-mesenchymal interactions.
Cell migration signals.
An unexpected finding of the topography transcriptome in fibroblasts is an elaborate variation in cell migration signals, many of which were first identified in the nervous system as axon guidance molecules (Fig. 2, gene names shown in purple). Many of these molecules are now appreciated as general cell migration guidance molecules, which can also pattern mesodermal and epidermal cell migration (16, 17). The topographically regulated guidance molecules we found in fibroblasts include semaphorins (SEMA3F, SEMA3C), receptors for semaphorins (neuropilin 2 and plexin C1), Slit proteins (SLIT2, SLIT3), ephrin B2, F-spondin, midkine, and pleiotrophin (Fig. 2). Many of these proteins were expressed constitutively in both adult and fetal cells and regardless of the proliferation state of fibroblasts (Fig. 2). The topographic patterning of many migration guidance molecules in fibroblasts indicates that fibroblasts may play a critical role in posting “road signs” that guide axons and possibly the migration of vasculature and many other cell types.
Expression of Genes Associated with Genetic Syndromes.
We observed that several genes defective in genetic syndromes were prominently expressed in fibroblasts originating from sites most affected in those diseases (summarized in Table 2, which is published as supporting information on the PNAS web site). As described above, genes underlying 6 of 10 types of EDS syndromes were identified by comparing genes expressed in dermal versus lung fibroblasts. Moreover, HoxA13 is expressed in toe and foreskin fibroblasts, and mutation of HOXA13 causes hand-foot-genital syndrome, characterized by hypoplastic distal phalanges and malformation of the urogenital system (OMIM, www.ncbi.nlm.nih.gov/omim). Desmoplakin I, a cytoskeletal linker protein mutated in striatal palmar-plantar keratoderma, is also highly expressed in toe fibroblasts. Similarly, emopamil binding protein and arylsulfatase C, two enzymes involved in epidermal cholesterol metabolism, were identified in the arm and cutaneous clusters, respectively. Mutation of emopamil binding protein causes X-linked Conradi–Hunerman syndrome, characterized by rhizomelic shortening of limbs and whorled ichthyosis, and mutation of arylsulfatase C causes X-linked generalized ichthyosis (OMIM, www.ncbi.nlm.nih.gov/omim). The correspondence of gene expression and phenotype may provide an explanation for the localization of various defects in these syndromes and suggests that genes that are topographically regulated in fibroblasts may present a fertile ground for finding genes underlying disorders and diseases of skin, the musculoskeletal system and even visceral organs.
Several of the proteins encoded by disease genes are thought to act in the epidermis but not in the dermis. For example, desmoplakin I has important structural functions connecting intermediate filaments to desmosomes in keratinocytes (OMIM, www.ncbi.nlm.nih.gov/omim). The parallel between the topographic regulation of genes in dermal fibroblasts and the pattern in which mutations in those genes affect the epidermis suggests that those genes may be expressed in the skin in a graded, segmental fashion that cuts across the germ layers.
Altered Lipid Metabolism in Serum-Starved Fetal Lung Fibroblasts.
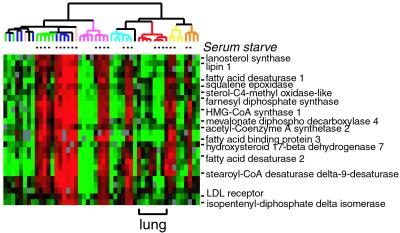
Although many genes are topographically regulated, a large group of genes was coordinately regulated in response to serum in all fibroblasts. Many features of this program have been described (6), and detailed analysis of the common response of fibroblasts to serum will be presented elsewhere. Because serum contains low density lipoprotein (LDL), fibroblasts grown in serum-containing media can take up LDL, suppressing endogenous production of cholesterol and fatty acids. When fibroblasts are cultured in limited serum (and thus low exogenous sterol), they induce expression of genes involved in cholesterol and fatty acid synthesis pathways (6). Sterol and fatty acid synthesis in mammalian cells is coordinately regulated at the transcriptional level by the sterol regulatory element-binding protein (SREBP) pathway (reviewed in ref. 18). SREBPs are transcriptional factors that are normally tethered to the endoplasmic reticulum; when sterol levels are low, regulated proteolysis of SREBP allows the NH2-terminal, active fragments of SREBP to enter the nucleus and activate the transcription of sterol and fatty acid biosynthetic genes (18). More recently, SREBPs have also been implicated in adipogenesis, but their exact roles are incompletely understood (19, 20).
We observed that all fibroblasts except fetal lung fibroblasts induced a large group of canonical SREBP target genes when placed in low-serum conditions (Fig. 3). This “lipid cluster” contains enzymes in the cholesterol biosynthetic pathway such as HMG-CoA synthase, enzymes for fatty acid biosynthesis such as sterol-CoA desaturase, and LDL receptor itself. The coordinated repression of the entire lipid cluster in fetal lung fibroblasts suggests that the regulation of SREBPs may be distinct in fetal lung fibroblasts. Moreover, we observed that lipin, a gene mutated in fatty liver degeneration (Fld) mice with partial lipodystrophy (21), is coordinately regulated with all of the SREBP target genes (Fig. 3). Transgenic mice overexpressing SREBP1c in white adipose tissue develop a syndrome of lipodystrophy, leptin deficiency, and insulin resistance, a phenotype very closely mimicked by the Fld mouse (20). These results suggest that lipin is either directly or indirectly regulated by SREBP, providing a link between SREBP function, adipogenesis, and insulin resistance.
Figure 3.
Coordinated variation in expression of genes involved in lipid and sterol metabolism. Note the correspondence between cultivation in low-serum (and thus low LDL) medium and activation of the lipid cluster genes in all the fibroblast cultures with the distinct exception of fetal lung fibroblasts. The order of samples and scale are the same as Fig. 1 B and C.
HOX Genes and Topographic Gene Expression.
The striking topographic differentiation of fibroblasts, observed in vitro, suggests that specification of the topographic program must involve some way of maintaining positional memory. It is notable in this connection that we found that many HOX genes were differentially expressed in fibroblasts derived from different anatomical sites (Fig. 2). The Hox genes encode a family of evolutionarily conserved transcription factors that determine positional identity along the anterior-posterior and secondary axes in animals (22, 23). During development, Hox genes are expressed in a nested, segmental pattern along the anterior-posterior axis in a sequence corresponding to their respective positions on the chromosome, a property called the colinearity rule (22).
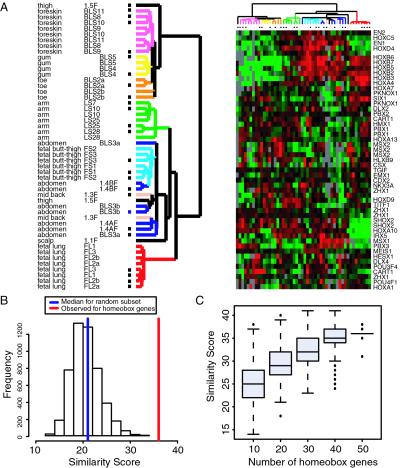
To assess a possible role for HOX genes in topographic differentiation, we first identified 51 genes encoding homeodomain transcription factors, with well-measured expression in these experiments. Hierarchical clustering of the cultured fibroblasts based solely on their patterns of expression of these 51 homeodomain genes recapitulated grouping of the fibroblasts according to their site of origin (Fig. 4A). Within the set of homeodomain transcription factors, the canonical HOX genes demonstrated significant variation depending on the fibroblast site of origin whereas the other genes, with a few exceptions, were not topographically regulated (Fig. 4B and Fig. 7, which is published as supporting information on the PNAS web site). Comparison with random sets of 51 well-measured genes from the fibroblast transcriptome confirmed that the ability of HOX genes to predict fibroblast origin is robust and highly statistically significant (P < 0.0002, Fig. 4 B and C). These results indicate that fibroblasts from each topographic site express a unique “Hox code,” thereby allowing the cells from the same site to be grouped together based simply on the expression pattern of the Hox genes.
Figure 4.
HOX genes and topographic differentiation. (A) Hierarchical clustering of fibroblast cultures based solely on expression of genes encoding homeodomain proteins reproduces the clustering by site of origin. Of 88 homeodomain-containing genes on the array, 51 were considered well measured as indicated by reference channel intensity over background ≥ 1.5-fold and no less than 80% informative data. Hierarchical clustering was performed with these 51 genes, and the result is displayed in the same format as in Fig. 1. Scale is the same as Fig. 1C. (B) Statistical significance of topographic clustering by homeobox genes. The 51 homeobox genes identified above were clustered by using Partitioning Around Medoids (PAM) with k = 6 clusters and 45 arrays (see Materials and Methods). The sites of origin of the fibroblast samples (abdominal skin, arm, fetal buttock thigh, fetal lung, foreskin, toe, and gum) were taken as the reference grouping of six clusters. The similarity score comparing the PAM clustering to the known site of origin is 36 of a maximum of 45. To assess the statistical significance of the similarity score, 5,000 sets of 51 random genes from a data set of 19,081 genes filtered as in A were subjected to the same analysis and the histogram of the similarity scores are shown. The median of the 5,000 similarity scores is shown in blue (21 of 45). None of the 5,000 trials achieved a score of 36; thus the P value is 0/5,000. (C) Robustness of topographic clustering. The same analysis in B was carried out for 500 of random subsets of 10, 20, 30, 40, or 50 homeobox genes. The distribution of the similarity scores is summarized by using boxplots. The central box in each plot represents the inter-quartile range (IQR), which is defined as the difference between the 75th and 25th percentiles. The line in the middle of the box represents the median. Extreme values greater than 1.5 IQR above the 75th percentile and less than 1.5 IQR below the 25th percentile were plotted individually. Site identity was reasonably recovered with as few as 10 homeobox genes, which is better than with random subsets of 51 genes (compare to median score of 21 in B).
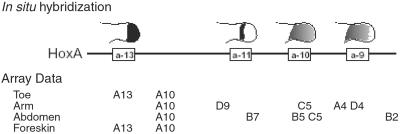
Because HOX genes are reused later in development to specify cell fate in contexts unrelated to positional identity, we investigated whether HOX genes observed in adult fibroblasts are related to the embryonic Hox code established along the antero-posterior axis. Comparing HOX genes expressed in fibroblasts from various sites revealed a rough correspondence to the colinearity rule (Fig. 8_A_, which is published as supporting information on the PNAS web site). To address this question at higher resolution, we examined HOX expression patterns on the limbs, where detailed embryology and in situ hybridizations had been performed in chicks and mice (23, 24). The 5′ HoxA genes are involved in patterning the posterior aspect of the animal and specify the proximal-distal axis of the limbs. HoxA10, A11, and A13 expression domains are initiated proximally in the stylopod, zeugopod, and autopod, respectively, which develop into the upper arm, forearm, and hand, respectively in the forelimb (Fig. 5). In two secondary axes sampled in our experiments, we observed the nested pattern of expression in HOXA genes following the proximal-distal axis. HOXA10 was expressed in fibroblasts from the upper arm and abdomen, and both HOXA10 and A13 were expressed in fibroblasts from the toe and foreskin. HOXC5, a marker of the upper limb (25), is expressed in arm fibroblasts but not toe fibroblasts (Fig. 5).
Figure 5.
HOX expression in adult fibroblasts and the embryonic Hox code. Comparison of Hox expression pattern in secondary axes. Schematic of expression domains of 5′ HoxA genes in the mouse limb bud at approximately 11.5 days postcoitum is shown on top (after ref. 31). The HOX genes up-regulated in fibroblasts from the indicated sites are shown below.
Comparison of the HOX expression domains identified in fibroblasts with phenotypes in knockout animals also reinforced the physiologic relevance of these expression domains. As noted previously, mutation of HOXA13 leads to hand-foot-genital syndrome in humans (OMIM, www.ncbi.nlm.nih.gov/omim), and mutation of HoxD9, which is expressed in upper arm fibroblasts, causes shortened humerus with misshapen deltoid tuberosity in mice (25). The coregulation of proximal-distal patterning in secondary axes such as limbs and genitalia had been demonstrated in mice; deletion of the posterior Hox genes causes dose-dependent shortening of toes and the genital ridge (26). Immunofluorescence microscopy confirmed that Hox protein was uniformly expressed in the appropriate fibroblasts and localized in the nuclei (Fig. 8_B_). Collectively, these results indicate that the HOX genes expressed in adult fibroblast maintained key features of the Hox code established during embryogenesis and thus is a bona fide representation of positional memory.
In Drosophila, Hox proteins act as transcription factors to induce segment-specific genes at successive stages of development (27). By analogy, we hypothesize that the site-specific combinations of Hox proteins in adult fibroblasts transcriptionally activate different genes to give rise to the topography transcriptome, which then endows fibroblasts with site-specific activities and inductive properties. In Drosophila and mammals, the Hox loci are epigenetically regulated by chromatin remodeling complexes that fix the transcriptional state of the genes once the Hox code is established (28). These mechanisms for transcriptional memory are consistent with the maintenance of HOX expression in fibroblasts after extensive culture in vitro. Whether the genes in the topography transcriptome are direct transcriptional targets for Hox proteins should be addressed in future experiments.
Concluding Remarks
It is now clear that the cells we traditionally call fibroblasts comprise a diverse class of distinct differentiated cell types. Because fibroblasts are frequently used in genetic and biochemical studies of human and mouse mutants, our results highlight the importance of using site-matched controls to focus on differences caused by the mutation or disease in question.
Using genome-wide expression profiling, we have shown that human fibroblasts exhibit topographic differentiation. The systematic differences among fibroblasts from different sites are so great that topographic differentiation is the principal source of variation in the genomic expression programs in a comparison of cultured cells that vary in donor source, site of origin, passage number, and even the presence or absence of serum. Indeed, gene expression differences among fibroblasts from different anatomic sites are comparable in scope and magnitude to the differences observed among different lineages of white blood cells (C. Palmer and P.O.B., unpublished observations). The ability of fibroblasts to maintain topographic differentiation in vitro is consistent with the evidence for mesenchymal determination in heterotopic epithelial-mesenchymal recombination experiments and suggests that topographic differentiation is cell autonomous or, at a minimum, requires only paracrine factors from other similarly fated fibroblasts (1).
Many intriguing issues are raised by the topographic classification of fibroblasts. In this study, the main branches in the hierarchical classification of transcription programs in these cells separate fibroblasts from cutaneous versus visceral (e.g., lung) tissues; this division may correspond to the initial separation of somatic and splanchnic mesoderm during embryogenesis. The topography transcriptome defined in this article is only a small glimpse of topographic differences in vivo as the fibroblasts would interact with other cell types and react to distinct physical environments imposed by the body site to elaborate additional site-specific programs. Nonetheless, our experimental approach may point to a systematic method for recognizing the fine specialization of physiological and regulatory function in stromal cells that have traditionally been considered homogenous.
In addition to their inductive roles in development, stromal cells play important roles in tissue repair after injury, participate in immune and inflammatory responses, and provide permissive and at times inductive environments for cancer development and metastasis (29). The site specificity of the molecular signals and extracellular proteins expressed by stromal fibroblasts raises the intriguing possibility that these signals may provide “home addresses” that can be monitored by epithelial cells to restrict their migration to, or their survival and proliferation at, ectopic sites. In such a scenario, invasion or metastasis of cancers cells might then require abrogation of these restrictions.
Effective strategies for tissue engineering or therapeutic use of embryonic stem cells will undoubtedly be advanced by a detailed understanding of the tissue-specific environments generated by stromal cells (30). The topographic differentiation of stromal cells such as fibroblasts points to the biologic specificity and regulatory complexity of these processes. The molecular “address code” of human fibroblasts described here provides part of a framework for systematic investigation of these questions.
Supplementary Material
Supporting Information
Acknowledgments
We thank A. Alizadeh, M. Diehn, and members of the Brown and Botstein labs for encouragement and support. We are grateful to A. Alizadeh, B. Hogan, and A. Oro for insightful critiques of the manuscript. This work was supported by the Howard Hughes Medical Institute and a grant from the National Cancer Institute (to P.O.B.). P.O.B. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
ECM
extracellular matrix
SAM
Significance Analysis of Microarrays
EDS
Ehler–Danlos disease
LDL
low density lipoprotein
SREBP
sterol regulatory element-binding protein
OMIM
Online Mendelian Inheritance in Man
References
- 1.Dhouailly D. In: Pattern Formation. Malincinski G, Bryant S, editors. New York: Macmillan; 1984. pp. 581–601. [Google Scholar]
- 2.Yamaguchi Y, Itami S, Tarutani M, Hosokawa K, Miura H, Yoshikawa K. J Invest Dermatol. 1999;112:483–488. doi: 10.1046/j.1523-1747.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Normand J, Karasek M A. In Vitro Cell Dev Biol Anim. 1995;31:447–455. doi: 10.1007/BF02634257. [DOI] [PubMed] [Google Scholar]
- 6.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 7.Scott M L, Fujita T, Liou H C, Nolan G P, Baltimore D. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 8.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman L, Rousseuw P J. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. [Google Scholar]
- 11.Mahlapuu M, Enerback S, Carlsson P. Development (Cambridge, UK) 2001;128:2397–2406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- 12.Costa R H, Kalinichenko V V, Lim L. Am J Physiol. 2001;280:L823–L838. doi: 10.1152/ajplung.2001.280.5.L823. [DOI] [PubMed] [Google Scholar]
- 13.Taipale J, Beachy P A. Nature (London) 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 14.Sanson B. EMBO Rep. 2001;2:1083–1088. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenn K S, Paus R. Physiol Rev. 2001;81:449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- 16.Fernandis A Z, Ganju R K. Sci. STKE. 2001. http://stke.sciencemag.org/cgi/content/full/sigtrans , http://stke.sciencemag.org/cgi/content/full/sigtrans;2001/91/pe1. ;2001/91/pe1. [DOI] [PubMed] [Google Scholar]
- 17.He Z, Wang K C, Koprivica V, Ming G, Song H J. Sci. STKE. 2002. http://stke.sciencemag.org/cgi/content/full/sigtrans , http://stke.sciencemag.org/cgi/content/full/sigtrans;2002/119/re1. ;2002/119/re1. [DOI] [PubMed] [Google Scholar]
- 18.Horton J D, Goldstein J L, Brown M S. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J B, Spiegelman B M. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 20.Shimomura I, Hammer R E, Richardson J A, Ikemoto S, Bashmakov Y, Goldstein J L, Brown M S. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterfy M, Phan J, Xu P, Reue K. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 22.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 23.Tabin C J. Development (Cambridge, UK) 1992;116:289–296. doi: 10.1242/dev.116.2.289. [DOI] [PubMed] [Google Scholar]
- 24.Nelson C E, Morgan B A, Burke A C, Laufer E, DiMambro E, Murtaugh L C, Gonzales E, Tessarollo L, Parada L F, Tabin C. Development (Cambridge, UK) 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 25.Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Dolle P, Chambon P. Development (Cambridge, UK) 1996;122:461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- 26.Zakany J, Fromental-Ramain C, Warot X, Duboule D. Proc Natl Acad Sci USA. 1997;94:13695–13700. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akam M. Curr Biol. 1998;8:R676–R678. doi: 10.1016/s0960-9822(98)70433-6. [DOI] [PubMed] [Google Scholar]
- 28.Francis N J, Kingston R E. Nat Rev Mol Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- 29.Elenbaas B, Weinberg R A. Exp Cell Res. 2001;264:169–184. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 30.Watt F M, Hogan B L. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 31.Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P. Development (Cambridge, UK) 1996;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information