T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites (original) (raw)
Abstract
A statistical analysis of 9000 flanking sequence tags characterizing transferred DNA (T-DNA) transformants in Arabidopsis sheds new light on T-DNA insertion by illegitimate recombination. T-DNA integration is favoured in plant DNA regions with an A-T-rich content. The formation of a short DNA duplex between the host DNA and the left end of the T-DNA sets the frame for the recombination. The sequence immediately downstream of the plant A-T-rich region is the master element for setting up the DNA duplex, and deletions into the left end of the integrated T-DNA depend on the location of a complementary sequence on the T-DNA. Recombination at the right end of the T-DNA with the host DNA involves another DNA duplex, 2–3 base pairs long, that preferentially includes a G close to the right end of the T-DNA.
Introduction
Transferred DNA (T-DNA) from Agrobacterium tumefaciens Ti plasmids is a widely used tool for genetic engineering and plant insertional mutagenesis (Galbiati et al., 2000). T-DNA is transferred into plant cell nuclei as a singlestranded molecule attached at the 5′ end to the protein VirD2 and coated with virulence protein E2. T-DNA integrates into the genome (Gelvin, 2000) by illegitimate recombination (Gheysen et al., 1991; Mayerhofer et al., 1991; Zupan et al., 2000) via a largely unknown mechanism. Characterization of a limited number of T-DNA insertions into genes showed an apparently even repartition along Arabidopsis thaliana chromosomes with no preferential integration into a specific gene structure (Azpiroz-Leehan and Feldmann, 1997). We have produced flanking sequence tags (FSTs) for >18 000 A. thaliana T-DNA transformants (Balzergue et al., 2001). This has allowed an in-depth analysis of the sequence specificity of T-DNA insertion sites (IS) and brings new insights into the integration process.
Results
Distribution of IS in the host genome
The FST distribution in the genome of A. thaliana is even throughout the five chromosomes. As an example, the FST distribution along chromosome 3 is given in Figure 1. FSTs are progressively less frequently observed towards the centromere, as shown in Figure 1 with the predicted genes [The Arabidopsis Genome Initiative (AGI), 2000]. About 40% of the integrations are in genes, i.e. in regions defined by the AGI-predicted genes plus 200 base pairs (bp) on each side of them and covering 54% of the genome. There is no apparent category of genes more or less prone to T-DNA insertion. We observed 121 FSTs per Mb in the 200 bp upstream of the start codon and 77 FSTs per Mb in the intergenic regions and 3′ UTR. In genes, FSTs are more frequently found in introns than in exons, with 43 and 33 FSTs, respectively, per Mb.
Figure 1.
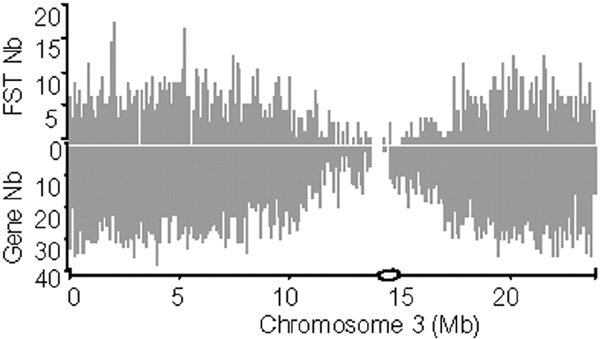
Gene and FST densities along chromosome 3 of A. thaliana. Each peak is a number of FSTs (above) or genes (below) per 100 kb. The centromeric region is represented by a circle on the abscissa.
T-DNA after integration
The sites of VirD2-mediated cleavage of the T-DNA have been determined both in vivo (Dürrenberger et al., 1989) and in vitro (Pansegrau et al., 1993). FSTs from the T-DNA left border (LB), at the 3′ end of the transferred singlestranded T-DNA, show that in 24% of the cases the T-DNA is integrated with a full-length LB (canonical insertion) (Figure 2A). The number of inserted T-DNAs with an LB truncated by 1–23 bases is relatively even and longer deletions are rare. The right border (RB) of the T-DNA (Figure 2A) is also sometimes fully conserved after integration (19%). The RB is frequently truncated between the second and fifth bases from the canonical IS (36%).
Figure 2.
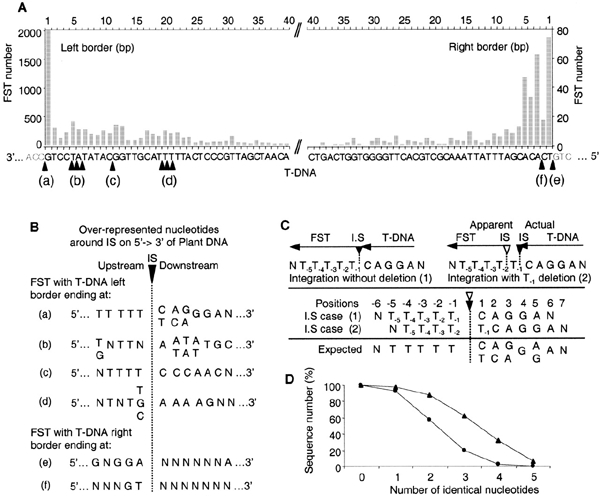
Deletions in the integrated T-DNA and A. thaliana sequences at IS. (A) Histogram of the number of FSTs classified on the basis of the ends of the sequence of the integrated T-DNA. 8525 and 394 FSTs were analysed for left and right borders, respectively. The canonical T-DNA sequence runs from (a) to (e). (B) The over-representation of nucleotides in the plant sequence before and after IS depends on the end of the integrated T-DNA. (a) to (f) indicate nicking sites as illustrated below the T-DNA sequence in (A). Over-represented nucleotides (shaded boxes in Table 1) are shown at five positions downstream and seven positions upstream of IS. (C) A deletion of 1 bp upstream of the IS can explain the over-representation of two different nucleotides at different positions in the case of a canonical integration (a). The sequence N(T)5CAGGAN is taken as an example of a plant sequence around an IS. (D) Identity scores between the complement of the canonical T-DNA left border, CAGGA and sequences downstream of IS (triangles) or randomly taken from the A. thaliana genome (circles).
Microsimilarity between the host genome and T-DNA borders
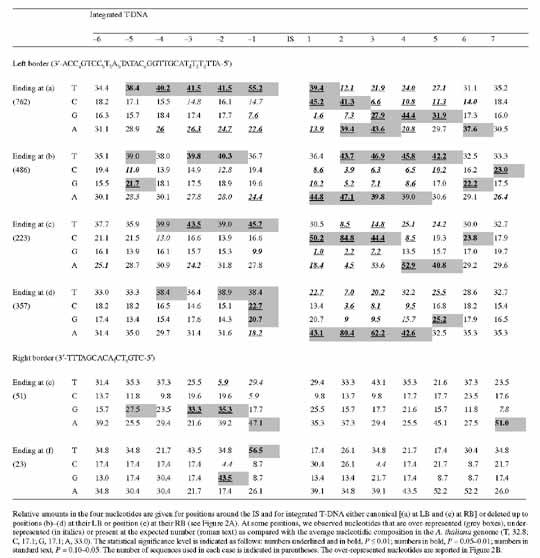
We characterized further the sequences upstream and downstream of 4430 IS. These two regions are defined with respect to the integrated T-DNA: the region upstream of the IS is the region that would be sequenced with a primer identical to a sequence in the LB, and the region downstream of the IS is the region that would be sequenced with a primer designed from the RB. In a region close to the IS, the nucleotide composition is different from the regional composition (Figure 3). We postulated that it might reflect the previously proposed role of microsimilarities (often defined as microhomologies) between host DNA and T-DNA sequences in the integration process (Tinland, 1996). This hypothesis, based on a small number of IS, can be tested with the larger set of IS sequences available: FSTs showing different lengths of deletion in the integrated T-DNA (Figure 2A) might exhibit modified microsimilarity when compared to canonical IS. Consistently, the most frequently observed sequence downstream of the plant IS (Table 1, Figure 2B) is related to the sequence at the end of the integrated T-DNA [indicated by arrows (a)–(d) below the T-DNA sequence in the abscissa in Figure 2A]. For instance, when the integrated T-DNA ends at position (c), (Figure 2A), the over-represented nucleotides in the plant IS indicate that the IS consensus sequence is CCCAAC (Table 1; Figure 2B downstream), whereas it is AAAAG when the integrated T-DNA ends at position (d). There is, in all cases, an imperfect but striking similarity between the complement of the 3′ sequence of integrated T-DNA and the consensus at the plant IS (Figure 2A and B). Thus, for canonical insertions [case (a) in Figure 2A and B], the consensus sequence 5′-(C/T)(A/C)(G/A)GGA-3′ in the plant genome has some similarity with the complement of the sequence 3′-GTCCT-5′ (i.e. CAGGA) at the end of the T-DNA. The occurrence observed for one nucleotide may be as high as 84.8% at the plant IS (Table 1). Nevertheless, the microcomplementarities observed between the T-DNA and plant DNA are not perfect, since, in many cases, two nucleotides may be over-represented at the same position. Indeed, as illustrated for the canonical insertion (Figure 2C), this may be explained by the alignment of two sequences (5′-CAGGAN-3′ and 5′-TCAGGA-3′), complementing the same sequence in the T-DNA LB (3′-GTCCT-5′), but shifted by one nucleotide. This strongly suggests a frequent deletion of one nucleotide downstream of the IS as a consequence of the integration. Inspection of individual sequences downstream of the canonical IS indicates that there is a clear shift due to the presence of one nucleotide before the microsimilarity in 53% of IS, either a T (35%), an A (11%), a C (6%) or a G (<1%). The relative nucleotidic representation of the first five positions upstream of the IS has been re-computed, taking into consideration, when necessary, the shift of one nucleotide between the apparent and actual IS. Corrected values are 71, 59, 39, 45 and 40% for C, A, G, G and A positions, respectively. Therefore, the consensus sequence of the microsimilarity, at the plant IS, for canonical integrations is clearly 5′-CAGGA-3′. Alignments with no accepted gap of each IS corresponding to canonical T-DNA insertions with the 5′-CAGGA-3′ sequence show that a majority of IS (63%) exhibits an identity of at least 50% with this sequence (Figure 2D). The same alignments with sequences randomly taken from the genome provide only 19% of sequences with this score. Thus, collectively, our data show that, whatever the position of the cut in the T-DNA, there is a microsimilarity between the integrated T-DNA border and the plant IS.
Figure 3.
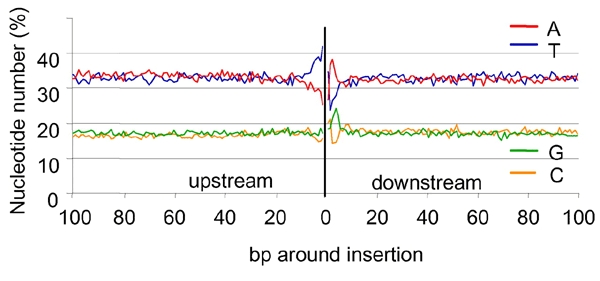
Relative base composition of the A. thaliana DNA at T-DNA IS. IS were mapped in the A. thaliana genome by sequence comparison between 4330 FSTs obtained from the T-DNA LB and not flanked by filler DNA.
Table 1.
Sequences around T-DNA IS in the A. thaliana genome
 |
|---|
Interestingly, the plant genome sequence upstream of the T-DNA IS shows an over-representation of the nucleotide T (Table 1). It is particularly striking when FSTs correspond to canonical insertions at the LB. In this case, upstream of the plant IS there is a highly significant occurrence of T at five contiguous positions that cannot be due to sequence matches between the T-DNA and the plant genome. If, as discussed above, the shift of one nucleotide introduced in the apparent IS by a deletion of one nucleotide is taken into consideration, the T representation becomes 38, 40, 45 and 73% at the −5 to −1 position upstream of IS, respectively. Thus, the T-rich region that frequently ends by a T is located immediately upstream of the microsimilarity region. Half of the five nucleotide sequences upstream of the IS contain at least three Ts.
Lastly, we searched for microsimilarities between the RB of the single-stranded T-DNA and the complement of the plant DNA. We observed a significant over-representation of Gs, in the plant DNA, at position −2 from the IS, either characterized by FSTs from the RB of canonical T-DNA insertions [(e) in Figure 2A and B] or FSTs from T-DNAs nicked between the second and the third bases [(f) in Figure 2A and B). These results show that in both cases the nucleotide G is significantly over-represented, in plant sequences, at 2 bp from the IS and it is preceded by an over-representation of A in the case where the T-DNA end is canonical, tttagcacaCT, and T when the T-DNA end is tttagcaCA.
Discussion
We have generated and analysed a large set of integrated T-DNAs and their respective pre-IS. We confirmed and further characterized the involvement of a microsimilarity between the T-DNA LB sequence and host DNA IS previously observed in a limited number of IS (Tinland et al., 1995) and proposed to be the docking force for integration (Tinland, 1996). Interestingly, we found that even in non-canonical insertions a microsimilarity with the host DNA is also present. Most of the microsimilarities involved the first 25 nucleotides of the LB, but some are observed up to the sequence of the oligonucleotide used to prime the FST sequencing. As a consequence of the small length of the microsimilarity and the large region of the T-DNA in which it may be found, the T-DNA may potentially integrate anywhere in the plant genome. However, we showed an over-representation of Ts at five positions immediately upstream of the IS, indicating that T-DNA integration is influenced by the nucleotide composition at the pre-IS independently of any microsimilarity. The preference for a T-rich context for T-DNA integration may explain favoured integrations in the gene region upstream of the start codon, as well as the higher density of FSTs found in introns than in exons. Our data have also increased our knowledge on the recombination reaction at the 5′ end of the T-DNA. Previous results (Tinland et al., 1995) indicated that an identity existed between at least the nucleotide linked to VirD2 and the last nucleotide of the plant IS. We extended these observations and demonstrated that the identity between the 5′ end of the T-DNA and the plant pre-IS often involved the last nucleotide but also a G located immediately downstream. Our data statistically support the model for T-DNA integration previously proposed by Tinland (1996). Taking all our new data into consideration, we propose a model that mainly differs from Tinland's model by the preference for T-DNA integration in the vicinity of a T-rich region (Figure 4). The following five steps are involved. (1) The integration process, often initiated by the 3′ (LB) of the T-DNA invading a poly T-rich site of the host DNA. (2) Upstream of the 3′ end of the singlestranded T-DNA, a more or less perfect duplex with the top strand of the host DNA is formed. Our findings are the first proof of a link between the location of the microsimilarity in the T-DNA and the cut in the LB of the integrated T-DNA. (3) After degradation of the 3′ end portion of the T-DNA downstream of the duplex, the ligation between the digested bottom strand of the host DNA and the 3′ end of the T-DNA is performed by host enzymes. We assume that the 1 bp deletion frequently observed in the plant DNA is a consequence of this digestion-ligation step. (4) A nick in the upper host DNA strand is created downstream of the duplex and used to initiate the synthesis of the complementary strand of the invading T-DNA. The imperfect matches in the duplex are detected and repaired by host enzymes, using the invading T-DNA sequence as a template. (5) The right end of the T-DNA is ligated to the bottom strand of the host DNA. The pairing frequently involves a G and another nucleotide upstream of it. (6) The top strand of the host DNA is degraded between the two microsimilarities and a ligation with the synthesized complementary T-DNA is made. This may result in a deletion of variable length in the host DNA. Out of 180 transformants for which we have FSTs from both sides of the integrated T-DNA, 88 have apparent deletions shorter than 150 bases.
Figure 4.
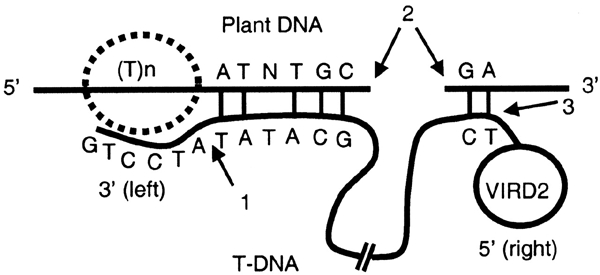
A model of the T-DNA integration process. The given example is for a putative T-DNA that, as a consequence of its integration is deleted by six nucleotides in the 3′ LB [position (b) in Figure 2A] but complete in its RB. For convenience, only the upper strand of the host DNA is represented. A T-rich region [T(n)] operates as a preferential site of entry of the T-DNA LB. Starting from its 3′ end, the T-DNA scans the plant DNA until it finds a microcomplementarity just downstream of the T-rich region. A nick (2) is generated in the host DNA downstream of the microcomplementarity-based duplex and used as a priming site to synthesize the complementary strand of the T-DNA until the 5′ RB covalently linked to VirD2 is reached. The integration process of the newly synthesized doublestranded DNA frequently brings about a deletion in the host DNA. Recombination between the host bottom strand and the T-DNA operates at sites 1 and 3 after the action of exonucleases on both ends of the T-DNA.
Some of the results presented here differ from previously published data, and one of the possible explanations is that for the first time we used a set of FSTs not biased by an analysis of a particular set of mutants. Secondly, we used a large set of data, enough to be statistically representative of the integration process.
Speculation
The insertion of T-DNA into the genome, may have recruited some of the cellular processes involved in illegitimate recombination (Tzfira and Citovsky, 2002). We demonstrate that the primary docking force of the T-DNA towards the plant IS may be a poly T-rich stretch in the plant DNA. An observed over-representation of T-rich sequences has been observed in other microsimilarity-mediated recombinations (Kohli et al., 1999). An A-T-rich sequence is a region with both a low DNA duplex stability (Breslauer et al., 1986) and a strong bending (Bolshoy et al., 1991). Bending rather than sequence itself has been shown to favour retroviral integration in vitro (Müller and Varmus, 1994) and P transposable element integration in the Drosophila genome (Liao et al., 2000). Recognition of a bended DNA region might, therefore, be a common feature in the integration of foreign DNA in eucaryote genomes.
Methods
The collection of T-DNA transformant lines has been generated with the A. thaliana ecotype Wassilewskija at the Institut National de la Recherche Agronomique (INRA Versailles), using the A. tumefaciens strain C58C1 (pMP90; Bechtold et al., 1993). The protocol used for obtaining FSTs is described in a previous paper (Balzergue et al., 2001), and details on FST sequences are available through the FLAGdb/FST database (Samson et al., 2002; http://genoplante-info.infobiogen.fr). The FST set analysed contained 8919 non-redundant sequences unequivocally mapped at only one locus of the plant genome. This FST set did not contain FSTs indicative of complex IS such as tandem insertions of two T-DNA. Only FSTs not flanked by filler DNA were used. About 20% of FSTs contain stretches of bases downstream of the end of the inserted T-DNA that do not match plant, plasmid or any known sequences. In most of cases, this filler DNA is shorter than 50 bases and its nucleotide composition is the same as the average in the A. thaliana genome. The presence of DNA filler at sites of non-homologous recombination is thought to be a consequence of DNA break repairing. BLAST programs (Altschul et al., 1997) were used to align FSTs with the five chromosomes of A. thaliana. The pseudo-molecules were downloaded from the TIGR site (http://www.tigr.org), with the associated coding sequence annotations (ID 68170, 51595, 68173, 68164 and 68172). A gene is interrupted by a T-DNA when a FST starts in the genomic region, including the predicted CDS (coding sequence) for this gene and 200 bp on each side.
Acknowledgments
The authors are grateful to Mickael Hodges and Ian Small for reading the manuscript. This work was supported by the French Génoplante Program.
References
- Altschul S.F., Gish W., Miller W., Myers E.W. and Lipman D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Azpiroz-Leehan R. and Feldmann K.A. (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet., 13, 152–156. [DOI] [PubMed] [Google Scholar]
- Balzergue S. et al. (2001) Improved PCR-Walking for largescale isolation of plant T-DNA borders. Biotechniques, 30, 496–504. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis J. and Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by filtration of adult Arabidopsis thaliana plants. CR Acad. Sci. (Paris), 316, 1194–1199. [Google Scholar]
- Bolshoy A., McNamara P., Harrington R.E. and Trifonov E.N. (1991) Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl Acad. Sci. USA, 88, 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer K.J., Frank R., Blocker H. and Marky L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrenberger F., Crameri A., Hohn B. and Koukolfkova-Nicola Z. (1989) Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc. Natl Acad. Sci. USA, 86, 9154–9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati M., Moreno M.A., Nadzan G., Zouyrelidou M. and Dellaporta S.L. (2000) Largescale T-DNA mutagenesis in Arabidopsis for functional genomic analysis. Funct. Integr. Genom., 1, 25–34. [DOI] [PubMed] [Google Scholar]
- Gelvin S.B. (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol., 51, 223–256. [DOI] [PubMed] [Google Scholar]
- Gheysen G., Villarroel R. and Van Montagu M. (1991) Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev., 5, 287–297. [DOI] [PubMed] [Google Scholar]
- Kohli A., Griffiths S., Palacios N., Twyman R.M., Vain P., Laurie D.A. and Christou P. (1999) Molecular characterization of transforming plasmid rearrangements in transgenic rice reveals a recombination hotspot in the CaMV 35S promoter and confirms the predominance of microhomology mediated recombination. Plant J., 17, 591–601. [DOI] [PubMed] [Google Scholar]
- Liao G.C., Rhem E.J. and Rubin G.M. (2000) Insertions site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 97, 3347–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer R. et al. (1991) T-DNA integration: a mode of illegitimate recombination in plants. EMBO J., 10, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.P. and Varmus H.E. (1994) DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J., 13, 4704–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Schoumacher F., Hohn B. and Lanka E. (1993) Sitespecific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc. Natl Acad. Sci. USA, 90, 11538–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M. and Lecharny A. (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res., 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative, AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Tinland B. (1996) The integration of T-DNA into plant genomes. Trends Plant Sci., 1, 178–184. [Google Scholar]
- Tinland B., Schoumacher F., Gloecker V., Bravo-Angel A.M. and Hohn B. (1995) The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J., 14, 3585–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T. and Citovsky V. (2002) Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol., 12, 121–129. [DOI] [PubMed] [Google Scholar]
- Zupan J., Muth T.R., Draper O. and Zambryski P. (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J., 23, 11–28. [DOI] [PubMed] [Google Scholar]