MetaCyc: a multiorganism database of metabolic pathways and enzymes (original) (raw)
Abstract
MetaCyc is a database of metabolic pathways and enzymes located at http://MetaCyc.org/. Its goal is to serve as a metabolic encyclopedia, containing a collection of non-redundant pathways central to small molecule metabolism, which have been reported in the experimental literature. Most of the pathways in MetaCyc occur in microorganisms and plants, although animal pathways are also represented. MetaCyc contains metabolic pathways, enzymatic reactions, enzymes, chemical compounds, genes and review-level comments. Enzyme information includes substrate specificity, kinetic properties, activators, inhibitors, cofactor requirements and links to sequence and structure databases. Data are curated from the primary literature by curators with expertise in biochemistry and molecular biology. MetaCyc serves as a readily accessible comprehensive resource on microbial and plant pathways for genome analysis, basic research, education, metabolic engineering and systems biology. Querying, visualization and curation of the database is supported by SRI's Pathway Tools software. The PathoLogic component of Pathway Tools is used in conjunction with MetaCyc to predict the metabolic network of an organism from its annotated genome. SRI and the European Bioinformatics Institute employed this tool to create pathway/genome databases (PGDBs) for 165 organisms, available at the BioCyc.org website. These PGDBs also include predicted operons and pathway hole fillers.
INTRODUCTION
MetaCyc is a reference database of small molecule metabolism that contains experimentally verified pathway and enzyme information curated from the scientific literature (1). A metabolic pathway in MetaCyc consists of reactions, enzymes, metabolites, information on feedback regulation and genes that encode the enzymes for each species (Figure 1). The current version of MetaCyc (9.5) contains 621 pathways from >500 species (Tables 1 and 2) ranging from microbes to plants and humans, with >90% of the information curated from >7300 research articles. MetaCyc can be searched and browsed using a web browser. Pathways are dynamically generated from the database and graphically displayed with hyperlinks to various pages detailing reactions, enzymes, genes and compounds from MetaCyc, as well as external databases such as Swiss-Prot and PubMed. It, therefore, serves as a readily accessible source of up-to-date, literature-curated information on metabolic pathways and enzymes to researchers for use in basic research and genome analysis, and to students and teachers for educational purposes. In addition, MetaCyc, in conjunction with the Pathway Tools software (2), can be used to predict metabolic networks from a list of annotated sequences resulting from genome or transcript sequencing (3–5). Those predicted networks can provide a knowledge framework onto which reaction flux models can be built.
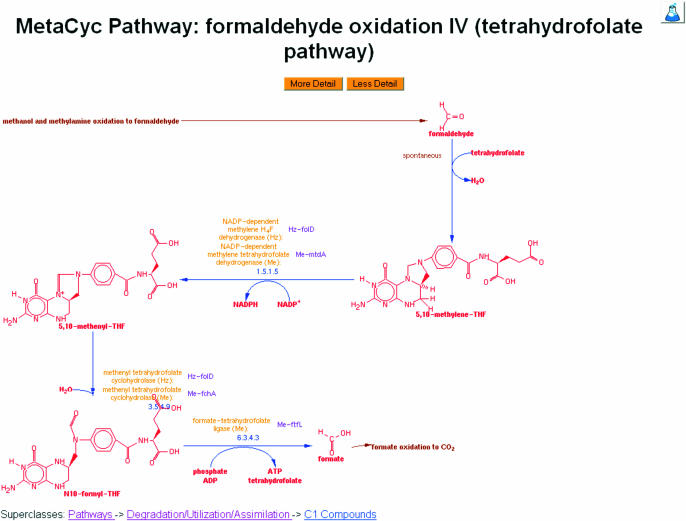
Figure 1.
A representative example of a pathway in MetaCyc. Pathways can be displayed at varying levels of detail. This pathway display depicts an intermediate level of detail including enzymes, EC numbers, genes and chemical structures of the main compounds. Notice the brown arrows that provide hyperlinks to related upstream and downstream pathways.
Table 1.
List of species that have five or more experimentally elucidated pathways represented in MetaCyc
| Bacteria | No. of pathways | Eukarya | No. of pathways | Archaea | No. of pathways |
|---|---|---|---|---|---|
| Escherichia coli | 179 | Arabidopsis thaliana | 116 | Sulfolobus solfataricus | 15 |
| Pseudomonas putida | 35 | Homo sapiens | 42 | Methanocaldococcus jannaschii | 5 |
| Bacillus subtilis | 24 | Glycine max | 36 | ||
| Pseudomonas aeruginosa | 19 | Saccharomyces cerevisiae | 31 | ||
| Mycobacterium tuberculosis | 17 | Pisum sativum | 20 | ||
| Salmonella typhimurium | 14 | Zea mays | 19 | ||
| Haemophilus influenzae | 13 | Solanum tuberosum | 12 | ||
| Deinococcus radiodurans | 12 | Cicer arietinum | 12 | ||
| Mycoplasma pneuomiae | 8 | Oryza sativa | 12 | ||
| Mycobacterium smegmatis | 8 | Spinacia oleraca | 11 | ||
| Sinorhizobium meliloti | 8 | Rattus norvegicus | 10 | ||
| Thauera aromatica | 7 | Hordeum vulgare | 8 | ||
| Klebsiella pneumoniae | 7 | Nicotiana tabacum | 8 | ||
| Lactococcus lactis | 6 | Lycopersicon esculentum | 7 | ||
| Corynebacterium glutamicum | 6 | Medicago sativa | 7 | ||
| Methylobacterium extorquens AM1 | 6 | Brassica napus | 6 | ||
| Pseudomonas fluorescens | 6 | Glycyrrhiza echinata | 6 | ||
| Thermotoga maritima | 6 | Triticum aestivum | 6 | ||
| Paracoccus denitrificans | 5 | Cucumis sativus | 5 | ||
| Arthrobacter globiformis | 5 | Ricinus communis | 5 | ||
| Mycoplasma capricolum | 5 | Pueraria montana | 5 | ||
| Bradyrhizobium japonicum | 5 | ||||
| Bacillus cereus | 5 |
Table 2.
The distribution of pathways in MetaCyc based on the taxonomic classification of associated species
| Bacteria | No. of pathways | Eukarya | No. of pathways | Archaea | No. of pathways |
|---|---|---|---|---|---|
| Proteobacteria | 598 | Viridiplantae | 396 | Euryarchaeota | 47 |
| Firmicutes | 169 | Metazoa | 61 | Crenarchaeota | 26 |
| Actinobacteria | 94 | Fungi | 58 | ||
| Deinococcus-Thermus | 18 | Euglenozoa | 4 | ||
| Cyanobacteria | 13 | ||||
| Thermotogae | 8 | ||||
| Planctomycetes | 6 | ||||
| Bacteroidetes/Chlorobi | 6 | ||||
| Nitrospirae | 5 | ||||
| Aquificae | 2 | ||||
| Chloroflexi | 2 | ||||
| Spirochaetes | 1 | ||||
| Chlamydiae | 1 | ||||
| Chrysiogenetes | 1 |
A number of major improvements made in the last 2 years are described in this article. There has been a significant increase in the content of the database, covering both primary metabolism and less common pathways, such as microbial degradation of environmental pollutants and plant secondary metabolism. Other improvements include reorganization and expansion of both the pathway and cellular component ontologies, and enhancement of the Web pages providing background information about MetaCyc, such as the user's guide at URL http://metacyc.org/MetaCycUserGuide.shtml. For a thorough discussion of the major differences between MetaCyc and other pathway databases please see http://metacyc.org/MetaCycUserGuide.shtml#otherpathwaydbs.
DATA CONTENT
As demonstrated in Table 3, there has been a significant increase in the number of database objects since the last Nucleic Acids Research publication 2 years ago (1). The number of metabolic pathways has increased by 26% from 491 to 621, while the number of enzymes, genes and citations has grown considerably more, by 75, 71 and 140%, respectively, owing to the fact that many existing pathways have been extensively edited and updated with comments, enzymes, genes and citations. There has also been a 128% increase in the number of organisms represented (currently at 506), reflecting the breadth of MetaCyc (Tables 1 and 2), and a 57% increase in the number of chemical compounds (currently 4620).
Table 3.
The size of MetaCyc as a function of time from its first release in 1999 to the latest release in 2005 (version 9.5)
| Database objects | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 |
|---|---|---|---|---|---|---|---|
| Metabolic pathways | 296 | 366 | 445 | 460 | 491 | 528 | 621 |
| Metabolic pathways with comments | 39 | 83 | 160 | 180 | 232 | 280 | 412 |
| Enzymatic reactions | 3779 | 4002 | 4218 | 4294 | 4817 | 4955 | 5428 |
| Enzymes | 82 | 344 | 1115 | 1267 | 1543 | 1940 | 2698 |
| Enzymes with comments | 75 | 234 | 1054 | 1123 | 1389 | 1716 | 2376 |
| Genes | 0 | 0 | 0 | 600 | 1554 | 1821 | 2662 |
| Compounds | 1949 | 2180 | 2335 | 2404 | 2951 | 3551 | 4620 |
| Literature citations | 184 | 604 | 2381 | 2718 | 3070 | 5050 | 7368 |
Data in MetaCyc are curated from the experimental literature, which is read and summarized by PhD-level curators. Curators at SRI cover microbial and animal pathways, while curators at Carnegie Institution cover pathways from higher plants. MetaCyc contains the full complement of EcoCyc metabolic pathways (6) and thus has most of the basic pathways of central and intermediary metabolism typical of enteric bacteria. Similarly, most of the pathways of central metabolism in higher plants are present in the database. Our current curation strategy focuses on both depth and breadth, by updating pathways that lack complete information, and adding new pathways outside of central metabolism.
The MetaCyc Curator's Guide for Pathway/Genome Databases, located at URL http://bioinformatics.ai.sri.com/ptools/curatorsguide.pdf, was developed to ensure the consistency of curation procedures. It documents the type of information that should be captured for each pathway, reaction, enzyme, gene and chemical compound. It also describes stylistic conventions. We recently revised it to explicitly define the organization of metabolic pathways in MetaCyc, and to provide detailed guidelines for defining pathway boundaries (the compounds with which a pathway should begin and end) and for defining links between pathways.
To ensure the accuracy and coverage of new and existing pathways, we are currently in the process of inviting outside experts to support the database as editors and/or curators in their fields of expertise.
Pathways, enzymes, reactions and compounds
Most of the pathways in MetaCyc occur in the microorganism and plant kingdoms, a manifestation of their metabolic diversity. Nevertheless, animal pathways are also represented. Since November 2003 we added >140 new pathways in the areas of small molecule intermediary metabolism, and the biosynthesis and degradation of natural environmental compounds, environmental pollutants, xenobiotics and compounds involved in general cellular processes and secondary metabolism. Of these 63 are plant pathways, many of which concern the production of compounds involved in cellular regulation processes (phytohormones) and defense mechanisms (phytoalexins and phytoanticipins), and plant secondary (or specialized) metabolites. To accommodate these new types of pathways, we expanded the pathway ontology to include categories for plant secondary metabolism, comprising 8 main classes and 26 subclasses (see http://biocyc.org/META/class-tree?object=Pathways). Selection criteria for the curation of these plant pathways include generality of occurrence across taxa, investigation in a model species like Arabidopsis and agronomic or medicinal importance. Arabidopsis pathways are exported to AraCyc, the Arabidopsis metabolism database (5) that was computationally predicted using MetaCyc as the reference database.
In mammalian metabolism we added multiple new pathways, including those describing human neurotransmitter biosynthesis (in collaboration with experimentalists in this field), drug metabolism, cholesterol biosynthesis, arsenate detoxification and glutathione metabolism. These pathways were either curated within MetaCyc, or propagated from the HumanCyc database (3). In addition, several existing pathways of intermediary metabolism were curated with rat enzymes and genes.
In parallel with curating new pathways, we extensively edited previously existing pathways. Approximately 60 microbial and 7 plant pathways have been updated and enhanced since 2003. One of our highest priorities is the curation of existing pathways that are in need of updating with enzymes, EC numbers, genes and comments with literature citations. While adding new pathways or revising existing pathways, we are also expanding coverage of pathway variants found in different organisms.
In addition to curation within MetaCyc, we continue to import pathways from other databases. At each quarterly release we propagate newly curated pathways from EcoCyc (6) and HumanCyc (3) into MetaCyc. We encourage outside curators of our BioCyc family of PGDBs to submit curated pathways to us for possible inclusion in MetaCyc. For example, we incorporated several new yeast pathways in collaboration with curators from the Saccharomyces Genome Database (SGD) (4). We minimize redundancy by associating several representative species with a pathway that is shared among them. We are in the process of refining the database by deleting pathways that are deemed redundant, dividing large pathways that contain overlapping sections into separate, smaller pathways, and assembling small, related pathways into superpathways, to give an overview of metabolic interrelationships.
A new pathway evidence code, EV-EXP-TAS (evidence-experimental-traceable author statement) was created to allow curators to cite review articles containing direct references to the primary literature in support of the pathways. We found this type of reference to be the most useful for large, complex pathways.
We are increasingly enhancing the quality of enzyme information in MetaCyc by adding more kinetic data, including _K_m values and optimal pH and temperature, and listing enzyme regulators, including activators, inhibitors, cofactors and alternative substrates. We have revised and extended the categories for enzyme regulators based on their kinetics. Activators are now classified as allosteric, nonallosteric or of unknown mechanism, and inhibitors are classified as competitive, noncompetitive, uncompetitive, allosteric, irreversible, none of the preceding or of unknown mechanism. Definitions of these types of regulators are provided in Appendix I of (7). It should be noted that while kinetic data are present for most enzymes curated within the last 2 years, there are still many enzymes in the database which lack kinetic data, either because they were curated earlier and have not yet been updated, or because no such data are available in the literature.
We introduced hyperlinks within pathway and enzyme comments, which can link commentary text to any data in the database. This dynamic linking greatly improves the ability of users to navigate between related database objects.
Our chemical library has grown significantly in the past 2 years, from 2951 to 4620 compounds. Currently, over 93% of our compounds have structures. In addition, last year we began adding stereochemistry representations to the structures.
Taxonomies
We significantly enhanced the cellular component ontology to annotate the subcellular locations of enzymes and to encode the cellular compartments involved in transport reactions. The ontology, which has been described recently (8), currently comprises 160 terms. More about the enhanced cell component ontology can be found at http://bioinformatics.ai.sri.com/CCO/.
MetaCyc is routinely updated with the latest data from the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB), which includes new and modified EC numbers. The last supplement to have been incorporated is supplement 10 (http://www.chem.qmul.ac.uk/iubmb/enzyme/supplements/sup2004/).
Links to other databases
While MetaCyc (unlike organism-specific PGDBs) does not contain sequence information, we use extensive linking to external amino acid and nucleotide sequence databases. Whenever possible, enzyme and gene entries include links to Swiss-Prot (9) and the Entrez Nucleotide and Gene databases. Arabidopsis genes are linked to TAIR (The Arabidopsis Information Resource). Enzymes are often linked to protein structure databases such as PDB (10) when applicable. In addition, whenever possible, literature references are linked to PubMed.
ENHANCEMENTS TO THE PATHWAY TOOLS SOFTWARE
The Pathway Tools software provides query and visualization services to users and editing functions to curators (2,11). Recent enhancements to the software that are relevant to MetaCyc users include the following:
- Improved displays: The pathway display algorithms have been modified to produce more compact pathway diagrams that are more likely to fit within a single page.
- Enzyme/gene naming: Protein and gene names within a pathway display are now labeled with the initials of an organism's genus and species name (e.g. an Escherichia coli enzyme and gene are written as ‘acetylornithine decarboxylase (Ec)’ and ‘Ec-argD’, respectively). This notation aids in identifying individual proteins and genes from multiple organisms that are assigned to the same pathway.
- Chemical drawing tools: The Pathway Tools software now includes interfaces for both the Marvin (http://www.chemaxon.com/marvin/) and JME (http://www.molinspiration.com/jme/) chemical drawing editors, permitting the user to enter or modify chemical structures within MetaCyc or other PGDBs.
- A new suite of comparative genomics tools is available in Pathway Tools in conjunction with the preceding expansion of the BioCyc database collection (see below). These tools include comparisons of the full pathway, reaction and metabolite sets present in a specified group of organisms; comparisons of genes associated with a single pathway or a single reaction across a specified group of organisms, including the operon distributions of those genes; and a comparative genome browser for visualizing chromosomal regions around a specified set of orthologous genes.
Expansion of BioCyc
A major application of MetaCyc is its use for predicting the metabolic pathways of an organism from its sequenced genome (12–14), using the PathoLogic program (15). We recently automated PathoLogic, allowing it to be applied to large numbers of genomes. Jointly with the European Bioinformatics Institute, we used this feature to expand our BioCyc collection of PGDBs to >200 organism-specific databases, each of which contains the predicted metabolic pathways of the organism, based on its annotated genome (16). These PGDBs are available for adoption and ongoing curation by the scientific community.
Each PGDB contains the genome of the organism, which is accessible using the new Pathway Tools genome browser (http://biocyc.org/ANTHRA/NEW-IMAGE?type=FULL-MAP&object=DNAA), predicted operons (only for bacteria) (17) and predicted metabolic pathways (15). In addition, since predicted pathways often contain pathway holes (reactions in a predicted metabolic pathway for which no enzymes have been identified in the sequenced genome), we applied our pathway-hole filler algorithm to all the BioCyc PGDBs. This algorithm searches the sequenced genome and identifies candidate genes for these missing enzymes (18).
All the PGDBs in the BioCyc collection can be used to analyze gene and protein expression data using the Omics Viewer, a Pathway Tools feature that allows expression data and metabolomics data to be painted onto the full metabolic network of an organism.
DATABASE AND SOFTWARE AVAILABILITY
MetaCyc is freely available via the Web at http://Metacyc.org/ (updated four times a year). It is also available for download, free of charge to non-profit organizations or for a fee to commercial institutions, as a stand-alone application program for Linux, Windows and Solaris workstations (updated two times a year). A set of flat data files that is updated four times a year is also available online at http://BioCyc.org/download.shtml.
Acknowledgments
We thank Teresa Steininger and Thomas Kilduff for their help with the catecholamine biosynthesis pathway, Eurie L. Hong and Rama Balakrishnan for the Saccharomyces pathways they submitted to MetaCyc, and Tanya Berardini, Leonore Reiser and Wolf Frommer for reviewing the cellular component ontology. We thank Sunita Reddy for contributions to the chemical compound data, and Aleksey Kleytman and Thomas Yan for curation assistance. This work was supported by grant R01-RR07861-01 from the NIH National Institute of General Medical Sciences, and by grant R01-HG02729-01 from the NIH National Human Genome Research Institute. P.Z. and S.Y.R. were supported in part by NSF grant DBI-0417062 and C.T. was supported, in part, by a gift from Pioneer Hi-Bred. Funding to pay the Open Access publication charges for this article was provided by grant GM70065 from the NIH National Institute of General Medical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Krieger C.J., Zhang P., Mueller L.A., Wang A., Paley S., Arnaud M., Pick J., Rhee S.Y., Karp P.D. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32:D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karp P.D., Paley S., Romero P. The pathway tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 3.Romero P., Wagg J., Green M.L., Kaiser D., Krummenacker M., Karp P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2004;6:R2.1–R2.17. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weng S., Dong Q., Balakrishnan R., Christie K., Costanzo M., Dolinski K., Dwight S.S., Engel S., Fisk D.G., Hong E., et al. Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res. 2003;31:216–218. doi: 10.1093/nar/gkg054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller L.A., Zhang P., Rhee S.Y. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SRI International. Pathway Tools User Guide. Menlo Park, CA: SRI International; 2005. [Google Scholar]
- 8.Zhang P., Foerster H., Tissier C.P., Mueller L., Paley S., Karp P.D., Rhee S.Y. MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol. 2005;138:27–37. doi: 10.1104/pp.105.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The Swiss-Prot protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karp P.D. Pathway Databases: a case study in computational symbolic theories. Science. 2001;293:2040–2044. doi: 10.1126/science.1064621. [DOI] [PubMed] [Google Scholar]
- 12.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 13.Wood D.W., Setubal J.C., Kaul R., Monks D.E., Kitajima J.P., Okura V.K., Zhou Y., Chen L., Wood G.E., Almeida N.F., Jr, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 14.Larsson P., Oyston P.C., Chain P., Chu M.C., Duffield M., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nature Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 15.Paley S.M., Karp P.D. Evaluation of computational metabolic-pathway predictions for H.pylori. Bioinformatics. 2002;18:715–724. doi: 10.1093/bioinformatics/18.5.715. [DOI] [PubMed] [Google Scholar]
- 16.Karp P.D., Ouzounis C.A., Moore-Kochlacs C., Goldovsky L., Kaipa P., Ahrén D., Tsoka S., Darzentas N., Kunin V., López-Bigas N. Expansion of the BioCyc collection of Pathway/Genome Databases to 160 Genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero P.R., Karp P.D. Using functional and organizational information to improve genome-wide computational prediction of transcription units on pathway-genome databases. Bioinformatics. 2004;20:709–717. doi: 10.1093/bioinformatics/btg471. [DOI] [PubMed] [Google Scholar]
- 18.Green M.L., Karp P.D. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]