Pseudomonas aeruginosa Displays Multiple Phenotypes during Development as a Biofilm (original) (raw)
Abstract
Complementary approaches were employed to characterize transitional episodes in Pseudomonas aeruginosa biofilm development using direct observation and whole-cell protein analysis. Microscopy and in situ reporter gene analysis were used to directly observe changes in biofilm physiology and to act as signposts to standardize protein collection for two-dimensional electrophoretic analysis and protein identification in chemostat and continuous-culture biofilm-grown populations. Using these approaches, we characterized five stages of biofilm development: (i) reversible attachment, (ii) irreversible attachment, (iii) maturation-1, (iv) maturation-2, and (v) dispersion. Biofilm cells were shown to change regulation of motility, alginate production, and quorum sensing during the process of development. The average difference in detectable protein regulation between each of the five stages of development was 35% (approximately 525 proteins). When planktonic cells were compared with maturation-2 stage biofilm cells, more than 800 proteins were shown to have a sixfold or greater change in expression level (over 50% of the proteome). This difference was higher than when planktonic P. aeruginosa were compared with planktonic cultures of Pseudomonas putida. Las quorum sensing was shown to play no role in early biofilm development but was important in later stages. Biofilm cells in the dispersion stage were more similar to planktonic bacteria than to maturation-2 stage bacteria. These results demonstrate that P. aeruginosa displays multiple phenotypes during biofilm development and that knowledge of stage-specific physiology may be important in detecting and controlling biofilm growth.
Recent investigations have been directed at determining the degree to which gene regulation during biofilm development controls the switch from planktonic to attached growth. Brözel and coworkers monitored changes in global gene expression patterns in attached Pseudomonas aeruginosa cells and found more than 11 proteins whose levels were altered during various stages of attachment (6). Genevaux and colleagues screened a library of Tn_10_ insertion mutants of Escherichia coli with altered adhesion abilities (18). Fifty adhesion-deficient mutants were isolated which showed less than 40% attachment compared to the wild type, and 22 mutants were found with an attachment of 40 to 75% compared to the wild type. The majority of these mutants were affected in motility. Prigent-Combaret and coworkers (42) carried out a screen in E. coli K-12 similar to that of Genevaux and revealed major changes in the patterns of gene expression during the switch from planktonic to attached growth. They found attachment-dependent regulation of gene expression in 38% of the generated lacZ gene fusions (out of 446 clones). More recently, it has been shown that in Pseudomonas putida, more than 30 genes and 40 gene products were altered within 6 h following attachment (47).
These results indicate that physiological changes in the transition from planktonic to attached cells are profound and undoubtedly complex. In addition, these results demonstrate that biofilm bacteria are physiologically different from planktonic bacteria, but much work is still needed to completely characterize the developmental process of biofilm formation. One of the stepping stones in achieving this goal is to characterize the complete suite of physiological changes that occur during transitional episodes in biofilm development, from initial attachment through detachment and reentry into planktonic growth.
While the reports above focus on initial attachment, the present study analyzed the physiological changes experienced by P. aeruginosa during all stages of biofilm development. Our strategy was to characterize transitional episodes in biofilm development through direct observation by microscopy, evaluating biofilm morphology, matrix polymer production, and activation of quorum sensing-regulated genes. Knowledge of the onset of physiological characteristics specific to each phase was used to determine the sampling times for protein analyses by two-dimensional gel electrophoresis. These analyses were performed by coordinating protein sampling with the onset of biofilm-specific characteristics such as activation of quorum sensing-regulated genes. Activation of quorum sensing was confirmed on the protein level by comparing protein patterns of P. aeruginosa strain PAO1 to the P. aeruginosa LasI mutant strain PAO-JP1, a strain deficient in production of the las quorum-sensing signal molecule 3O-C12-HSL (36). The study indicated that P. aeruginosa displays multiple phenotypes with distinct physiological characteristics (structural and metabolic changes) that can be correlated to the episodes of biofilm development.
MATERIALS AND METHODS
Bacterial strains and media.
The microorganisms used in this study are listed in Table 1. All planktonic cultures of P. aeruginosa and P. putida were grown at room temperature in glass chemostats (300 ml; flow rate 1.7 ml min−1) in minimal medium containing (per liter of H2O) 2.56 g of Na2HPO4, 2.08 g of KH2PO4, 1.0 g of NH4Cl, 0.132 g of CaCl2 · 2H2O, 0.5 g of MgSO4 · 7H2O, 0.1 mg of CuSO4 · 5H2O, 0.1 mg of ZnSO4 · H2O, 0.1 mg of FeSO4 · 7H2O, and 0.004 mg of MnCl2 · 4H2O (pH 7.0). Glutamic acid (130 mg/liter) was used as the sole carbon source. The residence time of P. aeruginosa cells in the chemostat was 170 min, in comparison to the doubling time (td) of 140 min in suspension.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Phenotype | Source or reference |
|---|---|---|---|
| P. aeruginosa | |||
| PAO1 | Wild type | Wild type | B. H. Holloway |
| PAO-JP1 | lasI::tet | LasI− | Pearson et al. (36) |
| PAO230 | lasB::lacZ | Wild-type PAO1 | H. P. Schweizer |
| PAO220 | rhlA::lacZ | Wild-type PAO1 | H. P. Schweizer |
| PA14 | Wild type | Wild type | O'Toole and Kolter (32) |
| PA257 | flgK::tet | FlgK−, nonmotile | O'Toole and Kolter (32) |
| P. putida | |||
| ATCC 39168 | Wild type | Wild type | Sauer and Camper (47) |
| Plasmids | |||
| pRP9-1 | GFP | Constitutive GFP expression | Davies et al. (9) |
Microscopic analysis.
A continuous-culture once-through flow cell was configured to observe the growth and development of biofilms attached to a glass substratum. The flow cell was constructed of polycarbonate containing a chamber 1.0 mm by 1.4 cm by 4.0 cm capped with a glass cover slip. Sterile minimal medium was pumped from a 10-liter vessel through silicone tubing to the flow cell using a Masterflex 8-roller-head peristaltic pump at a flow rate of 0.13 ml min−1. Flow through the chamber was laminar, with a Reynolds number of 0.17, having a fluid residence time of 4.3 min. Medium leaving the flow cell was discharged to an effluent reservoir via silicone tubing. The entire system was closed to the outside environment but maintained in equilibrium with atmospheric pressure by a 0.2-μm-pore-size gas-permeable filter fitted to each vessel.
Log-phase P. aeruginosa (approximately 108 CFU/ml) were inoculated as a 3.0-ml slug dose through a septum 4 cm upstream from the flow cell under flowing conditions. Cells attached to the inner surface of the glass cover slip were viewed by transmitted light or epi-UV illumination using an Olympus BX60 microscope and a 100× magnification A100PL objective lens or a 50× magnification ULWD MSPlan long working distance Olympus objective lens. All images were captured using a Magnafire cooled three-chip charge-coupled device (CCD) camera (Optronics Inc., Galena, Calif.) and stored as separate digital files for subsequent retrieval and analysis.
P. aeruginosa were grown in the flow cell for up to 12 days. Previous work in our laboratories has shown P. aeruginosa to develop steady-state biofilms following a continuous culture period of 7 to 9 days. Steady state is defined by no change in effluent cell counts (CFU) resulting from detached biofilm cells; in steady state, growth of the biofilm is balanced by detachment. Individual cell clusters were examined during the course of each experiment and assigned grid coordinates, which were reexamined periodically during the course of the experiments. Size measurements were taken of random cell clusters by locating the cluster nearest to a randomly selected microscope stage coordinate. Each cell cluster was measured to determine its height by focusing from the substratum through to the apex of the cluster, and its width by measurement at the base of the cell cluster using a stage micrometer. Cell clusters were defined as cells embedded within an exopolysaccharide matrix attached to the substratum and lacking motility; void areas within cell clusters were determined by the observation of free-swimming bacteria within a space inside a cell cluster.
Effect of flagella on biofilm development.
Cultures of the nonmotile P. aeruginosa strain PA257 (carrying a mutation in flgK [32]) and the parental P. aeruginosa strain PA14 were analyzed for their ability to attach to a glass substratum by continuously pumping cells from a chemostat at 0.13 ml min−1 through a flow cell (described above). Direct counts of total cells attached to 1.2 mm2 of the glass surface were taken at intervals for up to 3 days and recorded. Experiments were run in duplicate for the wild type and quadruplicate for the mutant.
Activation of lasB and rhlA during biofilm development.
To determine the timing of activation of the quorum-sensing regulons Las and Rhl, cultures of P. aeruginosa strain PAO220 and strain PAO230 were grown in minimal medium in flow cells (as described for microscopy above). Activity of β-galactosidase was measured following the addition of 0.04 g of methlyumbelliferyl β-d-galactopyranoside, dissolved in _N,N_-dimethylformamide, per ml to the influent medium reservoir. Expression of the lacZ reporter was visualized by examination under long-wave UV excitation according to the method of Davies et al. (10, 12). All samples were analyzed using a UV exposure time of 800 ms, and cells were illuminated only during the period of data collection (15-min intervals for the first 3 h and 30-min intervals for the remainder of the analysis period). Activation of the lasB and rhlA genes was checked in continuous cultures used for protein analysis (see below) in order to coordinate timing of sampling with the onset of these regulons.
Alginate determination by alcian blue staining.
In order to observe the presence and location of exopolysaccharide (EPS) produced by P. aeruginosa strain PAO1 carrying pRP9-1 and the LasI mutant strain PAO-JP1 carrying pRP9-1, cells constitutive for green fluorescent protein (GFP) production were grown as biofilms in the flow cell described above. GFP fluorescence under blue light excitation indicated the presence of cells within the biofilm. Alcian blue (0.2 g/liter) was added to the flow cell, and GFP fluorescence images of cell clusters were reexamined under transmitted light to detect alcian blue-stained EPS. Digital images were acquired and examined to evaluate the relationship between cell position and EPS location.
Biofilm growth for protein determinations.
The interior surfaces of silicone tubing were used to cultivate biofilms for protein sampling. A continuous-flow reactor designed for these experiments was configured with oxygen-permeable tubing within which the biofilms were cultivated. The tubing had a regular geometry throughout (size 15; Masterflex) with an internal volume of 25 ml (tubing length, 1 m). This system was chosen to reduce heterogeneities that could affect biofilm physiology while allowing sufficient biomass development to allow a minimum of 500 μg of crude protein extract per tube. Medium was pumped through the tubing via a Masterflex 8-roller-head pump to a closed effluent medium reservoir. The assembled system was sterilized by autoclaving prior to inoculation.
Cultures of P. aeruginosa were grown in chemostats prior to the inoculation of the silicone tubing by syringe injection. Seven milliliters of a chemostat-grown culture of P. aeruginosa was injected through the tubing wall and into the lumen and allowed to attach for 1 h before the flow of minimal medium (0.4 ml/min) was initiated. The residence time in the tubing was 62.5 min (less than the doubling time of P. aeruginosa in suspension), allowing only attached organisms to be retained within the tubing. A glass capillary having a square cross section was inserted at the midpoint of one silicone tube during each experiment and used for an in-line microscopic evaluation of the onset of biofilm morphology as described above. Sampling schedules for all protein analyses were based on results from these observations. After various times (up to 12 days), biofilm cells were harvested from the interior surface by pinching the tube along its entire length, resulting in extrusion of the cell material from the lumen. The resulting cell paste was collected on ice. Prior to sampling, the bulk liquid was purged from the tubing to prevent interference from detached cells.
Determination of protein yields.
To determine if P. aeruginosa was able to yield sufficient protein for analysis by two-dimensional polyacrylamide gel electrophoresis (PAGE) and subsequent mass spectrometry, planktonic P. aeruginosa cells were exposed to the interior surfaces of silicone tubing for 60 min, followed by the flow of fresh minimal medium. At various time points, the attached cells were harvested from the interior surface of silicone tubing and the protein yield was determined. Only a minimal increase in protein concentration was observed for a period of 4 to 6 h, followed by an increase in protein yield after this period (data not shown). The average biofilm protein yield harvested after 3 days of biofilm growth from one silicone tube was 3 mg, which increased to 10 mg after 6 days. A slight increase in protein yield was observed between 6 and 12 days of biofilm growth (data not shown).
Preparation of crude protein extract.
Chemostat- and biofilm-grown P. aeruginosa and P. putida cells were washed immediately after sampling by centrifugation at 12,000 × g for 10 min at 4°C and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), containing 0.3 mg of phenylmethylsulfonyl fluoride/ml (Boehringer Mannheim, Indianapolis, Ind.). All cell samples were lysed by sonication on ice using six 10-s bursts at 4 W (Cole Parmer Instr. Co., Vernon Hills, Ill.) and stored at −20°C until used for protein determination. Cell debris and unbroken cells were removed from all samples by centrifugation at 30,600 × g, 30 min, 4°C. Total protein concentration was determined by the modified method of Lowry et al. (40) using reagents from Sigma. Bovine serum albumin was used as the standard. Experiments for each time point were repeated at least five times.
Two-dimensional gel electrophoresis.
Two-dimensional gel electrophoresis was conducted according to the principles of O'Farrell (29) as outlined by Görg et al. (19) and as described in detail in Sauer and Camper (47) using a DALT vertical two-dimensional gel system from Amersham Pharmacia (Piscataway, N.J.). Two-dimensional gels were stained with Coomassie as described in Sauer and Camper (47) or with silver nitrate (3). Two-dimensional gels were repeated for each growth condition independently at least three times to confirm the reproducibility of the protein pattern under planktonic and attached growth conditions. Only differences in protein spots that were reproduced three times are described here.
Two-dimensional gel analysis.
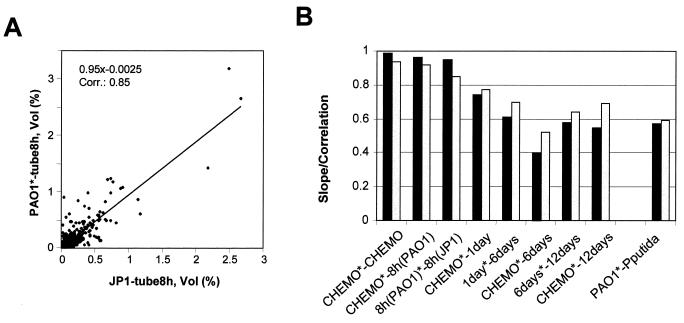
The software Melanie 3.0 from Genebio (Geneva, Switzerland) was used for two-dimensional gel analysis. Spot sizes were analyzed and compared using volume%. The similarities of protein profiles were determined using scatter plot graphics on pairs of protein spots that were found in two or more gels. Regression analysis and correlation coefficients between gels were then generated (see Fig. 8A). The relationship between two-dimensional gels is given by the linear dependence between the values of pairs of protein spots from one gel against the values in the other gel (Fig. 8A). The linear dependence is defined as the best-fit straight line through the pair values. The slope of the line and its offset define the fit. The correlation coefficient gives an approximation of the goodness of fit. For example, a linear dependence of 1.0 with a correlation of 1.0 indicates that all paired spot values are identical in two gels, while a linear dependence of 1.2 with a correlation of 0.95 indicates that there is a 95% chance that paired values are 20% higher in one gel. The two-dimensional gel comparisons and scatter plot analyses were carried out for both silver- and Coomassie-stained two-dimensional gels. All Coomassie and silver-stained two-dimensional gels were analyzed in duplicate for each time point.
FIG. 8.
Representative scatter plot graph (A) and analysis of two-dimensional gel similarities by scatter plot (B). Solid bars, linear dependence; shaded bars, correlation; ∗, reference gel for the scatter plot analysis; CHEMO, chemostat-grown P. aeruginosa strain PAO1; 8 h, 1 day, 6 days, and 12 days, P. aeruginosa strain PAO1 biofilm after 8 h, 1 day, 6 days, and 12 days of attachment time, respectively; tube 8 h, biofilm after 8 h of attachment time; JP1, P. aeruginosa strain PAO-JP1; PAO1, chemostat-grown P. aeruginosa strain PAO1; P. putida (ATCC 39168), chemostat grown. Vol (%), relative volume (according to Melanie 3.0 software description), volume divided by the total volume over the whole image.
Sample preparation for MS.
Protein spots of interest were excised from the gel, reduced, carboxymethylated, and digested in situ with trypsin as described by Wilms et al. (54) and Shevchenko et al. (48). After digestion overnight at 37°C, samples were centrifuged, and an aliquot of the supernatant was taken for analysis by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)-mass spectrometry (MS). The peptide solution was mixed with an equal volume of saturated _a_-cyano-4-hydroxy cinnamic acid and applied to a sample template of a MALDI-MS (Voyager EP; Perspective Biosystems, Foster City, Calif.). Data were obtained using the following parameters: linear mode, 25 kV accelerating voltage, 95% grid voltage, and 50 ns delay. Angiotensin and insulin were used as internal standards. Peptide mass fingerprints were searched using the programs ProFound at http://129.85.19.192/profound-bin/WebProFound.exe and Protein Prospector MS-FIT (http://prospector.ucsf.edu). Partial enzymatic cleavages leaving one cleavage site, carboxymethylation of cysteine, and acetylation of protein N terminus were considered in these searches. Sample preparation and MS analysis were performed by the Protein Core facility at Columbia University (New York, N.Y.).
RESULTS
Characterization of biofilm developmental cycle.
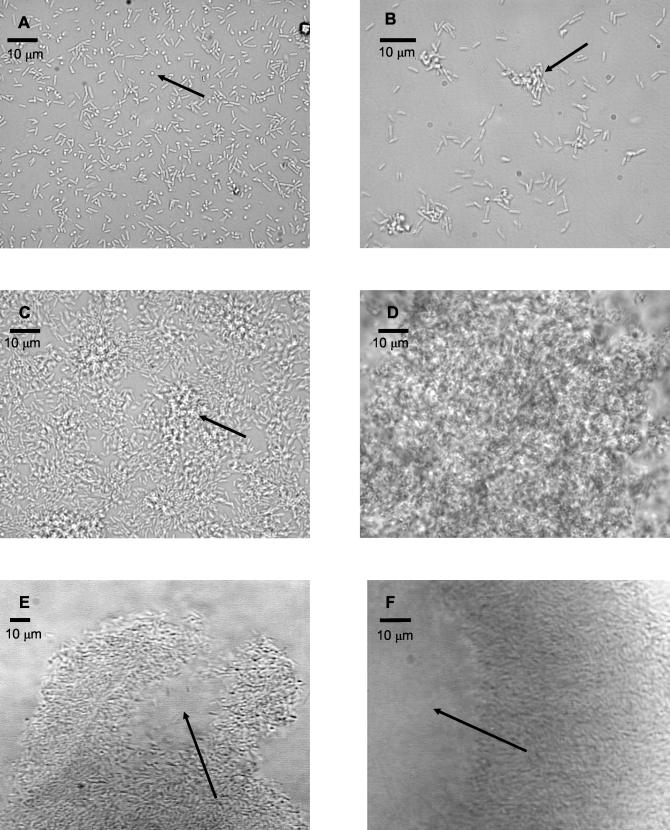
Biofilms of P. aeruginosa strain PAO1 were grown in continuous culture attached to glass in a flow cell fixed to a microscope stage and viewed continuously over the course of 12 days. Five distinct stages of development could be detected in this manner, each having reproducible characteristics (Fig. 1). During initial attachment, planktonic bacteria contacted the glass substratum, some via the cell pole, and became transiently fixed (Fig. 1A). The initial attachment was reversible, since some cells were observed to detach during this developmental stage or were observed to attach only briefly (not shown).
FIG. 1.
Transitional episodes in biofilm development by P. aeruginosa strain PAO1 examined by transmitted light microscopy. Each panel represents a distinct episode in biofilm development. (A) Reversible attachment. Initial event in biofilm development, bacteria are attached to substratum at cell pole (arrow). (B) irreversible attachment. Cells were cemented to the substratum and formed nascent cell clusters (arrow) with all cells in contact with the substratum. (C) Maturation-1. Cell clusters matured (arrow) and were several cells thick, embedded in the EPS matrix. (D) Maturation-2. Cell clusters reached maximum thickness, approximately 100 μm. (E and F) Dispersion. Cells evacuated interior portions of cell clusters (arrow), forming void spaces.
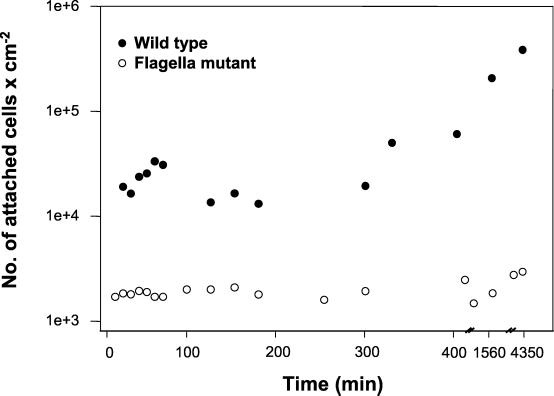
Under continuous biofilm culture conditions, the involvement of flagella in attachment to a glass surface was supported by the observation that some cells rotated about their polar axis while fixed to a spot on the glass surface (not shown). To test the importance of flagella on attachment to a glass surface in continuous flow, the attachment efficiency of the nonmotile P. aeruginosa strain PA257 (carrying a mutation in flgK) was compared to that of the parental strain PA14. Figure 2 shows that the flagellar mutant was unable to significantly increase in numbers at the glass surface of a flow cell compared to the flagellated wild type. Within the first 8 h the wild type and mutant increased in numbers at the glass surface by 6.3 × 104 cells per cm2 and 3.3 × 102 cells per cm2, respectively. Over the course of 3 days, the mutant increased by an average of 9.7 × 102 cells per cm2, while the parental strain increased in density to more than 4 × 105 cells per cm2. Although the increase in cell numbers was likely due to some growth at the glass surface, the differences were more likely due to the importance of flagella in the initial attachment phase of these organisms. Following the initial attachment, all cells that were able to remain at the surface were found to be attached via the side of the cell (see transition indicated by Fig. 1A and 1B). The nonflagellated mutant was observed to attach to the glass surface only along the side of the cell (not shown).
FIG. 2.
Effect of flagella on biofilm formation. Time course showing number of chemostat-grown cells attached to a glass substratum over a period of 3 days under conditions of continuous flow. •, P. aeruginosa strain PA14 (wild-type) cells attached at a constant rate, rapidly increasing the number of cells at the substratum. ○, mutants (P. aeruginosa strain PA257) defective in flagellum synthesis attached poorly and showed no significant increase in surface cell numbers over a period of 3 days.
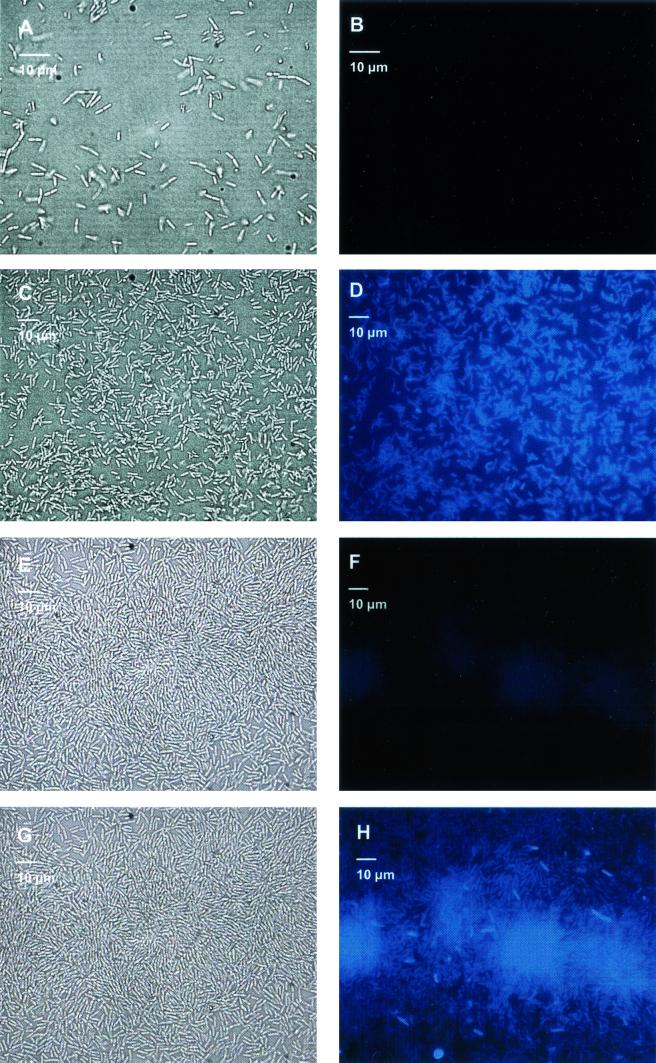
The second stage of development, irreversible attachment, was observed to occur when cell clusters commenced their development, as visualized by multiple cells in contact with one another and the substratum (Fig. 1B). During the second stage of development, motility ceased in attached cells, as observed microscopically (not shown). Furthermore, the Las quorum-sensing system became active, as determined by onset of reporter activity for the lasB gene (Fig. 3A to D). The lasB gene has been shown to be responsive to induction by PAI-1 autoinducer (38). The cell clusters which were formed during this biofilm developmental stage remained attached to the substratum through to the last stage of biofilm development (9 to 12 days of incubation).
FIG. 3.
Onset of quorum-sensing regulons in developing biofilm. (A) Transmitted light micrograph of P. aeruginosa strain PAO230 during the reversible attachment phase of biofilm development; (B) the same image under UV illumination demonstrated no activation of the Las regulon. (C) Transmitted light micrograph of P. aeruginosa strain PAO230 during the irreversible attachment phase of biofilm development; (D) the same image under UV illumination demonstrated activation of the Las regulon. (E) Transmitted light micrograph of P. aeruginosa strain PAO220 during the irreversible attachment phase of biofilm development; (F) the same image under UV illumination demonstrated no activation of the Rhl quorum-sensing regulon. (G) Transmitted light micrograph of P. aeruginosa strain PAO220 during the maturation-1 phase of biofilm development; (H) the same image under UV illumination demonstrated activation of the Rhl quorum-sensing regulon.
The third stage of development was observed when cell clusters became progressively layered (Fig. 1C). This was defined as the point in time at which cell clusters are thicker than 10 μm (not shown). In addition, this maturation-1 stage is accompanied by the activation of the Rhl quorum-sensing system. Activation of the Rhl quorum-sensing system was determined by the onset of reporter activity for the rhlA gene (see Fig. 3E to H), which was shown by Pearson et al. to be induced by the PAI-2 autoinducer (37).
As defined by Davies et al. (9) the penultimate stage in biofilm development is reached when cell clusters attain their maximum average thickness at approximately 100 μm. In the reactor system used for the current study, this stage, characterized as maturation-2 (see Fig. 1D), was shown to occur after 6 days of growth. During maturation-2, cells within clusters were observed microscopically to be nonmotile, the cell clusters reach their maximum dimensions, the majority of the cells are segregated within cell clusters, and clusters are displaced from the glass surface.
After 9 days, cell clusters were observed to undergo alterations in their structure due to the dispersion of bacteria from their interior portions (Fig. 1E and F). These bacteria were motile and were observed to swim away from the inner portions of the cell cluster through openings in the cluster and enter the bulk liquid. Figure 1E is an image of a cell cluster taken from the side (grown on the transverse wall of the flow cell), showing the opening through which the bacteria were observed to evacuate the cluster center. Bacteria remaining within the void space were motile. The ability of bacteria to swim freely within the void spaces as observed by microscopy indicated the absence of dense polymer or other gel-like material in the void space. Figure 1F shows the wall of a cell cluster after a complete dispersion event, where the remaining bacteria in the wall of the cell clusters are nonmotile.
Role of Las quorum sensing in biofilm development.
It has been observed previously that P. aeruginosa strain PAO1 defective in Las quorum sensing produced similar quantities of EPS as wild-type bacteria but that the mutants were unable to form normal biofilms (9). We suspected that the differences in biofilm architecture imparted by las activation were related to changes in the structure of the EPS and tested this hypothesis by staining growing biofilms of the lasI mutant and wild type with alcian blue, which binds to acidic polysaccharides. A difference in the location of stained acidic polysaccharide (presumably alginate) for the mutant and wild-type biofilm cultures was observed (not shown). The wild-type biofilm had EPS material located principally in the interstices between bacterial cells, which are well separated from one another (not shown). However, alcian blue staining of the lasI mutant biofilm showed the EPS material to be associated only with the cell surface. In contrast to the wild type, the mutant cells were closely packed and the spaces between bacteria were not filled with EPS (as they are in the wild-type biofilm).
Proteome analysis reveals differences in the protein profiles of strain PAO1 and strain PAO-JP1 in 1-day biofilms, but not in initially attached cells.
To investigate the degree to which biofilm stage-specific physiologies differ in each of the five stages of biofilm development (see Fig. 1 and Fig. 3), crude protein extracts were analyzed by two-dimensional gel electrophoresis. Sampling for these analyses was determined by timing the onset of the physiological characteristics specific to each phase observed during the microscopy and in situ reporter gene activation studies in the flow cells. This was achieved by following the development of biofilm within glass capillaries inserted within one of the lines of tubing during each experimental run. We observed no differences in the timing and events of biofilm development when comparing results from the flow cell experiments and the experiments run for the collection of samples for protein analysis.
The earliest times of onset observed for each of the biofilm characteristics were as follows: initial attachment, >0 min; irreversible attachment, 2 h; maturation-1, 3 days; maturation-2, 6 days; and dispersion, 9 to 12 days. These times were found to be reproducible with both the flow cells and the tube reactor used for growth of biofilms for protein analysis (data not shown). Microscopic observation demonstrated that bacteria having physiologies from more than one stage of biofilm could be present simultaneously within the biofilm. Therefore, the sampling schedule for protein analysis was chosen as follows: planktonic cells from the chemostat; 8-h biofilm, reversible attachment; 1-day biofilm, irreversible attachment; 3-day biofilm, maturation-1; 6-day biofilm, maturation-2; and 12-day biofilm, dispersion.
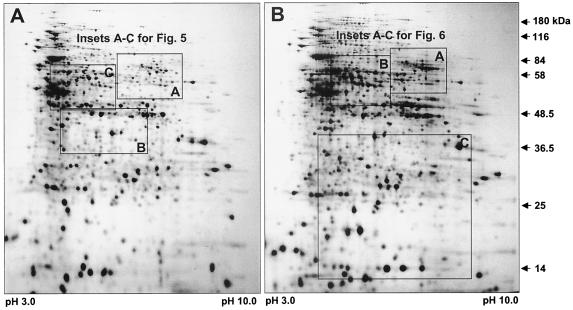
At these sampling times, the majority of biofilm cells were found to display the same phenotype. Representative examples of two-dimensional protein patterns of two biofilm developmental stages, irreversible attached biofilm cells (1 day) and biofilm cells in the dispersion stage (12 days), are shown in Fig. 4. Each two-dimensional gel profile resolved more than 1,500 distinct protein spots. Outlined areas in Fig. 4A indicate zones of the two-dimensional gels that are represented in Fig. 5, and outlined areas in 4B indicate zones represented in Fig. 6.
FIG. 4.
Two-dimensional images of crude protein extracts of P. aeruginosa grown as a 1-day biofilm (A) and as a 12-day biofilm (B). The crude protein extracts (200 μg) were separated on pH 3 to 10 nonlinear Immobiline Dry strips (Amersham Pharmacia, Piscataway, N.J.), followed by SDS-11% polyacrylamide gel electrophoresis. Gels were stained with silver nitrate (3). Outlined areas in panel A indicate zones of the two-dimensional gels that are represented in Fig. 5, and outlined areas in panel B indicate zones represented in Fig. 6.
FIG. 5.
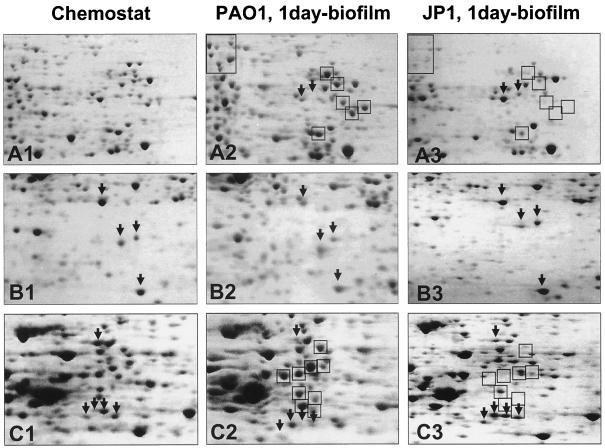
Enlarged partial two-dimensional gels showing crude protein extract of P. aeruginosa strain PAO1 grown planktonically (A1 to C1), attached for 1 day (A2 to C2) as well as P. aeruginosa JP1 after 1 day of attachment time (A3 to C3). Boxed protein spots indicate proteins that are produced at higher levels in 1-day P. aeruginosa strain PAO1 biofilm cells than in 1-day strain PAO-JP1 biofilm cells; arrows pointing downward indicate proteins that were absent or only weakly expressed in 1-day P. aeruginosa strain PAO1 biofilm cells compared to strain PAO-JP1. The crude protein extracts (500 μg) were extracted and separated on pH 3 to 10 nonlinear Immobiline Dry strips, followed by SDS-11% PAGE. The gels were stained with Coomassie brilliant blue R350, Dashed-box protein spots were identified by mass spectrometry (see Fig. 6 and Table 2).
FIG. 6.
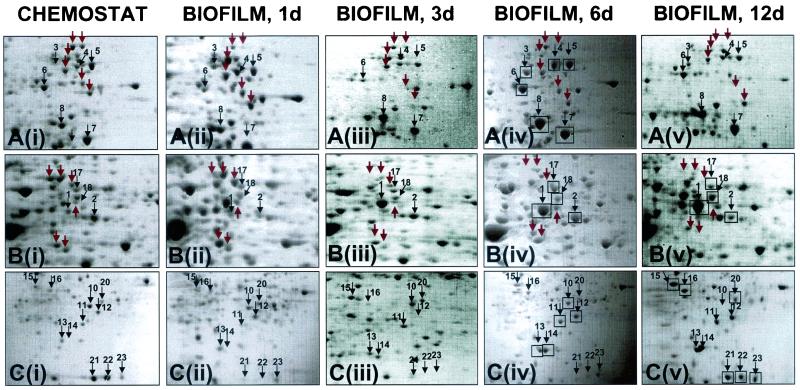
Enlarged partial two-dimensional gels showing crude protein extract of P. aeruginosa strain PAO1 grown planktonically in a chemostat [A(i) to C(i)] and for 1 day [A(ii) to C(ii)], 3 days [A(iii) to C(iii)], 6 days [A(iv) to C(iv)], and 12 days [A(v) to C(v)] in a biofilm. Sections A to C show an enlarged view of the two-dimensional images in Fig. 4. Proteins that were identified by MS analysis are indicated by boxes. The spot numbers correlate with the numbers given in Table 2. The gels were stained with Coomassie brilliant blue R-350.
Since in situ activation of the lasB reporter gene presented evidence of _las_-induced gene expression during irreversible attachment (Fig. 1 and 3), protein patterns of P. aeruginosa strain PAO1 were compared to those of the LasI mutant strain PAO-JP1 to confirm the onset of quorum sensing at the protein level. In addition, the comparison was used to confirm the sampling schedule for the protein analysis. Protein patterns of attached P. aeruginosa strain PAO1 cells were also compared to those of planktonic cells. Only a few differences in proteins between planktonic and attached P. aeruginosa strain PAO1 cells were visible within 8 h after initial attachment (see Fig. 8 for two-dimensional gel similarities). However, no differences in protein patterns were detected for P. aeruginosa strain PAO1 compared to strain PAO-JP1 when both were grown in the chemostat or as a biofilm for 8 h (as determined using two-dimensional gel analysis software Melanie 3.0; see Fig. 8 for two-dimensional gel similarities). This finding is consistent with the in situ activation of the lasB::lacZ reporter gene in strain PAO230, which did not occur until later in biofilm maturation (1 day) (see Fig. 3). These results demonstrated that cells of P. aeruginosa which are in the planktonic and early attachment stage did not display substantially different physiologies. It also demonstrates that quorum sensing does not appear to have an influence on this transitional stage of biofilm development.
In contrast, computer-assisted comparison between 1-day biofilm protein patterns for P. aeruginosa strain PAO1 and strain PAO-JP1 demonstrated differential expression of at least 50 protein spots (Fig. 5). This difference in protein patterns occurred after the Las quorum-sensing system had been activated in irreversibly attached cells (Fig. 3A). The results also confirmed the sampling schedule for protein analysis.
Several proteins were expressed at higher levels in P. aeruginosa strain PAO1 biofilm cells than in strain PAO-JP1 after the same attachment time. These proteins are indicated by boxes in Fig. 5. Some of these protein spots were present in the protein profiles of planktonic cells and P. aeruginosa strain PAO1 biofilm cells but absent in P. aeruginosa strain PAO-JP1 biofilm cells. In contrast, examples of protein spots that were absent or only weakly expressed in 1-day PAO1 biofilm cells but present in 1-day PAO-JP1 biofilm cells are indicated by downward-pointing arrows.
Interestingly, the protein patterns of PAO-JP1 biofilm cells were more similar to planktonic cells than to 1-day PAO1 biofilm cells (not shown). Proteins that were expressed at higher levels in P. aeruginosa strain PAO-JP1 were usually also expressed at higher levels in planktonic cultures compared to 1-day PAO1 biofilm cells. We also detected quorum-sensing-independent but probably surface-induced differential expression of proteins. Using the two-dimensional gel analysis software, we were able to identify 57 unique protein spots in 1-day biofilm protein patterns for both strains PAO1 and PAO-JP1 that were absent in the planktonic protein pattern and 48 protein spots that were unique for planktonic growth (e.g., see top of Fig. 5, enlarged sections A1 to A3).
Protein MS analysis indicates increased levels of amino acid biosynthetic proteins, membrane proteins, and oxidative stress response proteins.
Crude protein extracts obtained from P. aeruginosa strain PAO1 grown in a chemostat or attached to silicone tubing for 8 h or 1, 6, 9, and 12 days were analyzed by two-dimensional gel electrophoresis. Comparison of protein patterns of planktonic and biofilm cells after various attachment times revealed differential protein expression depending on the stage of biofilm development. These alterations in the protein patterns were visible as early as 8 h after initial attachment (not shown).
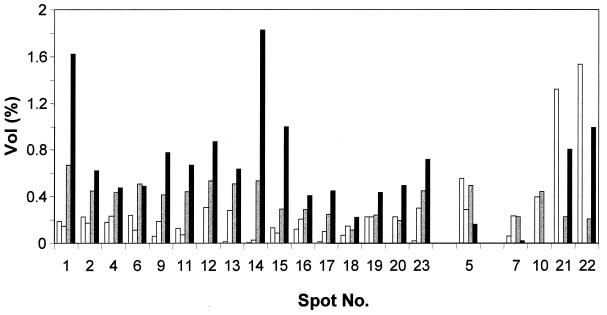
A detailed comparative view is shown in Fig. 6, presenting enlarged sections from the two-dimensional image in Fig. 4B. Most of the differentially expressed proteins are located in the neutral to basic pH range and showed an average molecular mass of 30 to 60 kDa. Using MALDI-TOF-mass spectrometry, 23 of the differentially expressed proteins (as indicated by boxes in Fig. 6, except protein 9) were identified. With few exceptions, the majority of the identified proteins were upregulated following adhesion to and growth on the silicone surface (Fig. 7). The average change in the protein expression pattern was 10-fold, but changes of up to 300-fold were detected. A 300-fold change was seen for spot 14, a secreted hemolysin-coregulated protein.
FIG. 7.
Change in spot intensity and differential expression of selected proteins over the course of biofilm development. From left to right in each group of four, bars represent spot intensity obtained under planktonic growth conditions and after 1 day, 6 days, and 12 days of biofilm development. The spot numbers correlate with the numbers given in Table 2 and Fig. 6. The protein spots are organized following their protein expression pattern (upregulated, downregulated, and varied expression). Vol (%), relative volume (according to Melanie 3.0 software description), volume divided by the total volume over the whole image.
Proteins showing differential regulation during the course of biofilm development fell into four general classes (Table 2). Class I included proteins that encode factors for metabolic processes, such as amino acid metabolism, carbon catabolism, and cofactor biosynthesis. The majority of these proteins were found to be upregulated following adhesion, even though the growth medium used for the chemostat and the biofilm tube reactor were identical. However, dihydrolipoamide dehydrogenase was downregulated, suggesting some degree of differential expression of metabolic proteins following attachment to this silicone surface (Fig. 6 and 7).
TABLE 2.
Identification of selected protein spots that were differentially expressed over the course of biofilm developmenta
| Class and function | Spot no. | Protein | Locus | Development stage of maximum protein concnb | Molecular mass (kDa) | pI | No. of matched peptides |
|---|---|---|---|---|---|---|---|
| Class I | |||||||
| Carbon catabolism and amino acid metabolism | 1 | Arginine deiminase | ArcA, PA5171 | 12d | 46.30 | 5.52 | 24 |
| 8 | Ornithine carbamoyltransferse, catabolic | ArcB, PA5172 | 12d | 37.97 | 6.14 | 17 | |
| 15 | Carbamate kinase | ArcC, PA5173 | 12d | 33.08 | 5.25 | 13 | |
| 2 | Fumarate hydratase C1 | PA4470 | 12d | 48.7 | 5.65 | 15 | |
| 3 | Class III aminotransferase, probable | PA2413 | 6d | 50.22 | 6.03 | 17 | |
| 4 | Acylase, probable | PA2385 | 12d | 84.04 | 6.64 | 27 | |
| 5 | Dihydrolipoamide dehydrogenase 3 | PA4829 | P | 50.02 | 6.48 | 19 | |
| 7 | Glyceraldehyde-3-phosphate dehydrogenase | PA3001 | 1d/6d | 50.11 | 7.74 | 15 | |
| 9 | Ketol-acid reductoisomerase | PA4694 | 12d | 36.43 | 5.57 | 19 | |
| 16 | UTP-glucose-1-phosphate uridyltransferase | PA2023 | 12d | 31.24 | 5.33 | 14 | |
| 17 | Succinate semialdehyde dehydrogenase | PA0265 | 12d | 51.62 | 5.59 | 18 | |
| Cofactor biosynthesis | 6 | l-Ornithine 5-monooxygenase (pyoverdine biosynthesis) | PA2386 | 6d | 49.47 | 6.03 | 24 |
| 15 | Thioredoxin reductase (pyrimidine biosynthesis) | PA2616 | 12d | 33.82 | 5.24 | 9 | |
| 20 | Adenylate kinase (purine biosynthesis) | PA3686 | 12d | 23.1 | 5.98 | 11 | |
| Class II (fatty acid, phospholipid, LPS) | 23 | β-Hydroxydecanoyl-ACP dehydrogenase | FabA, PA0085 | 12d | 18.75 | 6.41 | 3 |
| Class III (membrane proteins, transport) | 4 | Probable binding protein component of ABC transporter | PA4496 | 12d | 60.08 | 7.11 | 13 |
| 19 | Porin E1 | PA0291 | 12d | 49.67 | 8.67 | 23 | |
| 20 | Probable binding protein component of ABC transporter | PA5076 | 12d | 59.74 | 6.85 | 7 | |
| Class IV (adaptation, protection) | 11 | Alkyl hydroxyperoxide reductase subunit C | PA0139 | 12d | 20.54 | 5.89 | 7 |
| 12 | Superoxide dismutase | PA4468 | 12d | 22.61 | 5.81 | 5 | |
| 10 | Probable ribosomal protein L25 | PA4671 | 6d | 21.96 | 5.81 | 10 | |
| 14 | Secreted hemolysin-coregulated protein Hcp | PA0263 | 12d | 19.09 | 5.49 | 11 | |
| Class V (various) | 13 | Conserved hypothetical protein | PA0423 | 12d | 20.78 | 6.09 | 14 |
| 16 | Hypothetical protein | PA2395 | 12d | 31.07 | 5.44 | 10 | |
| 21, 22 | Helix-destabilizing protein of bacteriophage Pf1 | PA0720 | P/12d | 15.47 | 6.25 | 4 |
Class II included β-hydroxydecanoyl-acyl carrier protein (ACP) dehydrogenase, which is involved in various lipid biosynthesis reactions such as fatty acid, phospholipid, lipopolysaccharide, and acyl homoserine lactone (AHL) biosynthesis. Class III contained membrane proteins primarily involved in molecular transport, such as the bacterial extracellular solute-binding proteins (50) and porin E1 (OprE), which forms a small channel in the outer membrane (34). The membrane proteins were found to be upregulated following adhesion (Fig. 6 and 7).
Class IV included proteins involved in adaptation and protection. With the exception of the probable ribosomal protein L25, the proteins falling into this class, such as alkyl hydroxyperoxide reductase subunit C and superoxide dismutase, were elevated in expression following adhesion (Fig. 6 and 7). Other proteins found to be upregulated following adhesion were two hypothetical proteins. Differential expression was found for the helix-destabilizing protein of bacteriophage Pf1, which was found to be downregulated following attachment and upregulated in mature biofilms (Fig. 6 and 7). Unidentified proteins showing differential expression following adhesion were also visible (see Fig. 4 and Fig. 6 for comparison).
Multiple stages in the physiology of biofilm bacteria exist, as revealed by analysis of two-dimensional protein patterns.
As visible from Fig. 5 and 7, the protein patterns that were obtained from different stages of biofilm development were profoundly different. To analyze these differences, scatter plot analysis was used to evaluate similarities in protein spot intensities (Fig. 8A). A detailed description of scatter plot analysis is given in the Materials and Methods section.
By comparing protein profiles of replicate two-dimensional gels of chemostat-grown planktonic cells, the scatter plot analysis revealed a linear dependence of 0.99 and a correlation of 0.94 (Fig. 8B). These results indicated that most paired-spot values are identical between replicates and that very little experimental variation existed due to protein extraction, solubilization, sample loading, or staining. Similar results were obtained when gels of initially attached P. aeruginosa strain PAO1 cells (tube, 8 h) were compared to planktonic P. aeruginosa strain PAO1 cells or initially attached P. aeruginosa strain PAO-JP1 cells (Fig. 8). The high identity between the protein profiles of the two strains confirmed that during reversible initial attachment, the protein profiles are not influenced by quorum sensing.
When protein patterns of more mature P. aeruginosa strain PAO1 biofilms were compared to those of planktonic cells, the linear dependence as well as the correlation coefficient decreased (Fig. 8B). The greatest difference was observed when protein patterns of chemostat-grown and 6-day biofilm cells were compared (Fig. 8B), demonstrating a linear dependence of 0.4 and a correlation of 0.52. This finding indicated that almost all paired values are 60% higher in 6-day biofilm cells than in planktonic cells and that the similarity between these two protein patterns is close to 50%. This difference is mainly due to the high percentage of differentially expressed proteins (Table 3).
TABLE 3.
Differential expression patterns in planktonic cells and in biofilm cells after various time of biofilm growtha
| Expression pattern | % of proteins | |||||
|---|---|---|---|---|---|---|
| Chemo/1d | 1d/3d | 3d/6d | Chemo/6d | 1d/6d | 6d/12d | |
| Constant | 71 | 91 | 61 | 43 | 61 | 63.5 |
| Upregulated | 25 | 4.5 | 36 | 51 | 36 | 1.5 |
| Downregulated | 4 | 4.5 | 3 | 6 | 3 | 35 |
Within 6 days of biofilm growth, more than 50% of all the proteins were upregulated, less than 7% are downregulated, only 40% are constantly expressed compared to planktonic cells, and more than 150 Coomassie-detected protein spots (more than 300 protein spots by silver stain) were synthesized de novo. In contrast, between 6 and 12 days of biofilm formation, more than 60% of all the proteins are constantly expressed, while 35% are downregulated and less than 2% are upregulated, suggesting a reversion of the protein patterns of mature biofilms older than 6 days towards that of planktonic cells (Table 3, Fig. 8B). Thus, protein patterns of planktonic cells and 12-day biofilms were more similar than protein patterns of planktonic cells and 6-day biofilms or protein patterns of 1-day biofilms and 6-day biofilms. This finding indicated that a portion of biofilm bacteria are reverting to the planktonic phenotype (see Fig. 1). Over the developmental life cycle of P. aeruginosa from planktonic to the biofilm mode of growth, only 30% of all the proteins are expressed constantly.
As a point of reference, the protein patterns of two chemostat-grown Pseudomonas species, P. aeruginosa strain PAO1 and P. putida, were analyzed by scatter plot analysis as described above (Fig. 8). A linear dependence of 0.58 and a correlation of 0.59 were found. These values indicated that planktonic P. aeruginosa strain PAO1 and P. putida have more in common than planktonic P. aeruginosa does with 6-day biofilm P. aeruginosa. Correlation differences between P. aeruginosa strain PAO1 6- and 12-day biofilms as well as P. aeruginosa chemostat and 12-day biofilms were similar to those found between planktonic cultures of P. aeruginosa strain PAO1 and P. putida. This finding indicates that on the protein levels of planktonic, 6-day, and 12-day biofilm cells are as different as two different Pseudomonas strains and thus reflect different phenotypes.
DISCUSSION
The idea that P. aeruginosa displays multiple physiologies during development as a biofilm was proposed by Davies in 1996 (11), when it was noted that this organism progresses through multiple developmental stages, each having a unique phenotype. In a recent review, O'Toole et al. (30) further supported the idea that biofilm formation is a process of microbial development and is not unlike that observed in cell cycle-controlled swarmer-to-stalk cell transition in Caulobacter crescentus, sporulation in Bacillus subtilis, and fruiting-body formation by Myxococcus xanthus. This view is gaining increasing acceptance as studies on initial events in biofilm development reveal alterations in bacterial cell physiology that hint at changes that may occur throughout the developmental cycle (2, 7, 8, 10, 13, 15, 18, 24, 31, 32, 33, 41, 43, 45, 47).
In the current study, we have endeavored to show that biofilm bacteria can be complex, displaying at least five distinct physiologies over the course of development. To our knowledge, this is the first study to report on the regulation of proteins in a biofilm for all stages of development. Directly observable gene products were used as the basis for identifying transitional episodes during the maturation process. We focused on the expression of proteins that are unique at different stages of development by using internal markers involved in biofilm growth, such as activation of lasB. These markers were used as signposts to standardize biofilm collection points and ensure reproducibility for protein analyses.
Biofilms were grown in continuous-culture reactors (biofilm tube reactors) designed to reduce the impact of microenvironments on biofilm physiology and to provide large biomass yields necessary for two-dimensional PAGE analysis. These tube reactors were run at flow rates that washed out planktonic cells in less than one generation time. The wild-type P. aeruginosa strain PAO1 was used to characterize normal biofilm development, and results from the mutants were compared to this baseline information.
Direct observation of biofilm development revealed quantifiable transitional events which indicated significant episodes in the development of a bacterial biofilm population. Taken together, these were used to conveniently partition biofilm development into five distinct stages: (i) reversible attachment, (ii) irreversible attachment, (iii) maturation-1, (iv) maturation-2, and (v) dispersion. The developmental life cycle comes full circle when dispersed biofilm cells revert to the planktonic mode of growth. Bacteria within each of the stages of biofilm development are generally believed to be physiologically distinct from cells in other stages of development, but in a mature biofilm all stages of development may be present to some degree at the same time. Differences in biofilm developmental stages with respect to protein analysis and direct microscopic observation are summarized in Table 4. Changes in protein profiles are presumed to be directly related to phenotypic adaptations; some of these phenotypes were targeted for observation in flow cells. These results revealed a sequence of developmental episodes in biofilm growth demonstrating profound phenotypic alteration not generally attributed to bacteria.
TABLE 4.
Phenotypic characteristics of biofilm development
| Biofilm stage | Characteristics observed under the microscope | Protein profile |
|---|---|---|
| Planktonic | Motility | Difference compared to reversibly attached cells |
| Reversible attachment | Contact with surface via cell pole, transiently fixed to substratum | No difference between PAO1 and PAO-JP1; indication of differentially surface-induced gene expression (quorum sensing independent) |
| Irreversible attachment | Reorientation to longitudinal cell axis, cell cluster development, nonmotile, lasB activation | 50 proteins different between PAO1 and PAO-JP1: surface-induced regulation of >100 proteins |
| Maturation-1 | Layered cells in clusters, cluster thickness >10 μm rhlA activation | Profound difference in protein pattern compared to planktonic cells |
| Maturation-2 | Maximum cell cluster development, cluster thickness up to 100 μm, majority of cells displaced from substratum | Greatest difference in protein pattern compared to planktonic cells; protein pattern significantly different from maturation-1 stage; de novo synthesis of more than 100 proteins 50% of protein spots upregulated compared to planktonic cells |
| Dispersion | Alteration in cell cluster structure motile and nonmotile bacteria observed, formation of pores and channels, dispersion | Greater similarity to planktonic cells than to maturation-2 biofilm cells; 35% of proteins spots downregulated compared to mature biofilm cells |
The initial event in biofilm development occurs when contact is made between the surface of the cell and an interface. Using nonmotile mutants, a significant decrease in attachment efficiency compared to flagellated cultures was observed. Therefore, the presence of flagella during the planktonic phase of the biofilm growth cycle appears to enhance reversible attachment. A similar observation was made previously by O'Toole and colleagues for biofilms grown under static biofilm culture conditions. The authors demonstrated the importance of flagellar motility by comparing attachment of a motile and a nonmotile P. aeruginosa over time to PVC plastic under static biofilm culture conditions in the well of a microtiter dish (32). While the wild-type P. aeruginosa strain PA14 formed a dispersed monolayer under these growth conditions, only a few cells of the nonmotile P. aeruginosa strain PA257 (carrying a mutation in flgK) attached to the PVC plastic (32).
In continuous flow, irreversible attachment is indicated by the activation of the las regulon and commenced cell cluster development. As observed by microscopy, motility ceased during this developmental stage, possibly due to the loss of flagella. A similar observation was made for P. putida, where subtractive hybridization and immunoblot analyses indicated a switch from flagellar to type IV pilus-based twitching motility after attachment (47). Interestingly, biofilm developmental stages of the nonmotile P. aeruginosa strain PA257 preceded normally (not shown). In static biofilm systems, this developmental stage is indicated by the progressive formation of a denser monolayer of cells (as determined by discontinuously viewing small PVC plastic tabs that were previously incubated in microtiter dishes over time [32]). The formation of a dense monolayer was probably due to both growth at the PVC surface and continued attachment, since the static biofilm system was in a batch system. Within 7 h after initial attachment to the PVC plastic surface, the monolayer in the microtiter dish became punctuated with cell clusters (referred to by O'Toole and Kolter as microcolonies) that were 3 to 5 layers of cells thick (32). Other developmental stages using the static biofilm system were not described.
Microscopic analysis of biofilm development also suggested the importance of motility for the dispersion stage. Cells evacuating cell clusters showed motility, as observed by microscopy, while cells remaining in the walls of cell clusters were nonmotile (see Fig. 1). This finding implies that the flagella function in transport and initiation of cell-to-surface interactions and also in detachment from the biofilm. Similar observations using immunoblot analyses for flagellin were made for P. putida biofilm development (47).
Following reversible attachment, bacteria were able to alter the regulation of a number of genes which influenced the protein patterns derived from two-dimensional gel analysis. Among these are the genes that respond to Las quorum-sensing regulation. This has been demonstrated previously (49), where activation of the lasB gene was shown to occur within 24 h of initial attachment. Observations by microscopy demonstrated that the onset of lasB activity is correlated with the onset of initial development of biofilm cell clusters. Protein analysis revealed a change in the regulation of more than 50 spots, indicating a profound difference in physiology between reversibly and irreversibly attached cells (see reference 53 for quorum-sensing-regulated genes).
One protein responsive to regulation by quorum sensing was identified as l-ornithine 5-monooxygenase, a key enzyme in pyoverdine biosynthesis (36, 53). A second protein responsive to quorum sensing was identified as β-hydroxydecanoyl-ACP dehydrogenase, an enzyme that is part of the Fab pathway. The acyl-ACPs derived from the Fab pathway play central roles in biosynthetic pathways that depend on acyl transfer reactions and have been proposed to be acyl donors for the synthesis of acylated homoserine lactones (25, 51). Furthermore, analysis of protein patterns of irreversibly attached (1-day) and planktonic cells revealed the presence of quorum-sensing-independent but probably surface-induced differential expression of proteins.
Fifty-seven unique protein spots were identified in 1-day biofilm protein patterns for both P. aeruginosa strain PAO1 and strain PAO-JP1 that were absent in planktonic protein patterns, and 48 protein spots that were unique for planktonic growth were identified. These results imply that quorum sensing accounts for only a portion of the total number of genes whose regulation is altered during this stage of biofilm development and that the physiological change in attached bacteria is not due solely to induction by PAI-1 quorum sensing autoinducer (47). The implication of this result is that undiscovered biofilm regulons probably exist.
The next stage in biofilm development was maturation-1. At this stage, the second quorum-sensing regulon (rhlA) was shown to become active. During maturation-1, 36% of the approximately 1,500 genes detectable by two-dimensional PAGE analysis were shown to be upregulated, while only 3% were downregulated. This number exceeds what would be predicted for Rhl quorum sensing alone, and it hints that other undiscovered regulatory mechanisms are at work. Among the proteins showing significant upregulation during this stage of development are the Arc proteins, which are linked to anaerobic processes. This implies that during the maturation-1 stage of development, the biofilm becomes oxygen limited at least in select zones (presumably at or near the substratum). It is also during maturation-1 that evidence of matrix polymer production is detectable by alcian blue staining. Comparison of P. aeruginosa strain PAO-JP1 with the wild type showed that the Las quorum-sensing system is involved in the development of cell clusters (9). The change in architecture of the biofilm at this stage may influence the production of many of the proteins observed to differ in concentration during maturation-1.
The biofilm reaches its maximum thickness during the maturation-2 stage of development. This is also the point at which biofilm bacteria are profoundly different from planktonic bacteria with respect to the number of differentially expressed proteins. More than 50% of all detectable proteins undergo changes in regulation between planktonic growth and maturation-2 stage growth, with the majority being upregulated. Presumably, a large portion of the population is experiencing anaerobic or reduced oxygen conditions during this stage. Significantly, maturation-2 proteins are substantially different from maturation-1 proteins, with 39% of the total detectable complement (approximately 585 proteins) experiencing a sixfold or greater change in regulation. For comparative analysis, protein spots were considered significantly different if they displayed a sixfold or greater difference in volume as detected by the Melanie-3 software. Therefore, the numbers presented are minimal values for differences in protein concentrations for all proteins detected.
The final stage in the development of a biofilm is the dispersion stage. During this stage, bacteria within cell clusters can be seen to actively swim away from the interior portions of cell clusters, leaving behind structures that appear shell-like, with a hollow center and walls of nonmotile bacteria. Presumably, this dispersion process allows cells to swim back into the bulk liquid to gain better access to nutrients while at the same time allowing better access to nutrients for the cells that remain in the biofilm. Interestingly, the protein patterns for dispersion stage biofilms are actually closer to the patterns observed for planktonic bacteria than for maturation-2 stage cells. It is also worth noting that the transition to dispersion phase is the only episode in biofilm development where more proteins were downregulated than upregulated. Many of the bacteria that contributed to the protein samples and protein profiles in the dispersion stage may have been in a transition from a biofilm to a planktonic phenotype.
Of the proteins detected by two-dimensional gel electrophoresis, 23 that showed large changes in regulation at different time points during biofilm development were identified using MALDI-TOF-MS analysis. Among these are proteins involved in resistance to oxidative damage, EPS production, and aerobic and anaerobic metabolism. A similar finding was made by Prigent-Combaret and coworkers for E. coli K-12 biofilm cells (42). The authors demonstrated that biofilm formation triggered the differential expression of genes involved in EPS production, osmolarity, oxygen limitation, cell-to-cell signaling, and motility (42).
Certain of the proteins identified in the current study had been shown to be expressed in P. aeruginosa biofilm cells isolated from cystic fibrosis (CF) patients (20). These proteins included dihydrolipoamide dehydrogenase 3, ornithine carbamoyltransferse (ArcB), the ribosomal protein L25, superoxide dismutase (SodB), and alkyl hydroxyperoxide reductase, with SodB being produced only in the nonmucoid P. aeruginosa CF isolate and alkyl hydroxyperoxide reductase produced in both mucoid and nonmucoid isolates (20). Our finding and that by Hanna et al. (20) of increased superoxide dismutase production in biofilms contrasts with other reports which do not show evidence of transcription for this gene or presence of the protein (5, 17, 22).
Interestingly, the SodA concentration has been reported by Hassett et al. to be increased in alginate-producing bacteria (23). We detected an increase in the concentration of UTP-glucose-1-phosphate uridyltransferase in biofilm cells. This enzyme is involved in central intermediary metabolism, catalyzing the interconversions of pentose and glucuronate. The activity of this enzyme was shown to be necessary for exopolymer production in Lactobacillus casei CRL 87 and Streptococcus thermophilus (4, 14, 28) and may be active in the biosynthesis of biofilm matrix material in P. aeruginosa.
Evidence for changes in carbon and energy metabolism are given by the finding that proteins involved in amino acid metabolism and the tricarboxylic acid cycle, including succinate semialdehyde dehydrogenase, fumarate hydratase C1, and glyceraldehyde-3-phosphate dehydrogenase, were produced at higher levels in mature and late-stage biofilms. Our finding of increased glyceraldehyde-3-phosphate dehydrogenase levels is in contrast to a recent report by Whiteley and colleagues, where the gene for this enzyme was found to be downregulated fourfold in 5-day-old P. aeruginosa biofilm cells (52). While the majority of metabolic proteins were upregulated, dihydrolipoamide dehydrogenase 3 was downregulated following attachment. Dihydrolipoamide dehydrogenase is a flavoprotein component of multienzyme complexes catalyzing the oxidative decarboxylation of α-ketoacids in the Krebs cycle.
Interestingly, proteins involved in amino acid metabolism, including the proteins ArcA, ArcB, and ArcC were found to be upregulated following attachment. This arcCDAB operon encodes the enzymes of the arginine deiminase pathway in P. aeruginosa and was shown to be regulated by ANR, an analogue of Escherichia coli FNR, and by the regulatory protein ArgR, a helper to the anaerobic regulatory protein ANR (1, 26). ANR is necessary for denitrification, arginine deiminase activity, and cyanide production by P. aeruginosa under conditions of oxygen limitation (1). That the biofilm cells were deprived of oxygen is supported by the finding that the membrane protein E1 (OprE), an anaerobically induced porin under the control of sigma 54 (55), was also upregulated following attachment. Interestingly, the proteins for the arginine deiminase pathway were found to be downregulated in P. putida 6 h following initial attachment and upregulated again in later stages (47).
Besides the outer membrane protein E1, two other components of outer membrane proteins were found to be upregulated in mature biofilms. These two binding protein components of the ABC transporter belong to the family of extracellular solute-binding proteins of bacteria that serve as chemoreceptors, recognition constituents of transport systems, and initiators of signal transduction pathways (50). Eight classes of solute-binding proteins are known, with PA4496, an ABC transporter for small molecules such as dipeptides, belonging to cluster 5. Cluster 5 includes proteins that are specific for the transport of dipeptides, oligopeptides, and nickel. PA5076, a putative glutamine transporter, belongs to class 3. Class 3 includes proteins that are specific for histidine, lysine-arginine-ornithine, glutamine, octopine, nopaline, and basic amino acids.
Recently, two members of class 3 solute binding proteins have been characterized on the basis of adhesin function rather than involvement in solute uptake. These are the collagen binding protein Cnb from Lactobacilllus reuteri (44) and the PEB1 adhesin from Campylobacter jejuni (39). The function of outer membrane proteins and ABC transporter as adhesins is consistent with earlier reports showing that these cell components have a substantial influence on attachment (16, 27, 42, 47). Attachment to abiotic surfaces was shown in Escherichia coli to cause major changes in outer membrane protein composition (35). Using two-dimensional gel analysis of purified outer membrane proteins, Otto and coworkers demonstrated increased levels for 17 outer membrane proteins and decreased protein levels for 15. Proteins found to have a large decrease in concentration following attachment were identified as OmpA, OmpX, TolC, and the lipoprotein Slp.
While we were able to detect outer membrane proteins by two-dimensional PAGE, we were unable to detect proteins from the inner membrane. This was probably due to the higher hydrophobicity of inner membrane proteins resulting in poor solubilization during sample preparation and isoelectric focusing (21). In addition, the analysis of membrane proteins from total cell extracts by two-dimensional gel electrophoresis may be hampered by other components such as lipids, which may interfere with solubilization in the rehydration buffer (45).
In this work, we have characterized five stages of biofilm development, each with a distinct protein pattern and observable phenotype, and each different from planktonic bacteria. The changes detected in the protein patterns between biofilm developmental stages were greater than anticipated (see reference 52 for comparison) and suggest that the P. aeruginosa biofilm growth cycle is both complex and highly regulated. The revelation that changes in protein regulation at each stage of biofilm development were as profound as the differences observed for different (but related) species at the same stage of biofilm development has important implications for designing rational and effective methods for biofilm detection and control. The discovery of regulatory systems responsible for key transitional episodes in biofilm development will be an important step in achieving this goal.
Acknowledgments
We gratefully acknowledge the technical assistance of Henning Seedorf from Montana State University-Bozeman and Kristin Davies from the State University of New York at Binghamton. We are also grateful to M. Parsek, H. P. Schweizer, J. Pearson, and G. A. O'Toole for assistance in providing strains.
This work was supported by a grant from the Research Foundation of the State of New York, through cooperative agreements DC04173-01 and DC02148 between the National Institutes of Health and Allegheny Singer Research Institute, through cooperative agreement EEC-897039 between the National Science Foundation and Montana State University, by the industrial partners of the Center for Biofilm Engineering, and by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Arai, H., T. Kodama, and Y. Igarashi. 1997. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol. Microbiol. 25**:**1141-1148. [DOI] [PubMed] [Google Scholar]
- 2.Belas, R., M. Simon, and M. Silverman. 1986. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J. Bacteriol. 167**:**210-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8**:**93-99. [Google Scholar]
- 4.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67**:**3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bollinger, N., D. J. Hassett, B. H. Iglewski, J. W. Costerton, and T. R. McDermott. 2001. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183**:**1990-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brõzel, V. S., G. M. Strydom, and T. E. E. Cloete. 1995. A method for the study of de novo protein synthesis in Pseudomonas aeruginosa after attachment. Biofouling 8**:**195-210. [Google Scholar]
- 7.Dagastino, L., A. E. Goodman, and K. C. Marshall. 1991. Physiological responses induced in bacteria adhering to surfaces. Biofouling 4**:**113-119. [Google Scholar]
- 8.Dalton, H. M., and P. E. March. 1998. Molecular genetics of bacterial attachment and biofouling. Biotechnology 9**:**252-255. [DOI] [PubMed] [Google Scholar]
- 9.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280**:**295-298. [DOI] [PubMed] [Google Scholar]
- 10.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61**:**860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, D. G. 1996. Regulation of alginate biosynthesis in Pseudomonas aeruginosa biofilms. Ph.D. thesis. Montana State University-Bozeman, Bozeman, Mont.
- 12.Davies, D. G., A. M. Charabarty, and G. G. Geesey. 1993. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 59**:**1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeFlaun, M. F., S. R. Oppenheimer, S. Streger, C. W. Condee, and M. Fletcher. 1999. Alterations in adhesion, transport, and membrane characteristics in an adhesion-deficient pseudomonad. Appl. Environ. Microbiol. 65**:**759-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degeest, B., F. Vaningelgem, A. P. Laws, and, L. De Vuyst. 2001. UDP-_N_-acetylglucosamine 4-epimerase activity indicates the presence of _N_-acetylgalactosamine in exopolysaccharides of Streptococcus thermophilus strains. Appl. Environ. Microbiol. 67**:**3976-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekkers, L. C., A. J. van der Bij, I. H. Mulders, C. C. Phoelich, R. A. Wentwoord, D. C. Glandorf, C. A. Wijffelman, and B. J. Lugtenberg. 1998. Role of the O-antigen of lipopolysaccharide, and possible roles of growth rate and of NADH:ubiquinone oxidoreductase (nuo) in competitive tomato root-tip colonization by Pseudomonas fluorescens WCS365. Mol. Plant-Microbe Interact. 11**:**763-771. [DOI] [PubMed] [Google Scholar]
- 16.Espinosa-Urgel, M., A. Salido, and J. L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182**:**2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frederick, J. R., J. G. Elkins, N. Bollinger, D. J. Hassett, and T. R. McDermott. 2001. Factors affecting catalase expression in Pseudomonas aeruginosa biofilms and planktonic cells. Appl. Environ. Microbiol. 67**:**1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172**:**1-8. [DOI] [PubMed] [Google Scholar]
- 19.Görg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 6**:**1037-1053. [DOI] [PubMed] [Google Scholar]
- 20.Hanna, S. L., N. E. Sherman, M. T. Kinter, and J. B. Goldberg. 2000. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by two-dimensional gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology 146**:**2495-2508. [DOI] [PubMed] [Google Scholar]
- 21.Harry, J. L., M. R. Wilkins, B. R. Herbert, N. H. Packer, A. A. Gooley, and K. L. Williams. 2000. Proteomics: capacity versus utility. Electrophoresis 6**:**1071-1081. [DOI] [PubMed] [Google Scholar]
- 22.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34**:**1082-1093. [DOI] [PubMed] [Google Scholar]
- 23.Hassett, D. J., W. A. Woodruff, D. J. Wozniak, M. L. Vasil, M. S. Cohen, and D. E. Ohman. 1993. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J. Bacteriol. 175**:**7658-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korber, D. R., J. R Lawrence, M. J. Hendry, and D. E. Caldwell. 1993. Analysis of spatial variability within mot+ and mot− Pseudomonas fluorescens biofilms using representative elements. Biofouling 7**:**339-358. [Google Scholar]
- 25.Kutchma, A. J., T. T. Hoang, and H. P. Schweizer. 1999. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD). J. Bacteriol. 181**:**5498-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181**:**2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthysse, A. G., H. A. Yarnall, and N. Young. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178**:**5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mozzi, F., G. Rollan, G. S. de Giori, and G. Font de Valdez. 2001. Effect of galactose and glucose on the exopolysaccharide production and the activities of biosynthetic enzymes in Lactobacillus casei CRL 87. J. Appl. Microbiol. 91**:**160-167. [DOI] [PubMed] [Google Scholar]
- 29.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250**:**4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 30.O'Toole, G. A., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54**:**49-79. [DOI] [PubMed] [Google Scholar]
- 31.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182**:**425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30**:**295-304. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28**:**419-461. [DOI] [PubMed] [Google Scholar]
- 34.Obara, M., and T. Nakae. 1992. Porin of Pseudomonas aeruginosa forms low conductance ion channel in planar lipid bilayers. Biochem. Biophys. Res. Commun. 186**:**645-651. [DOI] [PubMed] [Google Scholar]
- 35.Otto, K., J. Norbeck, T. Larsson, K. A. Karlsson, and M. Hermansson. 2001. Adhesion of type 1-fimbriated Escherichia coli to abiotic surfaces leads to altered composition of outer membrane proteins. J. Bacteriol. 183**:**2445-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179**:**5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92**:**1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91**:**197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pei, Z., C. Burucoa, B. Grignon, S. Baqar, X. Z. Huang, D. J. Kopecko, A. L. Bourgeois, J. L. Fauchere, and M. J. Blaser. 1998. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66**:**938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson, G. L. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83**:**346-356. [DOI] [PubMed] [Google Scholar]
- 41.Pratt, L., A, and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30**:**285-293. [DOI] [PubMed] [Google Scholar]
- 42.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181**:**5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Herva, J. J., D. Reniero, E. Galli, and J. L. Ramos. 1999. Cell envelope mutants of Pseudomonas putida: physiological characterization and analysis of their ability to survive in soil. Environ. Microbiol. 1**:**479-488. [DOI] [PubMed] [Google Scholar]
- 44.Roos, S., P. Aleljung, N. Robert, B. Lee, T. Wadstrom, M. Lindberg, and H. Jonsson. 1996. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol. Lett. 144**:**33-38. [DOI] [PubMed] [Google Scholar]
- 45.Santoni, V., M. Molloy, and T. Rabilloud. 2000. Membrane proteins and proteomics: un amour impossible? Electrophoresis 21**:**1054-1070. [DOI] [PubMed] [Google Scholar]
- 46.Santos, R., M. E. Callow, and T. R. Bott. 1991. The structure of Pseudomonas fluorescens biofilms in contact with flowing systems. Biofouling 4**:**319-336. [Google Scholar]
- 47.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183**:**6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68**:**850-858. [DOI] [PubMed] [Google Scholar]
- 49.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407**:**762-764. [DOI] [PubMed] [Google Scholar]
- 50.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57**:**320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Val, D. L., and J. E. Cronan. 1998. In vivo evidence that _S_-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer synthases. J. Bacteriol. 180**:**2644-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413**:**860-864. [DOI] [PubMed] [Google Scholar]
- 53.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96**:**13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379**:**466-469. [DOI] [PubMed] [Google Scholar]
- 55.Yamano, Y., T. Nishikawa, and Y. Komatsu. 1993. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosa PAO1. Mol. Microbiol. 8**:**993-1004. [DOI] [PubMed] [Google Scholar]