RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae (original) (raw)
Abstract
We identified and mapped RNA-binding sites of yeast Saccharomyces cerevisiae translation initiation factor eIF4G1 and examined their importance for eIF4G1 function in vitro and in vivo. Yeast eIF4G1 binds to single-stranded RNA with three different sites, the regions of amino acids 1–82 (N terminus), 492–539 (middle), and 883–952 (C terminus). The middle and C-terminal RNA-binding sites represent RS (arginine and serine)-rich domains; the N-terminal site is asparagine-, glutamine- and glycine-rich. The three RNA-binding sites have similar affinity for single-stranded RNA, whereas the affinity for single-stranded RNA full-length eIF4G1 is about 100-fold higher (approximate _K_d of 5 × 10−8 M). Replacement of the arginine residues in the middle RS site by alanine residues abolishes its RNA-binding activity. Deletion of individual RNA-binding sites shows that eIF4G1 molecules lacking one binding site are still active in supporting growth of yeast cells and translation in vitro, whereas eIF4G1 molecules lacking two or all three RNA-binding sites are strongly impaired or inactive. These data suggest that RNA-binding activity is required for eIF4G1 function.
Keywords: Cell-free translation, initiation of translation, RNA binding, RS domain, yeast
INTRODUCTION
Initiation of translation of most eukaryotic mRNAs involves recognition of the cap structure m7GpppN (where N is any nucleotide and m is a methyl group) at the 5′ end by the cap-binding protein eukaryotic Initiation Factor 4E (eIF4E) bound to the anchor protein eIF4G (for reviews, see Sachs et al. 1997; Gingras et al. 1999), recruitment of eIF4A and eIF4B, melting of RNA secondary structure close to the cap structure, binding of the 40S ribosomal subunit and associated initiation factors (43S initiation complex) to the 5′ end of the mRNA, and scanning of the mRNA in the 3′ direction by the 43S initiation complex for the AUG initiation codon (Kozak 1989). AUG recognition leads to hydrolysis of GTP bound to eIF2 in the initiation complex, release of initiation factors from the 40S ribosomal subunit, and binding of a 60S ribosomal subunit to form an 80S initiation complex competent to translate the open reading frame of the mRNA (for reviews, see Pain 1996; Hershey and Merrick 2000; Dever 2002).
Initiation factor eIF4G plays a central role in mRNA binding to the 40S ribosomal subunit in the cap structure dependent initiation pathway. Mammalian eIF4G1 and its homolog, eIF4G2, contain domains interacting with eIF4E, eIF4A, eIF3, poly(A)-binding protein (Pabp1), and the kinase MNK1 (Lamphear et al. 1995; Mader et al. 1995; Imataka and Sonenberg 1997; Gradi et al. 1998; Imataka et al. 1998; Pyronnet et al. 1999). The RNA helicase eIF4A associated with eIF4G unwinds RNA secondary structure in the vicinity of the cap structure. This reaction is dependent on ATP and ATP hydrolysis and is stimulated by eIF4B. Interaction of eIF4G with the multisubunit factor eIF3 bound to the 40S ribosomal subunit then promotes binding of the 43S preinitiation complex to the 5′ end of the mRNA (for reviews, see Hentze 1997; Morley et al. 1997). In accordance with its important function in initiation of translation, the assembly of the multiprotein complex at the 5′ end of mRNA is regulated by intra- and extracellular signals through modulation of the interaction between eIF4E and eIF4G via eIF4E-binding proteins (for a review, see Gingras et al. 2001).
The Saccharomyces cerevisiae homologs of mammalian eIF4G1 and eIF4G2, yeast eIF4G1 and eIF4G2, are encoded by the genes TIF4631 and TIF4632 (Goyer et al. 1993) and are 50% identical at the amino acid level. Yeast cells with a disrupted TIF4631 gene display a slow growth phenotype (Lanker et al. 1992), whereas a double TIF4631 and TIF4632 gene disruption is lethal for yeast cells (Goyer et al. 1993). Like mammalian eIF4G, yeast eIF4G has been shown to interact with eIF4E (Mader et al. 1995; Altmann et al. 1997), eIF4A (Dominguez et al. 1999, 2001; Neff and Sachs 1999), and Pabp1, when the latter is bound to poly(A) (Tarun and Sachs 1996).
Besides interacting indirectly with the 5′ end of mRNA through eIF4E and with the 3′ end through Pabp1, eIF4G may also interact directly with mRNA or ribosomal RNA sequences. Indeed, a nucleic-acid-binding domain was found in wheat eIFiso4G (Kim et al. 1999). Furthermore, putative RNA-binding motifs were identified in yeast eIF4G by amino acid sequence inspection (Goyer et al. 1993). Originally, candidate sequences included a putative RNA recognition motif (RRM) (amino acids 646–776) and two arginine- and serine-rich (RS) domains (amino acids 488–553 and 873–908). RS domains consist of repeated arginine–serine dipeptides (and often contain additional repeats) and may be involved in protein–protein binding or mediate protein–nucleic acid interactions (Birney et al. 1993). RS domains are highly basic and therefore believed to interact with the negatively charged phosphate backbone of nucleic acids (Birney et al. 1993). Arginine residues contribute significantly to the interaction of RS domains with RNA, as they can form a network of hydrogen bonds with the sugar–phosphate backbone and the bases of RNA (Burd and Dreyfuss 1994).
The domain of yeast eIF4G containing the putative RRM (Goyer et al. 1993) was more recently shown to represent the eIF4A-binding site (Dominguez et al. 1999; Neff and Sachs 1999) and the homologous domain in mammalian eIF4G2, apart from interacting with eIF4A (Lamphear et al. 1995), was demonstrated to bind to the internal ribosome entry site (IRES) of EMC viral RNA (Pestova et al. 1996). Structural analysis revealed that this domain folds like a HEAT repeat (five antiparallel pairs of α-helices) and not like an RRM (Marcotrigiano et al. 2001). Because the region containing the HEAT repeat is well conserved between mammalian and yeast eIF4G at the amino acid level (Marcotrigiano et al. 2001) and serves as a binding site for both yeast and mammalian eIF4A (Dominguez et al. 2001), we assume that yeast eIF4G also contains a HEAT repeat.
In this work, we mapped RNA-binding sites on eIF4G1 and examined the importance of RNA-binding activity of eIF4G1 for translation in vitro and growth of yeast cells.
RESULTS
Identification of RNA-binding sites of eIF4G1
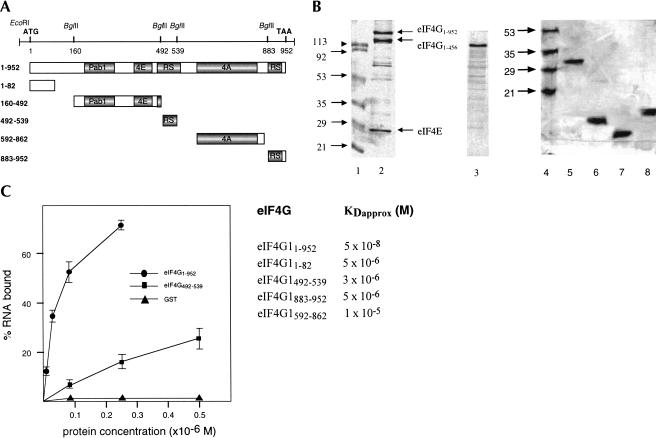
The role played by eIF4G in initiation of translation very likely requires its direct interaction with mRNA and/or ribosomal RNA. To test full-length eIF4G1 and parts of the protein for RNA-binding activity, we expressed different fragments as glutathione S-transferase (GST) fusion proteins in Escherichia coli, isolated the fusion proteins by glutathione S-Sepharose affinity chromatography, cleaved the GST part from the hybrid proteins with protease, and purified the eIF4G1 moiety by fast performance liquid chromatography (FPLC) chromatography. Most of the fragments were subcloned using in-frame _Bgl_II restriction sites (see Materials and Methods; Fig. 1A). The purified recombinant proteins are shown in Figure 1B. We could not obtain homogenous full-length eIF4G1 protein but were able to reduce the amount of degradation products by coexpressing eIF4E (Fig. 1B, lane 2). The predicted molecular weight of yeast eIF4G1 is 107 kD, but the protein migrates much slower on sodium dodecyl sulphate (SDS) polyacrylamide gels (Goyer et al. 1993). The smaller band of the doublet represents eIF4G11–456 as determined by mass analysis (results not shown). This fragment migrates aberrantly and was shown earlier to be inactive in eIF4A binding (Dominguez et al. 2001). Because its C terminus lies in the eIF4E-binding site, this fragment is expected not to bind eIF4E. We do not know whether eIF4G11–456 is generated by proteolysis or premature termination of translation in E. coli. Because the fusion protein eIF4G1160–492 (predicted molecular weight of 66 kD) also migrates slower than expected on SDS gels at about 100 kD (Fig. 1B, lane 3), sequences responsible for aberrant migration of eIF4G1 must be located in this region. Interestingly, abnormal migration of eIF4G on SDS gels was also observed for mammalian eIF4G and was shown to be due to the N-terminal domain of the protein (Yan et al. 1992; Lamphear et al. 1995). Other fragments of eIF4G1 migrate as expected: eIF4G1592–862 at about 30 kD (Fig. 1B, lane 5), eIF4G1883–952 at about 8 kD (Fig. 1B, lane 6), eIF4G1492–539 at about 6 kD (Fig. 1B, lane 7) and eIF4G11–82 at about 9 kD (Fig. 1B, lane 8). As a negative control in RNA-binding assays, GST was expressed from the vector pGEX-1λT and purified on glutathione S-Sepharose (not shown).
FIGURE 1.
Identification of eIF4G1 RNA-binding sites. (A) Schematic representation of yeast eIF4G1 and eIF4G1 fragments. The DNA with the relevant restriction sites used for subcloning is shown on top. ATG and TAA indicate start and end of the open reading frame. The numbers indicate amino acid positions. Pab1, poly(A)-binding protein 1 binding domain; 4E, eIF4E-binding domain; RS, arginine- and serine-rich domain; 4A, eIF4A-binding domain. (B) About 1–2 μg of each protein were fractionated on 15% (lanes 1_–_3) and 20% (lanes 4_–_8) SDS polyacrylamide gels and stained with Coomassie blue. (Lanes 1,4) molecular weight markers (numbers indicate kilodaltons), (lane 2) eIF4G11–952, (lane 3) eIF4G1160–492, (lane 5) eIF4G1592–862, (lane 6) eIF4G1883–952, (lane 7) eIF4G1492–539, (lane 8) eIF4G11–82. (C) Filter-binding assay. On the left, the titration curves of three of the proteins are shown. Proteins were titrated with 0.25 pmole [32P]-labeled 38-nt RNA in a 20-μL reaction volume containing buffer A. On the _right, K_Dapprox. values for eIF4G1 proteins are shown. For estimation of protein concentration of full-length eIF4G1, only the slowest migrating band (B, lane 3) was included.
To test for RNA-binding activity of eIF4G1 and eIF4G1 fragments, we first used a filter-binding assay. Proteins were incubated with radioactively labeled single-stranded RNA and the reaction mixtures passed through nitrocellulose membranes (which retain protein). Radioactivity retained on the filters after washing represents RNA bound to protein. Typical titration curves for three of the proteins are shown in Figure 1C. We cleaved the GST moiety from the fusion proteins for these experiments; however, control experiments showed that GST fusion proteins behave identically in RNA-binding experiments. Full-length eIF4G11–952 showed the highest affinity for RNA with an estimated _K_D (_K_Dapprox.) of 5 × 10−8 M whereas the fragments eIF4G11–82, eIF4G1492–539, and eIF4G1883–952 show about 100-fold lower affinity. The fragments eIF4G1160–492, eIF4G1542–883 (not shown), and eIF4G1592–862, as well as GST (used as negative control) did not bind significant amounts of RNA. The presence of eIF4E in full-length eIF4G1 had no effect on RNA-binding activity as eIF4G1 expressed in its absence has the same affinity for RNA (result not shown).
We can exclude RNA degradation as the cause for low RNA-binding activity, because we checked the integrity of RNA after incubation by fractionating incubation mixtures on denaturing urea gels and exposing gels to X-ray film. A major band migrating at the position of intact RNA was observed after incubation, indicating little or no degradation of RNA in the filter-binding assay. Proteins that bind RNA with high affinity might retain bacterial RNA during their purification from E. coli; however, none of the purified proteins shown in Figure 1B carried nucleic acids, as revealed by UV light absorption at 260 and 280 nm.
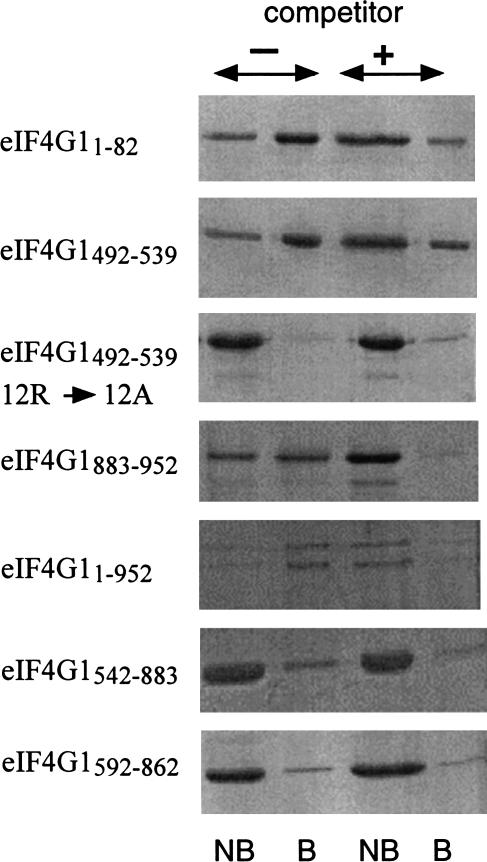
We verified the RNA-binding activity of eIF4G1 and eIF4G1 fragments by a different method and chose adsorption to poly(U)-agarose (Kim et al. 1999), which has the advantage over the filter-binding assay of allowing visualization of proteins bound to RNA. We tested full-length eIF4G11–952 and the fragments eIF4G11–82, eIF4G1492–539, and eIF4G1883–952 carrying RNA-binding sites as determined by filter-binding assays. The fragments eIF4G1542–883 and a shorter version, eIF4G1592–862, which lacks the last few arginine and serine residues of the middle RS domain, were used as negative controls. To test whether arginines in the RS domains are involved in RNA binding, we also analyzed the fragment eIF4G1492–539/12R→A with all 12 arginine residues replaced by alanine residues (Fig. 2). We used GST fusion proteins in this experiment but obtained the same results with cleaved proteins. Specific binding to the poly(U) part of poly(U)-agarose was verified by competition of binding with excess free poly(U). All three eIF4G1 fragments carrying RNA-binding sites as determined in the filter-binding assay as well as full-length eIF4G1 bound to poly(U)-agarose and binding is competed by free poly(U). The full-length preparation of eIF4G1 used for these experiments also contained the fragment eIF4G11–456. This fragment contained the N-terminal RNA-binding site and bound to poly(U)-agarose. In contrast, the binding of eIF4G1542–883 and eIF4G1592–862 to poly(U)-agarose was very low and not influenced by free poly(U). The same was true for eIF4G1492–539/12R→A, supporting the hypothesis that arginine residues contribute to the ability of the protein to bind to RNA.
FIGURE 2.
Binding of eIF4G1 and eIF4G1 fragments to poly(U) agarose. Purified GST-eIF4G1 fusion proteins were bound to poly(U) agarose for 30 min at 4°C, the resin washed extensively with buffer, proteins bound to the matrix eluted by boiling with SDS sample buffer, fractionated on 17.5% SDS polyacrylamide gels, and stained with Coomassie blue. Where indicated, the protein was preincubated for 15 min with soluble poly(U). For each fusion protein, unbound protein (NB) and protein bound to the matrix (B) in the absence (first and second lanes) and in the presence (third and fourth lanes) of soluble poly(U) is shown.
We conclude from these results that eIF4G1 contains at least three sites, amino acids 1–82, 492–539, and 883–952, able to interact with RNA. The two latter sites correspond to the arginine- and serine-rich regions originally identified as candidate RNA-binding sites by inspection of the eIF4G1 amino acid sequence (Goyer et al. 1993).
RNA-binding sites of eIF4G1 are required for growth of yeast cells
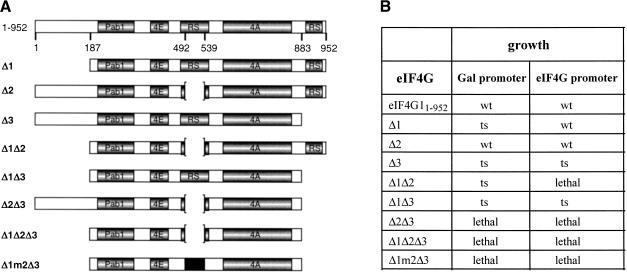
To test whether regions of eIF4G1 identified as RNA-binding sites are required for growth, we expressed eIF4G1 proteins containing deletions of single or multiple RNA-binding sites in yeast cells with disrupted TIF4631 and TIF4632 genes (see Materials and Methods). The mutants Δ1 [deletion of amino acids (aa) 1–186], Δ2 (deletion of aa 492–539), Δ3 (deletion of aa 844–952), combinations of two or three deletions (Δ1Δ2, Δ1Δ3, Δ2Δ3, Δ1Δ2Δ3), and combinations of deletions of N- and C-terminal sites with mutations of arginines to alanines in the middle domain (Δ1m2Δ3) were expressed either from the galactose inducible GAL1/10 promoter or from the eIF4G promoter, and we tested whether they could replace wild-type eIF4G1 in supporting growth of cells (Fig. 3). Deletion of all three RNA-binding sites (Δ1Δ2Δ3), deletion of the N- and C-terminal RNA-binding sites, and replacement of arginine residues by alanines in the remaining middle RNA-binding site (Δ1m2Δ3) or deletion of both the middle and C-terminal RNA-binding sites (Δ2Δ3) were lethal for yeast cells (Fig. 3B). eIF4G1 molecules missing one of the RNA-binding sites (Δ1, Δ2, or Δ3) supported growth. Deletion of the C-terminal RNA-binding site (Δ3) led to a temperature-sensitive (ts) phenotype, deletion of the N-terminal domain (Δ1) conferred a ts phenotype only when expressed from the GAL1/10 promoter, and deletion of the middle RNA-binding site (Δ2) produced no visible phenotype. Mutant eIF4G1 with both the N- and C-terminal RNA-binding sites deleted (Δ1Δ3) was still able to support growth of cells but conferred a ts phenotype, whereas Δ1Δ2 was lethal when expressed from the eIF4G promoter but partially functional (ts) when expressed from the GAL1/10 promoter. The different phenotypes of Δ2 and Δ1Δ2 when expressed from either the GAL1/10 or eIF4G promoter might reflect differences in expression levels.
FIGURE 3.
In vivo phenotype of eIF4G1 deletion mutants. (A) Schematic representation of yeast eIF4G1 and eIF4G1 fragments. Pab1, poly(A) binding protein binding domain; 4E, eIF4E-binding domain; RS, arginine- and serine-rich domains; 4A, eIF4A-binding domain. (B) The phenotype observed on plates of yeast strains expressing the various eIF4G1 deletion proteins as only source of eIF4G1 (either under the Gal1/10 or the eIF4G promoter) is indicated. wt, wild-type growth; ts, temperature sensitive (no growth at 37°C, normal growth at 30°C).
To test whether lethal phenotypes might be due to lack of expression of the corresponding proteins, their expression from the GAL1/10 promoter and from eIF4G1 promoter in cells coexpressing wild-type eIF4G1 from its own promoter was analyzed by Western blotting. The expression of the nonlethal mutant proteins was also analyzed after FOA selection (results not shown). All mutant eIF4G1 proteins could be detected at similar or somewhat reduced levels (compared to wild-type eIF4G1) except Δ1Δ2 and Δ1Δ2Δ3 expressed from the eIF4G1 promoter, which were not detected. We conclude that Δ1Δ2Δ3 is a nonfunctional protein, as expression from the GAL1/10 promoter leads to lethal phenotype whereas Δ1Δ2 is still partially active and the lethal phenotype (when expressed from the eIF4G1 promoter) due to lack of or too low expression.
These results suggest that RNA-binding sites of eIF4G1 are essential for growth. Deletion of all three RNA-binding sites (Δ1Δ2Δ3) inactivates eIF4G1. Deletion of two RNA-binding sites inactivates eIF4G1 either fully (Δ2Δ3) or partially (Δ1Δ2 and Δ1Δ3). Deletion of one RNA-binding site has either no effect (Δ2) or leads to partial inactivation (Δ1, Δ3). This suggests that at least one RNA-binding site (middle or C-terminal) is required and two RNA-binding sites are sufficient for eIF4G1 function in vivo.
RNA-binding sites of eIF4G1 are required for translation in vitro
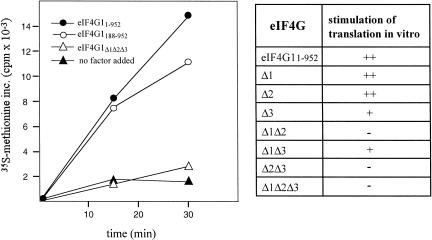
We next asked whether mutant eIF4G1 proteins lacking RNA-binding sites show a defect in supporting translation initiation in vitro. For these experiments we used an eIF4G-dependent yeast extract. This extract is inactive in translation due to two mutations in eIF4G1 that lead to loss of interaction of eIF4G1 with eIF4A (Dominguez et al. 2001). Addition of eIF4G1 restores translation of total yeast RNA as shown by the stimulation of methionine incorporation into protein. The eIF4G1 deletion mutants shown in Figure 3A were expressed as GST fusion proteins together with eIF4E in E. coli, proteins purified by affinity chromatography as described (see Materials and Methods) and tested in the eIF4G-dependent cell free translation system (Fig. 4). We had found earlier that the presence of eIF4E in eIF4G1 preparations does not measurably influence the activity of eIF4G1 in this in vitro translation system (Dominguez et al. 2001). A range of concentrations of each protein was measured to determine the optimal concentration for stimulation of translation. Mutant proteins with single deletions Δ1 and Δ2 were active in stimulating translation, whereas Δ3 and Δ1Δ3 were only weakly active. The mutant proteins Δ1Δ2, Δ2Δ3, and Δ1Δ2Δ3 were inactive for in vitro translation. These data show that the requirements for individual RNA-binding sites for growth of yeast cells and translation in vitro are qualitatively very similar.
FIGURE 4.
In vitro translation of GST-eIF4G1 fusion proteins in an eIF4G-dependent extract. Methionine incorporation at 25°C was measured as described previously (Dominguez et al. 1999) using 10-μL reaction mixtures containing 5 μg total yeast RNA as mRNA source and 3.6 μCi 35S-methionine. The kinetics of methionine incorporation for four proteins is shown on the left. On the right, stimulation of translation is indicated by ++ (like GST-eIF4G11–952 and GST-eIF4G1188–952); +, intermediate; -, like “no factor added.”
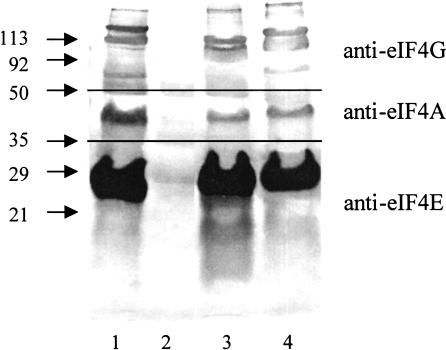
We tested whether eIF4G1 deletion mutants inactive in translation initiation in vitro still interact with eIF4E and eIF4A. GST-eIF4G1 fusions proteins (coexpressed with eIF4E in E. coli) bound to glutathione-Sepharose were incubated with purified eIF4A, eluted by boiling with SDS sample buffer, fractionated on SDS gels, and analyzed by Western blotting, using antibodies against eIF4E, eIF4G, and eIF4A (Fig. 5). All purified mutant proteins bound to eIF4E and eIF4A, whereas GST did not interact with eIF4E or eIF4A (only the proteins inactive in in vitro translation are shown in Fig. 5). The eIF4A band in this experiment is not particularly strong but nontheless specific: Reaction of GST-eIF4G11–952 with anti-eIF4A antibody gave no signal (result not shown). This indicates that the mutant eIF4G1 proteins are correctly folded and capable of interacting with other initiation factors.
FIGURE 5.
Interaction of GST-eIF4G1 proteins with eIF4E and eIF4A. Approximately 40 pmole of the eIF4G1 proteins (coexpressed with eIF4E in E. coli and purified on glutathione-Sepharose) were diluted to 1.5 mL in TBS/NaCl/Triton and bound on 20 μL of glutathione-Sepharose for 2 h at 4°C. The resin was washed twice with TBS and once with buffer A. The resin was resuspended in 100 μL of buffer B, incubated with 0.2 μg of purified eIF4A (expressed in E. coli) for 1 h at 4°C, washed four times with buffer B, and transferred into fresh Eppendorf tubes. Proteins bound were eluted by boiling with SDS sample buffer, fractionated on SDS polyacrylamide gels, and analyzed by Western blotting, using antibodies against eIF4E, eIF4G, and eIF4A as described (Berset et al. 1998). (Upper panel) anti-eIF4G antibody, (middle panel) anti-eIF4A antibody, (lower panel) anti-eIF4E antibody. (Lane 1) eIF4G11–952, (lane 2) GST, (lane 3) GST-eIF4G1 Δ1Δ2Δ3, (lane 4) GST-eIF4G1 Δ1m2Δ3.
RNA-binding activity of eIF4G1 deletion mutants
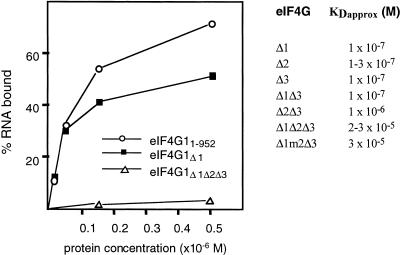
To examine the effect of removing RNA-binding sites on overall RNA-binding activity of eIF4G1, deletion mutants were tested for RNA binding in vitro in the filter-binding assay and approximate _K_D values calculated (Fig. 6). Full-length eIF4G1, Δ1, Δ2, Δ3, and Δ1Δ3 bound RNA efficiently (_K_Dapprox. around 1 × 10−7 M), whereas the mutant Δ2Δ3 had a 10-fold lower affinity for RNA and Δ1Δ2Δ3 and Δ1m2Δ3 were strongly impaired in RNA binding (_K_Dapprox. around 2–3 × 10−5 M). We did not calculate the _K_Dapprox. for Δ1Δ2 because this protein preparation contained fragments that made protein concentration determination and therefore _K_Dapprox. calculation unreliable.
FIGURE 6.
Filter-binding assay. The titration curves of three of the GST-eIF4G1 fusion proteins tested are shown on the left. Proteins were titrated against 0.25 pmole [32P|-labeled 38-nt RNA in a 20-μL reaction volume containing buffer A. _K_Dapprox. values for GST-eIF4G1 fusion proteins are shown on the right.
We conclude from these data that RNA-binding activities and activities in translation initiation in vitro of the deletion proteins correlate well with RNA-binding activity, indicating that RNA-binding activity is required for eIF4G1 activity in vitro and in vivo.
DISCUSSION
In this work, we show that yeast eIF4G1 contains three RNA-binding sites. The N-terminal region of eIF4G1, amino acids 1–82, contains 12 glutamine, 12 asparagine, 10 glycine, 6 arginine, and 6 tyrosine residues. They are arranged as glycine cluster, RGG-, and RG-motifs. Glycine-rich clusters are common in RNA-binding proteins and are composed of glycine–glycine dipeptides interspersed with arginine and aromatic residues (RGG or GAR domains; Birney et al. 1993). The RGG motif is ubiquitous in proteins involved in splicing or pre-rRNA processing, in RNA helicases, and hnRNPs (Ghisolfi et al. 1992; Kiledjian and Dreyfuss 1992; Birney et al. 1993). RNA-binding activity of the N-terminal part of eIF4G1 is supported by amino acid sequence comparison with other proteins implicated in RNA binding. A search for related sequences revealed homology with Trypanosoma brucei nucleolar RNA-binding protein Nopp44/46 (TrEMBL, Q26711) (36% identity in a 64 amino acid sequence), which also carries RGG motifs.
Full-length eIF4G2 shows similar RNA-binding activity like eIF4G1 (our unpubl. result). Even though we have not tested fragments of eIF4G2 for RNA-binding activity, we anticipate that eIF4G2 also contains three RNA-binding sites. The amino acid sequences of the middle and C-terminal arginine- and serine-rich RNA-binding sites are well conserved between eIF4G1 and eIF4G2, 62% identity for eIF4G1492–539 compared to eIF4G2462–503 and 38% for eIF4G1883–952 compared to eIF4G2854–914 (sequence identity determined with the program Bestfit from the GCG-Wisconsin package). It is interesting to note that eIF4G1492–539 is located close to the eIF4E-binding domain. It is possible that this RNA-binding site contributes to the stability of the complex formed by eIF4G, eIF4E (interacting with the cap structure of mRNA), and mRNA. The N-terminal parts of eIF4G1 and eIF4G2 are not very strongly conserved (28 % identity), but basic amino acid residues and asparagine residues are conserved in number and position.
The eIF4G1 fragment containing the putative HEAT repeat motif, eIF4G1542–883, showed very low RNA-binding activity in our assay. Interestingly, the homologous domain in mammalian eIF4G binds to the IRES of EMC viral RNA (Pestova et al. 1996; Marcotrigiano et al. 2001) and this binding is strongly stimulated by eIF4A (Lomakin et al. 2000). Apparently, this domain recognizes specific RNA sequences. We tested whether the binding of eIF4A to eIF4G1542–883 might enhance the affinity of eIF4G1 for RNA, perhaps through a change of conformation. However, eIF4A did not influence the RNA-binding activity of eIF4G1542–883 or of full-length eIF4G1. Besides synthetic RNA, we also tested diverse in vitro transcribed RNAs for binding to our eIF4G1 preparations and did not observe significantly different affinities (results not shown). The questions of whether certain RNA sequences show different affinty for eIF4G and of whether eIF4G1542–883 might bind certain sequences, perhaps IRES sequences, remain therefore open.
In the filter-binding assays, proteins carrying single RNA-binding sites (eIF4G11–82, eIF4G1492–539, and eIF4G1883–952) bound RNA with about 100-fold lower affinity than full-length eIF4G1 protein. This difference may be due to binding of two or three domains to a single RNA molecule or a conformational change in the protein leading to enhanced affinity of individual RNA-binding sites.
In vivo, eIF4G1 mutants carrying two of the three RNA-binding sites were fully (Δ1, Δ2) or partially functional (Δ3), whereas proteins containing only one RNA-binding site were either nonfunctional (Δ2Δ3) or partially functional (Δ1Δ2, Δ1Δ3). To support growth of yeast cells, eIF4G1 containing only the C-terminal RNA-binding site (mutant Δ1Δ2) requires expression from the GAL1/10 promoter (presumably overexpression) and eIF4G1 containing only the middle RNA-binding site (mutant Δ1Δ3) is unable to support growth at 37°C. Mutant eIF4G1 proteins lacking all three RNA-binding sites (Δ1Δ2Δ3) or eIF4G1 with N- and C-terminal RNA-binding sites deleted and argine residues in the remaining middle site replaced by alanine (Δ1m2Δ3) are not functional. These data show that two RNA-binding sites in eIF4G1 (one of them being the C-terminal RNA-binding site) are required for normal growth of yeast cells.
In in vitro translation experiments, the activities of mutant eIF4G1 proteins closely resemble their activities in supporting growth of yeast cells. This correlation indicates that the inability of mutant eIF4G1 proteins to support growth is due to their inability to support translation initiation. The defect in translation initiation is not caused by a defect in binding to eIF4A or eIF4E (Fig. 5) but may be due to reduced ability to bind RNA. This view is supported by the correlation of RNA-binding activity of mutant proteins with their activities in supporting growth and translation initiation in vitro.
It remains to be elucidated whether RNA-binding sites of eIF4G1 bind messenger RNA and/or ribosomal RNA in vivo. Binding of eIF4G1 to the latter would contribute to the interaction between ribosomes and mRNA. With three distinct RNA-binding sites, eIF4G1 could bind to an mRNA molecule at several contact points simultaneously and bring it into a specific conformation to allow the binding of the 40S ribosomal subunit.
MATERIALS AND METHODS
Plasmid construction
Fragments of eIF4G1
GST fusion proteins were expressed in E. coli from the vectors pGEX-1λT, pGEX-2T, or pGEX-6P-2 (Pharmacia Biotech). The complete TIF4631 open reading frame on a 3266-bp DNA _Eco_RI fragment was subcloned from the plasmid pCG8.4 (Goyer et al. 1993). eIF4G1160–492, eIF4G1492–539, and eIF4G1542–883 were subcloned as _Bgl_II fragments into the _Bam_HI site and eIF4G1883–952 as _Bgl_II/_Eco_RI fragment into the _Bam_HI/_Eco_RI sites of pGEX-1λT. The cDNA encoding eIF4G11–952 with six histidine residues at the carboxy terminus (introduced by PCR) was subcloned as _Eco_RI fragment into pGEX-6-P2 to obtain pGEX6-TIF4631. eIF4G1592–863 and eIF4G1863–952 were obtained by PCR amplification using primer pairs 5′-CCGGATCCCTAGCCCCTGACGGAAAGACC-3′; 5′-CCGAATTCTCACTTAGGACCGTTGTCCTTCT-3′ and 5′-CCGGATCCACCATTCAACAGATTCATGAGG-3′; and 5′-CCG AATTCTTACTCTTCGTCATCACTTTCTCC-3′ and subcloning of the _Bam_HI-_Eco_RI-digested DNA fragments into pGEX-6-P2.
eIF4G11–82 was obtained by exonuclease III treatment of pGEX6-TIF4631 (Henikoff 1987, 1990). Constructs were sequenced and transformed into the E. coli strain BL21 carrying a plasmid encoding a rare arginyl-tRNA and conferring chloramphenicol resistance (Saxena and Walker 1992).
Deletion and mutation of RNA-binding sites
The N-terminal deletion eIF4G1187–952 (Δ1) was obtained by PCR amplification from TIF4631 DNA using the upstream primer 5′-CCGGATCCATAATGCCACTAATGACTCTAAGGCC-3′ (which introduces a new start ATG at amino acid position 186) and the downstream primer 5′-GAATTAGCTCTGTCATCC-3′ and subcloning of the _Bam_HI-_Hpa_I fragment (a unique _Hpa_I site is located at the site encoding amino acid 298 of TIF4631 DNA) into the unique sites of pGEX6-TIF4631. The C-terminal deletion eIF4G11–843 (Δ3) was obtained by exonuclease III treatment of pGEX6-TIF4631. The combined N-terminal and C-terminal deletion mutant eIF4G1187–843 (Δ1Δ3) was obtained by inserting the _Hpa_I-_Eco_RI fragment of pGEX6-TIF4631/Δ844–952 into pGEX6-TIF4631/Δ1–186. The triple deletion mutant pGEX6-TIF4631 Δ1–186/Δ 493–541/Δ844–952 was obtained by _Bgl_II digestion and religation of pGEX6-TIF4631 Δ1–186/Δ844–952. The mutant carrying the combined double N-terminal and C-terminal deletion and the replacement of 12 arginine residues by alanine in the central RNA-binding domain of pGEX6-TIF4631 was obtained with two partially complementary oligonucleotides: 5′-AATAG ATCTGCAGATTCTGGCGCATTCGGCAATAATTCTAGTGCAG GCCATGACTTTGCAAATACCTCAGTGGCAAATATGGATGAC GC-3′ and 5′-ATAAGATCTATTAGATGCCGCGTCATTCATT GCCTTTGATGCTGCCTTTGATGAAGTTGCTGAATTAGCTGC GTCATCCATATTTGCC-3′. An 18-bp duplex was formed after annealing of the 3′ ends of both oligonucleotides. Single-strand regions were filled in with T4 DNA polymerase and dNTPs and a 162-bp double-stranded DNA fragment obtained (the corresponding arginine residues of Tif4631p located at positions 493, 497, 504, 509, 514, 519, 523, 528, 529, 532, 537, and 538 were replaced by alanines) that was digested with _Bgl_II and inserted in the correct orientation in pGEX6-TIF4631 Δ1–186/Δ493–543/Δ844–952. The construct was called pGEX6-TIF4631 Δ1m2Δ3.
For expression in yeast cells, constructs were subcloned as _Bam_HI-_Eco_RI or _Eco_RI fragments into the unique sites of the vectors p301 (TRP1 gene as selectable marker; GAL1/10 promoter) and pRS313 (HIS3 gene as selectable marker; Sikorski and Hieter 1989) carrying the complete 5′ UTR of TIF4631 from bp −522 to +1 (Goyer et al. 1993). For expression in E. coli, the constructs were introduced into pGEX-6P-2 for production of GST fusion proteins and eIF4G1 proteins coexpressed with eIF4E (from a pT74E-derivative plasmid conferring kanamycin-resistance; Edery et al. 1988).
Growth of E. coli strains
E. coli strains carrying pGEX plasmids were grown in LB medium (bacto-tryptone 1%, NaCl 1%, yeast extract 0.5%, 50 μg/mL ampicillin, 30 μg/mL chloramphenicol) overnight at 37°C, diluted 1:50 into 1 L of fresh LB medium, grown at 37°C until the A600 reached 0.5–1 and the expression of fusion proteins induced with 0.5–1 mM isopropyl β-D-thiogalactoside for 4 h. Cells were harvested and resuspended in 15 mL of ice-cold phosphate-buffered saline (PBS; 10 mM Na2HPO4, 1.8 mM KH2PO4 at pH 7.3, 140 mM NaCl, 2.7 mM KCl) or in TBS/NaCl/Triton (10 mM Tris-HCl at pH 7.5, 650 mM NaCl, 0.2% Triton X-100) and lysed with a French Press at 1000 psi. The suspension was centrifuged at 7650_g_ for 15 min at 4°C.
Protein purification
GSH-Sepharose affinity chromatography
Fifteen milliliters of cleared lysate were incubated with 0.5 mL glutathione (GSH) Sepharose 4B (Amersham Pharmacia Biotech; 0.25-mL slurry equilibrated with TBS) with gentle agitation for 4 h to overnight at 8°C. After centrifugation at 350_g_ for 5 min, the unbound fraction was disposed. The slurry was washed with 15 mL TBS, transferred to an Eppendorf tube, and washed twice with 1 mL TBS. Then the slurry was incubated with 0.5 mL GSH elution buffer (100 mM Tris-HCl at pH 9.0, 120 mM NaCl, 10 mM GSH, adjusted to pH 9) for 30 min at 8°C, centrifuged for 5 min at 350_g_, and the proteins collected from the supernatant. When GST fusion proteins were used for RNA binding or in vitro translation experiments after this step, they were dialyzed against buffer B (20 mM HEPES at pH 7.4, 1 mM MgCl2, 100 mM KCl) and, when necessary, concentrated by Centricon centrifugation (Amicon). Glycerol (10% final concentration) and β-mercaptoethanol (7 mM final concentration) were added, and aliquots were frozen in liquid nitrogen and kept at −70°C.
Ni-chelate affinity chromatography
For the purification of double-tagged eIF4G11–952 carrying GST at the N terminus and six histidine residues at the C terminus, the cleared lysates were first incubated with glutathione Sepharose 4B and eIF4G11–952 isolated as described above. The GSH eluates were combined and LiCl was added to a final concentration of 0.3 M. The eluate was then incubated with 30 mL Ni-NTA Agarose (Qiagen) overnight at 8°C, centrifuged at 350_g_ for 5 min, and washed three times with 1 mL TBS. The protein was eluted in two steps at 8°C with 80 mL 250 mM imidazole (in TBS) for 2 h. The eluates were combined and dialyzed against buffer B (20 mM HEPES-KOH at pH 7.4, 1 mM MgCl2, 100 mM KCl) and frozen in 10% glycerol at −70°C.
DEAE treatment
Approximately 0.5 mL of DEAE resin were washed with 10 mL ddH2O, mixed with protein diluted 1:10 with buffer A (30 mM HEPES-KOH at pH 7.4, 100 mMKAc2, 2 mM MgAc2), and incubated with the DEAE matrix for 30 min at 8°C on a shaker. After centrifugation at 350_g_ for 5 min, the proteins were collected from the supernatant.
Gel filtration
Proteins were purified by gel filtration on a Superdex S75 (16/60) column (Pharmacia) in FPLC buffer (20 mM Tris-HCl at pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.005% NaN3). Fractions containing the proteins were identified by SDS-polyacrylamide gel electrophoresis and pooled. The pooled fractions were dialyzed against buffer A, when necessary concentrated by Centricon centrifugation (Amicon), and kept in 10% glycerol at −70°C or at −20°C.
Polyacrylamide gel electrophoresis and Western blot analysis
Proteins were fractionated on 15% SDS-polyacrylamide gels (Anderson et al. 1973) and stained with Coomassie blue R-250. Western blotting was done as described (Berset et al. 1998).
Protein concentration determination
The concentrations of eIF4G11–82 and eIF4G1883–952 were measured by absorbance spectrometry. The concentrations of other protein preparations were measured with the Bradford assay or estimated on gels stained with Coomassie blue using bovine serum albumin (BSA) as standard. Dilutions of protein preparations were prepared in buffer B containing 20 μg/mL BSA.
RNA labeling
One-hundred picomoles of synthetic RNA (5′-(AAAAC)4 AAAAUAGCACCGUAAAGC-3′, 38 nt; all synthetic RNA oligonucleotides were obtained from W.C. Merrick) were labeled with 10 units T4 polynucleotide kinase (PNK) in PNK buffer (50 mM Tris-HCl at pH 8.2, 10 mM MgCl2, 0.1 mM EDTA, 5 mM dithiothreitol, 0.1 mM spermidine) and 10 pmole [γ-32P]-ATP (≈ 30 μCi). This reaction mixture was incubated for 30 min at 37°C. Subsequently, phenol/chloroform extraction and ethanol precipitation were carried out and the labeled RNA was purified on a G25-Sepharose column to remove unincorporated [γ-32P]-ATP. The specific activity of the labeled RNA was about 0.3 μCi/pmole RNA.
Filter-binding assay
Two-hundred-fifty to 300 fmole (12.5 nM to 15 nM final concentration) of [32P]-RNA was incubated with eIF4G1 in a 20-μL reaction mixture containing buffer A. The binding partners were incubated for 2 min at 0°C and filtered through a nitrocellulose filter (type HA 0.45 μm; Millipore) prewetted with 35 μL buffer A. The filters were washed with 600 μL buffer A, dried, and counted in 3-mL scintillation cocktail (IRGA-SAFE PLUS, Packard). Triple values for each protein concentration were measured. The _K_Dapprox was calculated according to the formula _K_D = ([protein]free • [RNA]free)/[complex], where [protein]free corresponds to [protein]tot − [complex], [RNA]free corresponds to [RNA]tot − [complex], and [complex] corresponds to [RNA]tot • bound cpm on the filter/total cpm input (total RNA).
Optimal conditions for RNA binding in our filter-binding assay were tested: eIF4G1492–539, eIF4G1883–952, and eIF4G11–952 bound similar amounts of RNA at 0°C and at 37°C after 10 min of incubation. The concentration of KCl (0 mM, 50 mM, or 100 mM) in the buffer had little or no influence on the amount of RNA bound. The kinetics of RNA binding were fast, as there was no significant change in binding between 1, 3, 5, and 10 min of incubation at 0°C.
The integrity of RNA after incubation with the different proteins was checked by fractionation of the reaction mixtures on a 12% denaturing polyacrylamide gel (12% acrylamide/bisacrylamide, 19:1, 40% urea) in 90 mM Tris-borate (pH 8.0), 2 mM EDTA. The gel was dried and exposed to an X-ray film.
Protein binding to poly(U)
Forty milligrams of poly(U)-agarose (dried; from Sigma) were resuspended in 2 mL dH2O and washed twice with 50 mM Tris-HCl (pH 7.5), 50 mM NaCl. One hundred microliters of poly(U)-agarose suspension were incubated with about 1 μg of eIF4G1 proteins (100–200 ng of full-length eIF4G1) on ice for 1 h. Where indicated, incubation was done in the presence of 200 μg/mL poly(U) (Sigma). After incubation, the resin was washed three times with 200 μL of 50 mM Tris-HCl at pH 7.5, 50 mM NaCl, and bound protein was eluted by boiling for 1 min in 10 μL SDS sample buffer.
Phenotype of eIF4G1 RNA-binding mutants
The plasmids carrying the eIF4G1 mutants were transformed into yeast strain CBY19 (tif4631::LEU2 tif4632::ura3 〈YCp50-TIF4631; URA3〉; Berset et al. 1998). CBY19 transformants were plated on fluoro-orotic acid (FOA) plates containing 0.7 % FOA to eliminate the plasmid expressing wild-type eIF4G. CBY19 transformants that had lost the URA3 plasmid were grown at 25°, 30°, and 37°C to analyze for a temperature- or cold-sensitive phenotype.
Cell-free translation
Preparation of yeast translation systems and translation reactions were done according to Blum et al. (1989) using the eIF4G-dependent yeast extract described by Dominguez et al. (2001).
Acknowledgments
We thank Elisabeth Kislig and Sandra Nansoz for excellent technical assistance. This work was supported by grant 31-45528.95 of the Swiss National Science Foundation and a grant to C.B. from the Novartis Foundation, Basel, Switzerland.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
- 5-FOA, fluoro-orotic acid
- Ac, acetate
- eIF, eukaryotic initiation factor
- EMC, encephalomyocarditis
- FPLC, fast performance liquid chromatography
- GSH, glutathione
- GST, glutathione-S-transferase
- IRES, internal ribosome entry site
- PBS, phosphate-buffered saline
- SDS, sodium dodecyl sulfate
- TBS, tris-buffered saline
- UTR, untranslated region
REFERENCES
- Altmann, M., Schmitz, N., Berset, C., and Trachsel H. 1997. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 16**:** 1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, C.W., Baum, P.R., and Gesteland, R.F. 1973. Processing of adenovirus 2-induced proteins. J. Virol. 12**:** 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset, C., Trachsel, H., and Altmann, M. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 95**:** 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Kumar, S., and Krainer, A.R. 1993. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21**:** 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, S., Mueller, M., Schmid, S.R., Linder, P., and Trachsel, H. 1989. Translation in Saccharomyces cerevisiae: Initiation factor 4A-dependent cell-free system. Proc. Natl. Acad. Sci. 86**:** 6043–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G. and Dreyfuss, G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265**:** 615–621. [DOI] [PubMed] [Google Scholar]
- Dever, T.E. 2002. Gene-specific regulation by general translation factors. Cell 108**:** 545–556. [DOI] [PubMed] [Google Scholar]
- Dominguez, D., Altmann, M., Benz, J., Baumann, U., and Trachsel, H. 1999. Interaction of translation initiation factor eIF4G with eIF4A in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274**:** 26720–26726. [DOI] [PubMed] [Google Scholar]
- Dominguez, D., Kislig, E., Altmann, M., and Trachsel, H. 2001. Structural and functional similarities between the central eukaryotic initiation factor (eIF)4A-binding domain of mammalian eIF4G and the eIF4A-binding domain of yeast eIF4G. Biochem J. 355**:** 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery, I., Altmann, M., and Sonenberg, N. 1988. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene 74**:** 517–525. [DOI] [PubMed] [Google Scholar]
- Ghisolfi, L., Joseph, G., Amalric, F., and Erard M. 1992. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 267**:** 2955–2959. [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68**:** 913–963. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 15**:** 807–826. [DOI] [PubMed] [Google Scholar]
- Goyer, C., Altmann, M., Lee, H.S., Blanc, A., Deshmukh, M., Woolford, J.L., Trachsel, H., and Sonenberg, N. 1993. TIF4631 and _TIF4632_—Two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor-4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 13**:** 4860–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi, A., Imataka, H., Svitkin, Y.V., Rom, E., Raught, B., Morino, S., and Sonenberg, N. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18**:** 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. 1987. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 155**:** 156–165. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. 1990. Ordered deletions for DNA sequencing and in vitro mutagenesis by polymerase extension and exonuclease III gapping of circular templates. Nucleic Acids Res. 18**:** 2961–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W. 1997. eIF4G: A multipurpose ribosome adapter? Science 275**:** 500–501. [DOI] [PubMed] [Google Scholar]
- Hershey, J.W.B. and Merrick, W.C. 2000. Pathway and mechanism of initiation of protein synthesis. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 615–635. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Imataka, H. and Sonenberg, N. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17**:** 6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka, H., Gradi, A., and Sonenberg. N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17**:** 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian, M. and Dreyfuss, G. 1992. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 11**:** 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.Y., Takahashi, K., Nguyen, T.B., Roberts, J.K., and Webster, C. 1999. Identification of a nucleic acid binding domain in eukaryotic initiation factor eIFiso4G from wheat. J. Biol. Chem. 274**:** 10603–10608. [DOI] [PubMed] [Google Scholar]
- Kozak, M. 1989. The scanning model for translation: an update. J. Cell. Biol. 108**:** 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear, B.J., Kirchweger, R., Skern, T., and Rhoads, R.E. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases—Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270**:** 21975–21983. [DOI] [PubMed] [Google Scholar]
- Lanker, S., Müller, P.P., Altmann, M., Goyer, C., Sonenberg, N., and Trachsel, H. 1992. Interactions of the eIF-4F Subunits in the Yeast Saccharomyces cerevisiae. J. Biol. Chem. 267**:** 21167–21171. [PubMed] [Google Scholar]
- Lomakin, I.B., Hellen, C.U., and Pestova, T.V. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20**:** 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader, S., Lee, H., Pause, A., and Sonenberg, N. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15**:** 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J., Lomakin, I.B., Sonenberg, N., Pestova, T.V., Hellen, C.U., and Burley, S.K. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7**:** 193–203. [DOI] [PubMed] [Google Scholar]
- Morley, S.J., Curtis, P.S., and Pain, V.M. 1997. eIF4G: Translation’s mystery factor begins to yield its secrets. RNA 3**:** 1085–1104. [PMC free article] [PubMed] [Google Scholar]
- Neff, C.L. and Sachs, A.B. 1999. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol. Cell. Biol. 19**:** 5557–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain, V.M. 1996. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236**:** 747–771. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U., and Shatsky, I.N. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16**:** 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet, S., Imataka, H., Gingras, A.C., Fukunaga, R., Hunter, T., and Sonenberg, N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 18**:** 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A.B., Sarnow, P., and Hentze, M.W. 1997. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell 89**:** 831–838. [DOI] [PubMed] [Google Scholar]
- Saxena, P. and Walker, J.R. 1992. Expression of argU, the Escherichia coli gene coding for a rare arginine tRNA. J. Bacteriol. 174**:** 1956–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122**:** 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, S.J. and Sachs, A.B. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15**:** 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Yan, R.Q., Rychlik, W., Etchison, D., and Rhoads R.E. 1992. Amino acid sequence of the human protein synthesis initiation factor-eIF-4gamma. J. Biol. Chem. 267**:** 23226–23231. [PubMed] [Google Scholar]