Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs (original) (raw)
Abstract
The factors regulating the expression of microRNAs (miRNAs), a ubiquitous family of ~22-nt noncoding regulatory RNAs, remain undefined. However, it is known that miRNAs are first transcribed as a largely unstructured precursor, termed a primary miRNA (pri-miRNA), which is sequentially processed in the nucleus, to give the ~65-nt pre-miRNA hairpin intermediate, and then in the cytoplasm, to give the mature miRNA. Here we have sought to identify the RNA polymerase responsible for miRNA transcription and to define the structure of a full-length human miRNA. We show that the pri-miRNA precursors for nine human miRNAs are both capped and polyadenylated and report the sequence of the full-length, ~3433-nt pri-miR-21 RNA. This pri-miR-21 gene sequence is flanked 5′ by a promoter element able to transcribe heterologous mRNAs and 3′ by a consensus polyadenylation sequence. Nuclear processing of pri-miRNAs was found to be efficient, thus largely preventing the nuclear export of full-length pri-miRNAs. Nevertheless, an intact miRNA stem–loop precursor located in the 3′ UTR of a protein coding gene only moderately inhibited expression of the linked open reading frame, probably because the 3′ truncated mRNA could still be exported and expressed. Together, these data show that human pri-miRNAs are not only structurally similar to mRNAs but can, in fact, function both as pri-miRNAs and mRNAs.
Keywords: microRNAs, RNA interference, RNA processing
INTRODUCTION
MicroRNAs (miRNAs) are ~22-nt noncoding RNAs expressed in a wide range of eukaryotic organisms (for review, see Bartel 2004). Although humans are believed to express over 200 distinct miRNAs, little is known about their biological functions. However, several plant and invertebrate miRNAs have been shown to inhibit the expression of mRNAs bearing complementary target sites. In Caenorhabditis elegans, expression of the miRNAs let-7 and lin-4 is developmentally regulated, and loss of let-7 or lin-4 function disrupts normal larval development (Lee et al. 1993; Reinhart et al. 2000). The observed developmental or tissue-specific expression patterns of many vertebrate miRNAs (Lagos-Quintana et al. 2002; Houbaviy et al. 2003; Chen et al. 2004) may imply an analogous role in regulating human development or cellular differentiation.
miRNAs are initially expressed as part of one arm of an imperfect ~80-nt RNA hairpin that, in turn, forms part of a longer transcript termed a primary miRNA (pri-miRNA) (Lee et al. 2003). The first step in miRNA biogenesis is the excision of the upper part of this RNA hairpin by the nuclear RNase III enzyme Drosha to produce an ~65-nt intermediate, termed a pre-miRNA (Lee et al. 2002; Zeng and Cullen 2003). Pre-miRNAs, which form short RNA hairpins bearing a 2-nt 3′ overhang, are then bound by the nuclear export factor Exportin 5, which transports them to the cytoplasm (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). Here, a second RNase III enzyme termed Dicer removes the terminal loop of the pre-miRNA to generate an ~20-bp RNA duplex with 2-nt 3′ overhangs (Grishok et al. 2001; Hutvágner et al. 2001; Lee et al. 2003). The mature miRNA, which forms one strand of this duplex, is then incorporated into a large protein complex, termed the RNA induced silencing complex (RISC), where it functions to guide RISC to complementary mRNA targets (Hammond et al. 2000; Martinez et al. 2002; Schwarz et al. 2002).
Although the RNA processing pathway that gives rise to mature miRNAs is increasingly well understood, transcriptional regulation of miRNA genes has been barely studied, and it has, in fact, remained unclear which RNA polymerase is responsible for miRNA transcription, although RNA polymerase II (pol II) and RNA polymerase III (pol III) are obvious candidates (Bartel 2004). Pol II is responsible for mRNA transcription and also transcribes several small, noncoding RNAs, including four of the small nuclear RNAs, while Pol III transcribes a range of short, noncoding RNAs, including tRNAs and 5S rRNA.
Several observations suggest that pol II may be the RNA polymerase responsible for miRNA transcription. These observations include the finding that miRNA expression is often developmentally regulated, a characteristic of many pol II dependent genes, and the observation that primary miRNA transcripts can be quite long (>1000 nt), as determined by RT-PCR (Lee et al. 2003) and by inspection of EST databases. Moreover, in the case of the two C. elegans miRNA genes lsy-6 and let-7, it has been reported that fusion of their predicted promoter elements to the green fluorescent protein (gfp) gene gives rise to gfp expression in the predicted nematode tissues (Johnson et al. 2003; Johnston and Hobert 2003). Finally, candidate pri-miRNA precursors have been described for Arabidopsis miR-172 (Aukerman and Sakai 2003), C. elegans let-7 (Bracht et al. 2004), and human miR-155 (Tam 2001; Lagos-Quintana et al. 2002). While these candidate pri-miRNA precursors show the characteristics of pol II transcripts, including evidence of splicing and the presence of a 3′ poly(A) tail, their ability to give rise to a mature miRNA in vivo has not been directly addressed.
Analysis of the genomic localization of known human miRNAs has revealed that the majority are in intergenic regions, and sometimes in clusters of several miRNAs, and therefore must depend on their own promoters (Lagos-Quintana et al. 2003). However, ~25% of human miRNA genes are located within known protein coding genes primarily, but not invariably, within introns. This location could imply that these miRNAs are excised from intron lariats derived from the splicing of the pre-mRNAs transcribed from these flanking genes, as previously reported for some small nucleolar RNAs (Weinstein and Steitz 1999). However, as a number of these intronic miRNAs are found in the antisense orientation, relative to the surrounding gene (Lagos-Quintana et al. 2003), this localization does not prove that miRNAs can be derived from pre-mRNAs. Moreover, the fact that mature human miRNAs can be ectopically expressed using either pol II– or pol III–based expression plasmids (Zeng and Cullen 2003; Chen et al. 2004) indicates that miRNA genes are not dependent on a specific polymerase, such as pol II, for their appropriate processing and expression in vivo.
In this report, we have examined several isolated or clustered human miRNAs and find that they are derived from capped, polyadenylated pri-miRNA precursors. In the case of the human miR-21 miRNA, we have cloned the entire ~3433-nt pri-miRNA transcript as well as the flanking promoter element. We show that mature miR-21 is indeed processed from this long pri-miRNA and not from a smaller RNA transcribed from a cryptic internal promoter element, and we further demonstrate that the miR-21 promoter can be used to express a protein-coding mRNA in human cells. Finally, we demonstrate that the presence of a miRNA gene within the 3′ untranslated region (3′ UTR) of an mRNA, as seen with a small number of human miRNAs, results in a surprisingly modest inhibition of the expression of the linked open reading frame. When considered together with earlier work, these data argue that RNA polymerase II is likely to be the major, and possibly the only, polymerase involved in human miRNA transcription.
RESULTS
Human Pri-miRNAs are polyadenylated and capped
A defining characteristic of almost all eukaryotic mRNAs is that they are terminally modified by addition of a 5′ 7-methyl guanylate (m7G) cap and a 3′ poly(A) tail. Evidence showing that pri-miRNAs are polyadenylated and capped would therefore argue strongly in favor of pol II as the relevant polymerase.
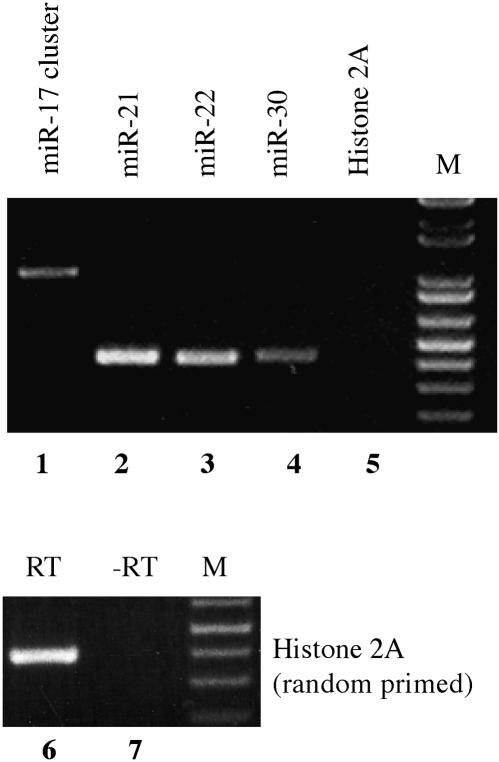
The miRNAs analyzed in this experiment were the isolated miRNAs miR-21, miR-22, and miR-30 and the miR-17/miR-18/miR-19a/miR-20/miR-19b-1/miR-92–1 miRNA cluster. These miRNAs have been previously shown to be expressed at readily detectable levels in HeLa cells (Lagos-Quintana et al. 2001). For analysis of polyadenylation status, we used a HeLa cell cDNA preparation that had been generated using an oligo-dT primer, and unique PCR primers targeted to sequences ~170 bp 5′ and 3′ to the predicted miRNA stem–loops. As a negative control, we used PCR primers specific for histone H2A mRNA, which is highly unusual in that it does not contain a poly(A) tail (Dominski et al. 2003). As shown in Figure 1, we detected amplified DNA fragments of the expected size for miR-21, miR-22, miR-30, and the miR-17 miRNA cluster but consistently failed to detect any signal using the histone H2A–specific primers, although these gave a readily detectable signal when random primed cDNA was tested (Fig. 1). While we cannot exclude the possibility that oligo-dT priming of cDNA synthesis occurred at an internal, 3′ flanking stretch of A residues, rather than at an authentic poly(A) tail, these data nevertheless strongly suggest that these four human pri-miRNAs are polyadenylated.
FIGURE 1.
Polyadenylation of pri-miRNA transcripts. A HeLa cDNA preparation, obtained by oligo(dT) affinity purification of total HeLa cell RNA followed by oligo(dT) primed reverse transcription, was subjected to PCR using primers flanking the human miR-21, miR-22, or miR-30 miRNA stem–loop or the miR-17 miRNA cluster. Primers were designed to sit down ~170 bp 5′ and 3′ to the predicted miRNA stem–loops and were, therefore, expected to give rise to ~410-bp fragments for miR-21, miR-22, and miR-30, or a 1000-bp fragment for the miR-17 cluster. Histone H2A mRNA, which is not polyadenylated, served as a negative control. (Lanes 6,7) To confirm that the Histone 2A primers were functional, PCR was also performed using random primed HeLa cDNA. M, DNA size markers.
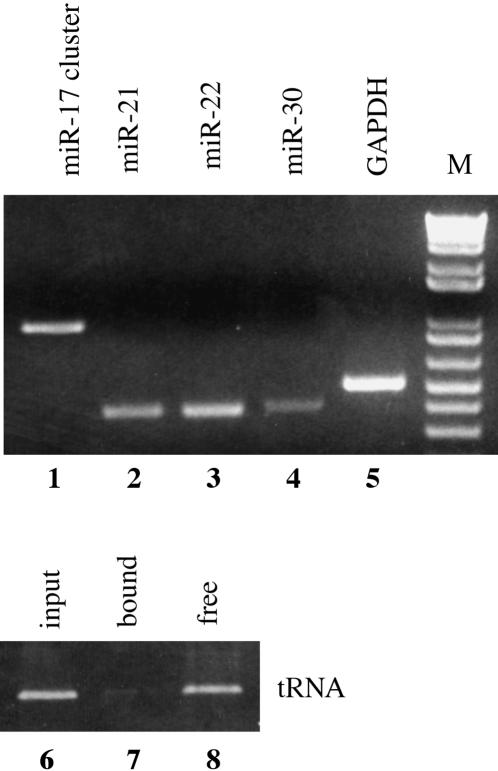
To test whether pri-miRNAs are also capped, we purified m7G capped RNA from total HeLa cell RNA using beads loaded with a mutant form of the cap-binding protein eIF4E that binds 5′ m7G capped RNAs efficiently (Choi and Hagedorn 2003). The resultant RNA preparation was subjected to reverse transcription using random primers and then analyzed by PCR. Human glyceraldehyde phosphate dehydrogenase (GAPDH) mRNA served as a positive control, while a human alanine tRNA was used as a negative control. As shown in Figure 2, miR-21, miR-22, miR-30, the miR-17 cluster and the GAPDH mRNA were all found to bind to the eIF4E beads (lanes 1–5). In contrast, the alanine tRNA did not bind to these beads and instead was found in the unbound, supernatant fraction (Fig. 2, lanes 6–8). We therefore conclude that pri-miRNAs bear a terminal m7G cap moiety.
FIGURE 2.
Pri-miRNA transcripts are capped. Capped RNAs were purified from total HeLa cell mRNA using beads loaded with a high-affinity mutant form of eIF4E, as previously described (Choi and Hagedorn 2003). The recovered capped RNAs were reverse transcribed using random primers and then subjected to PCR using the primers described in Figure 1 or using primers specific for human GAPDH mRNA or alanine tRNA. In lanes 6_–_8, the input, bound and free, supernatant alanine tRNAs were also analyzed.
Cloning and characterization of a full-length pri-miRNA
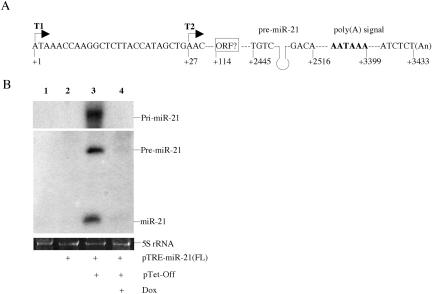
To unambiguously confirm that the mature miR-21 miRNA is indeed derived from a polyadenylated pri-miRNA precursor, we cloned the full-length pri-miR-21 transcript by a combination of PCR of human genomic DNA and RACE and mapped the 5′ and 3′ ends of the pri-miR-21 transcript. Although our data (not shown) indicated a heterogenous transcription start site, the location of the most 5′ major transcription start site (T1 in Fig. 3A) predicts a 3433-nt pri-miRNA precursor in which the predicted miR-21 RNA stem–loop occupies residues +2445 to +2516. Residues +3394 to +3399 form a consensus “AAUAAA” polyadenylation signal, while the polyadenylation site was mapped to residue +3433 (Fig. 3A). No evidence of splicing of this pri-miRNA was obtained. Analysis of open reading frames (ORFs) within this 3433-nt sequence failed to identify any ORF longer than 124 amino acids, although this ORF was located proximal to the transcription start site, beginning at residue +114, and shows significant homology to a proposed 180-amino-acid human protein of unknown function (accession number BAC05246). We therefore do not currently know whether the pri-miR-21 transcript also can function as an mRNA.
FIGURE 3.
Characterization of the full-length pri-miR-21 RNA transcript. (A) Schematic representation of the miR-21 gene. The 3′-end of the pri-miR-21 transcript was determined by RACE, while two approximate transcription start sites, indicated by T1 and T2, were mapped by RACE and by primer extension. The pri-miR-21 RNA poly(A) signal, the cleavage site used for polyadenylation and a possible 124-amino-acid ORF are also indicated. (B) The full-length pri-miR-21 RNA was placed under the transcriptional control of an inducible TRE-CMV promoter and transfected into 293T cells in the presence or absence of the pTet-Off activator plasmid. Dox was used to inhibit TRE-CMV-driven transcription. Expression of miR-21 was detected by Northern blot. 5S rRNA was utilized as a loading control.
To demonstrate that the ~3433-nt pri-miRNA can indeed function as the precursor for mature miR-21, we expressed the full-length 3433-nt pri-miR-21 RNA, linked to its own 3′ genomic polyadenylation site, under the control of a tetracycline regulatory element/cytomegalovirus immediate early promoter fusion (TRE-CMV). As shown in Figure 3B, the predicted full-length ~3.5-kb pri-miR-21, 60-nt pre-miR-21, and mature 22-nt miR-21 RNA (Zeng and Cullen 2003) were readily detected in transfected human 293T cells, but only in the presence of the coexpressed “Tet-Off” activator protein and in the absence of doxycycline (Dox), which blocks the ability of the Tet-Off activator to bind the TRE. Because expression of these miR-21 RNAs was dependent on Tet-Off function, these data indicate that the pre-miR-21 and mature miR-21 RNAs must derive from transcripts that initiate in the TRE-CMV promoter and are not processed from short-lived transcripts derived from a cryptic promoter located within the full-length pri-miR-21 RNA. These data therefore not only define the full-length pri-miR-21 precursor but also show that it can indeed serve as a precursor for the embedded mature miRNA.
As noted above, it has previously been reported that the predicted promoter elements for the C. elegans miRNAs lsy-6 and let-7 can transcribe an mRNA that is translated in vivo (Johnson et al. 2003; Johnston and Hobert 2003). The ability of these promoter elements to transcribe a functional mRNA strongly suggests that they are able to recruit pol II. To address this question for the human miR-21 gene, we PCR cloned a 1008-bp human genomic DNA fragment extending from −959 to +49 relative to the T1 pri-miR-21 transcription start site (Fig. 3A). Analysis of the sequence of this candidate promoter element identified a candidate “CCAAT” box transcription control element located ~200 nt 5′ to the T1 transcription start site but did not identify a “TATA” box at the predicted 18–26-bp distance 5′ to T1. Curiously, however, the T1 transcription start site was itself located within a sequence that appears similar to a “TATA” box; i.e. TTA/ATAAA.
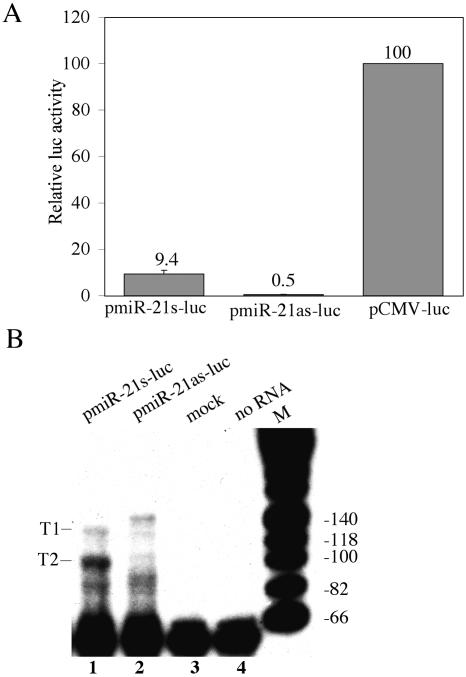
To test whether these 5′ flanking sequences can indeed direct the transcription of an mRNA, we inserted this ~1-kb DNA fragment in either the sense or antisense orientation 5′ to a firefly luciferase indicator gene. As shown in Figure 4A, the sense orientation gave rise to readily detectable levels of firefly luciferase activity in transfected 293T cells, while the antisense orientation was inactive. We therefore conclude that the sequences located 5′ to the pri-miR-21 transcription unit can function as an mRNA promoter.
FIGURE 4.
Characterization of the pri-miR-21 promoter. (A) miR-21 promoter-driven luc expression. Plasmids pmiR-21s-luc, pmiR-21as-luc, and pCMV-luc were cotransfected into 293T cells along with a Renilla luciferase internal control plasmid. Induced luciferase activities were determined at 48 h after transfection and normalized to the Renilla luciferase internal control (average of three independent experiments). Data are presented relative to the firefly luciferase activity detected in cultures transfected with pCMV-luc, which was set at 100. (B) Primer extension analysis using a luciferase gene-specific primer and RNA recovered from 293T cells transfected with pmiR-21s-luc or pmiR-21as-luc or was mock transfected. Major (T2) and minor (T1) extension products observed in the pmiR-21s-luc transfected cells are indicated and are mapped to the underlying miR-21 promoter sequence in Figure 3A.
Although RACE represents a very sensitive technology to map the 5′ end of an RNA, it can be somewhat imprecise. Efforts to fine map the 5′ ends of endogenous pri-miR-21 RNAs using primer extension were unfortunately unsuccessful, most likely because of the very low expression of pri-miR-21 in HeLa cells. To more accurately determine the 5′ ends of the pri-miR-21 precursor RNA, we therefore performed primer extension using a primer specific for the firefly luciferase gene and RNA derived from 293T cells transfected with pmiR-21s-luc or pmiR-21as-luc (Fig. 4B). These data identified two transcription start sites in the pmiR-21s-luc transfected cells that were absent in control cells. Alignment of these start sites with the pre-miR-21 gene sequence indicated a minor transcription start site (T1) approximately coincident with the +1 start site identified by RACE and a major transcription start site (T2) at approximately +27 relative to the +1 start site identified by RACE. Of note, the T2 transcription start site is located 22 nt 3′ to the possible “TATA” box described above that underlies the T1 start site. Importantly, as the T1 start site identified by primer extension and the start site identified by RACE appear to be similar, these data suggest that the pmiR-21s-luc expression plasmid is probably initiating transcription of luciferase mRNAs at the same sites utilized by the endogenous pri-miR-21 RNA precursor.
Relatively little full-length pri-miRNA reaches the cytoplasm
A small number of human miRNA stem–loops are predicted to be located in the 3′ UTR of a protein coding gene. Examples include the human follistatin-related protein (FRP) gene and the BHRF1 gene found in Epstein Barr virus (Tanaka et al. 1998; Pfeffer et al. 2004). Excision of the pre-miRNA stem–loop from the longer pri-miRNA precursor by Drosha is known to occur in the nucleus (Lee et al. 2003). If this processing event is efficient, little or none of the overlapping mRNA would be able to reach the cytoplasm and be expressed. We therefore asked whether pri-miRNAs are confined to the nucleus or whether some proportion of these capped, polyadenylated transcripts can reach the cytoplasm.
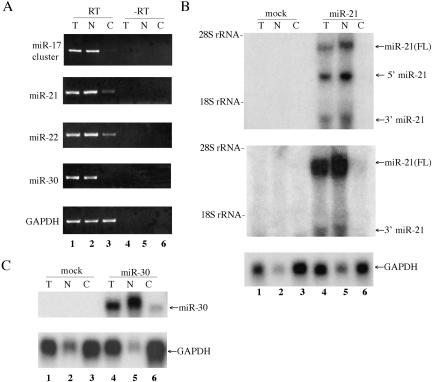
To address this question, we prepared total, nuclear, and cytoplasmic RNA fractions from HeLa cells and then analyzed the subcellular localization of endogenous pri-miRNAs by generating cDNA by oligo-dT primed reverse transcription followed by PCR using specific primers. As shown in Figure 5A, GAPDH mRNA, which served as a positive control, was readily detectable in both the nucleus and cytoplasm. In contrast, all the pri-miRNAs were localized predominantly to the nucleus. More specifically, little or none of the pri-miRNA derived from the miR-17 cluster or the miR-30 gene was detected in the cytoplasm, while modest levels of the miR-21 and miR-22 pri-miRNA were observed.
FIGURE 5.
Primary miRNA transcripts are largely confined to the nucleus. (A) Distribution of endogenous pri-miRNAs. HeLa cell RNA was isolated from the total (T) cell, or from the nucleus (N) and cytoplasm (C), and was then subjected to reverse transcription using oligo(dT) primers. PCR was performed using the primers described in Figure 1. RNA that had not been subjected to reverse transcription served as a negative control. (B) Distribution of overex-pressed pri-miR21 RNA. 293T cells were mock transfected or were transfected with pTRE-miR-21(FL) in the presence of the pTet-Off activator plasmid. RNA was isolated at ~36 h after transfection and subjected to Northern analysis using nick-translated miR-21 specific probes, derived from nucleotides +2234 to +2648 (upper panel) or +2904 to +3283 (middle panel) of the predicted pri-miR-21 RNA, or using a probe specific for the endogenous GAPDH mRNA as a loading control (lower panel). (C) Similar to B, except that 293T cells were transfected with pTRE-miR-30, which encodes an artificial pri-miR-30 RNA. The probe used was derived from the 414-bp miR-30 DNA fragment present in pTRE-miR-30, which extends both 5′ and 3′ to the predicted pre-miRNA processing sites.
To further address the question of whether pri-miRNAs can reach the cytoplasm, we used Northern blot analysis to determine the subcellular localization of ectopically expressed pri-miR-21 RNA in 293T cells that had been transfected with pTRE-miR-21(FL), which expresses a full-length miR-21 pri-miRNA transcript. As shown in Figure 5B (upper panels), we again observed only minimal levels of the pri-miR-21 RNA in the cytoplasm. However, high levels of nuclear pri-miR-21 RNA (Fig. 5B, upper panels) and of a control, cytoplasmic GAPDH mRNA (Fig. 5B, lower panel) were detected.
The Northern analysis of the RNAs expressed from pTRE-miR-21(FL) shown in Figure 5B (upper panel), which used a probe specific to residues +2234 to +2648 of the predicted pri-miR-21 RNA (Fig. 3A), revealed the expression of not only the expected ~3.5-kb full-length pri-miRNA but also two additional RNAs of ~2.4 kb and ~1.0 kb that were largely confined to the nucleus. We hypothesized that these represent, respectively, the 5′ and 3′ cleavage products that remain after excision of the 60-nt pre-miR-21 RNA from the initial pri-miRNA transcript by Drosha (Fig. 3B). To confirm this hypothesis, we repeated the Northern analysis shown in Figure 5B (upper panel), using a probe specific for nucleotides +2904 to +3283. This probe annealed to the full-length ~3.5-kb pri-miR-21 RNA and to the predicted ~1.0-kb 3′ flanking RNA fragment but did not recognize the ~2.4-kb RNA fragment (Fig. 3B, middle panel). These data therefore strongly suggest that the ~2.4-kb and ~1.0-kb RNA fragments seen in the nuclear fraction in Figure 5B (upper panel) indeed result from Drosha processing of the pri-miR-21 RNA precursor.
To extend this analysis, we also analyzed 293T cells transfected with a second plasmid, pTRE-miR-30, that expresses an artificial pri-miR-30 RNA precursor. This plasmid transcribes a 414-bp genomic human DNA fragment, centered on the ~80-nt miR-30 RNA stem–loop structure, that is processed to yield high levels of mature miR-30 (Zeng and Cullen 2003). As shown in Figure 5C, the encoded artificial pri-miR-30 precursor was detected at high levels in the nucleus of transfected 293T cells but at only very low levels in the cytoplasmic fraction. We therefore conclude that the presence of a miRNA precursor stem–loop in cis can induce efficient nuclear miRNA processing and, presumably as a result, inhibit the nuclear export of the full-length pri-miRNA.
A single transcript can function as both a pri-miRNA and an mRNA
We next asked whether the presence of a miRNA stem–loop in cis would, in fact, inhibit the expression of a linked protein coding gene. For this purpose, we obtained an expression plasmid (pTRE-luc) consisting of the firefly luciferase indicator gene under the control of the regulatable TRE-CMV promoter, and we then inserted the 414-bp genomic human DNA fragment centered on the predicted miR-30 stem–loop, used in Figure 5C, into the 3′ UTR of pTRE-luc in either the sense (pTRE-luc-miR-30) or antisense (pTRE-luc-03-Rim) orientation.
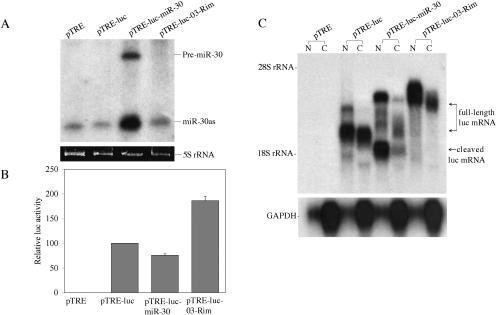
To confirm that pTRE-luc-miR-30 alone was indeed able to give rise to a mature miRNA, we transfected 293T cells with each of these plasmids and detected miR-30 transcripts by Northern analysis. As shown in Figure 6A, and previously reported (Zeng et al. 2002), low levels of the mature miR-30as miRNA were constitutively detected in 293T cells. However, far higher levels of pre-miR-30 and of mature miR-30as were detected in the pTRE-luc-miR-30, but not the pTRE-luc-03-Rim, transfected cells. Note that miR-30 gives rise to two mature microRNAs, termed miR-30 and miR-30as, derived from both arms of the pre-miR-30 precursor (Zeng et al. 2002).
FIGURE 6.
A pri-miRNA can function as an mRNA. (A) 293T cells were cotransfected with the indicated pTRE-based expression plasmid and with the pTet-Off activator plasmid. Pre-miR-30 and miR-30as expression was detected at 60 h after transfection by Northern blot. 5S rRNA was utilized as a loading control. (B) In parallel, 293T cells were transfected with the indicated pTRE-based expression plasmid together with pTet-Off and a Renilla luciferase expression plasmid. At 48 h after transfection, firefly and Renilla luciferase activities were measured. Data were normalized to the Renilla luciferase internal control and are presented relative to the activity detected in the pTRE-luc transfected culture, which was set at 100. Average of three experiments with standard deviation indicated. (C) Northern analysis using nuclear (N) or cytoplasmic (C) RNA fractions obtained from 293T cells transfected with the indicated pTRE-based expression plasmids. The probe used in the upper panel is specific to the firefly luciferase gene. The predicted full-length and 3′ truncated luc mRNAs expressed in each culture are indicated. The blot was stripped and reprobed with a probe specific for the endogenous GAPDH mRNA (lower panel).
As the pri-miR-30 RNA expressed from pTRE-luc-miR-30 is apparently efficiently processed to a mature miRNA, we expected that the level of luciferase activity observed in pTRE-luc-miR-30 transfected cells would be substantially lower than that detected in cells transfected with pTRE-luc or pTRE-luc-03-Rim. In fact, however, the level of luciferase activity in the pTRE-luc-miR-30 transfected cells was only reduced by twofold or less (Fig. 6B).
To further address why pTRE-luc-miR-30 can give rise to an mRNA transcript that appears to be both processed into a pre-miRNA in the nucleus and translated in the cytoplasm, we performed a Northern analysis using a luc specific probe and nuclear and cytoplasmic RNA fractions derived from 293T cells transfected with the pTRE-luc derivatives. As shown in Figure 6C (lanes 3,4), pTRE-luc gave rise to a single major mRNA species that was detected at comparable levels in the nucleus and cytoplasm. Similarly, pTRE-luc-03-Rim gave rise to a single major mRNA species that was, because of the antisense insertion of the 414-bp miR-30 sequence, significantly larger in size (Fig. 6C, lanes 7,8). Finally, pTRE-luc-miR-30 gave rise to two major RNA species in the nuclear fraction, the less intense of which was identical in size to the pTRE-luc-03-Rim transcript, and therefore likely represents the initial transcript. The second major RNA species, in contrast, migrated at the size predicted for a pTRE-luc-miR-30 transcript that had been cleaved in the introduced miR-30 stem-loop (Fig. 6C, lanes 5,6). Consistent with the data presented in Figure 5, very little of the full-length pTRE-luc-miR-30 “pri-miRNA” transcript reached the cytoplasm of the transfected cells, especially when contrasted with pTRE-luc-03-Rim transfected cells (cf. Fig. 6C, lanes 5,6 and lanes 7,8). Indeed, quantification of the relative level of this RNA by phosphor-imager showed an ~93% drop in the level from lane 8 to lane 6. However, we did detect a significant level of a shorter luc RNA that we believe represents a Drosha cleaved derivative of this full-length mRNA. As this cleaved mRNA would retain the complete luciferase open reading frame, it appears possible that it contributes to the observed level of luciferase expression despite the lack of a poly(A) tail. Quantification of this truncated RNA (Fig. 6C, lane 6) indicates a cytoplasmic expression level equivalent to ~24% of the level of the full-length luciferase mRNA encoded by pTRE-luc-03-Rim (lane 8). The combined level of the full-length and truncated cytoplasmic pTRE-luc-miR-30 mRNA would therefore be equivalent to ~31% of the observed level of the cytoplasmic full-length pTRE-luc-03-Rim mRNA, thus potentially explaining why the level of luc enzyme activity is only modestly reduced in 293T cells transfected with the former (Fig. 6B).
DISCUSSION
In this manuscript, we have sought to address two questions: First, are human pri-miRNAs transcribed by RNA polymerase II? Second, if pri-miRNAs are indeed structurally similar to mRNAs, can they in fact simultaneously function as both pri-miRNAs and mRNAs?
The evidence presented strongly suggests that pol II is indeed the major, and possibly the only, mediator of human miRNA transcription. Analysis of nine human miRNAs, three isolated and six in a miRNA cluster, indicated that their pri-miRNA precursors are both capped and polyadenylated (Figs. 1, 2). Cloning of the full-length pri-miRNA for human miR-21 identified a transcript of ~3433 nt that was shown to indeed function as the precursor for mature miR-21 expression (Fig. 3). Moreover, this intronless transcript was flanked 5′ by a promoter element that proved able to transcribe a functional mRNA in transfected cells (Fig. 4) and was flanked 3′ by a consensus mRNA polyadenylation signal (Fig. 3A). The structure of the pri-miR-21 RNA reported here is somewhat analogous to the previously reported structure of the pri-miR-172 or “EAT” RNA expressed by Arabidopsis (Aukerman and Sakai 2003), the pri-miR-155 or “BIC” RNA expressed in humans (Tam 2001), and the pri-let-7 RNA expressed in C. elegans (Bracht et al. 2004), with the exception that these other RNAs are all spliced (trans-spliced in the case of pri-let-7), while pri-miR-21 is not. All four pri-miRNAs are, however, polyadenylated, and all four seem to be noncoding, although this remains uncertain in the case of pri-miR-21. Nevertheless, these data in total suggest that pri-miRNAs and mRNAs are structurally equivalent.
Analysis of the genomic context of the pri-miR-21 transcription unit showed, perhaps surprisingly, that the pri-miR-21 gene overlaps with the 3′ end of the gene encoding human Vacuole Membrane Protein 1 (VMP-1), which consists of 11 coding exons in the same genomic orientation (Dusetti et al. 2002). Exon 10 of VMP-1 coincides with residues −527 to −425, relative to the pri-miR-21 T1 transcription start-site shown in Figure 3A, while the final exon 11 begins at residue +947 and is polyadenylated at position +1524, +1768, or, predominantly, +2271. This last polyadenylation site is only 174 nt 5′ to the beginning of the miR-21 stem-loop (Fig. 3A). No VMP-1 mRNA extending beyond +2271 and/or polyadenylated at the poly(A) site present at +3433, which is used by the pri-miR-21 RNA, was observed in the EST database.
The presence of three poly(A) sites used by the VMP-1 mRNA within the body of the pri-miR-21 precursor RNA raises the question of how these poly(A) sites are avoided during pri-miR-21 transcription. Evidence that they are not effectively utilized includes the high level of the ~3.4-kb pri-miR-21 RNA observed in transfected cells (Figs. 3B, 5B), which is clearly inconsistent with efficient utilization of these sites. ESTs derived from the predicted full-length pri-miR-21 transcript are also readily found in the EST database, including one EST (BC053563) that extends from +45 to +3433 and is therefore almost full length. Inspection of the sequence of the three poly(A) sites used by VMP-1 mRNA shows that the minor sites at +1524 and +1768 are nonconsensus, in that they both contain a “TATAAA,” instead of an “AATAAA,” polyadenylation signal. Moreover, all three sites appear to differ from efficiently utilized poly(A) sites in being flanked 3′ by sequences that are relatively poor in G/U residues (Proudfoot et al. 2002). Finally, we note that the pri-miR-21 RNA transcript is unspliced, while the VMP-1 mRNA contains 10 introns. Splicing is known to enhance the efficiency of polyadenylation (Proudfoot et al. 2002) and might therefore permit the use of “weak” poly(A) sites in the VMP-1 mRNA that are not effectively recognized when present in the intronless pri-miR-21 transcript. A possible alternative hypothesis, i.e., that transcription of the pri-miR-21 precursor actually initiates 3′ to the VMP-1 poly(A) site located at +2271, would appear to be excluded by the data shown in Figure 3B and, as noted above, is not supported by the EST database.
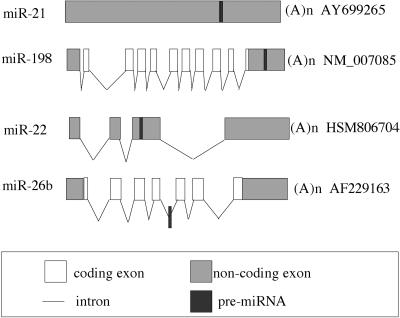
Genomic analyses of vertebrate miRNAs have mapped the majority to genomic regions that do not coincide with known transcripts (Lagos-Quintana et al. 2003; Bartel 2004). For those that do coincide with known genes, the majority are found within introns, with ~75% in the sense orientation and ~25% in the antisense orientation (Fig. 7). This implies, at least in the latter cases, that the pri-miRNA is not, in fact, excised from an intron lariat and instead must derive from an unknown, overlapping RNA. Nevertheless, the fact that the majority of intronic miRNA genes are found in the same orientation as the overlapping mRNA does suggest that some proteins and miRNAs may be coordinately expressed.
FIGURE 7.
Location of microRNA stem–loops in RNA transcripts. The figure shows the relative position of miRNA stem–loops in known human RNA transcripts. The miR-21 miRNA analyzed in this manuscript is transcribed as part of an unspliced, apparently noncoding RNA, while miR-22 is found in an exonic location in a spliced non-coding RNA. miRNAs located in protein coding genes are mostly found in introns, as shown here for miR-26b, although a location in the 3′ UTR of an mRNA, as shown here for miR-198 and the human gene FRP, is occasionally observed. The figure is not drawn to scale. The exon/intron structure of candidate pri-miRNAs was generated by aligning the sequence of putative pri-miRNAs with the corresponding genomic DNA sequence. The pri-miR-21 RNA shown was characterized in this manuscript. GenBank accession numbers of all the candidate pri-miRNA transcripts are shown.
In addition to intronic locations, a small number of miRNAs are found in exonic locations, including in the UTRs of mRNAs. These include miR-198, found in the 3′ UTR of the human FRP gene (Fig. 7; Tanaka et al. 1998), and miR-BHRF1-1 and miR-BHRF1-2, both found in the UTRs of the Epstein-Barr Virus BHRF1 gene (Pfeffer et al. 2004). The pri-miR-21 RNA may also fall into this category, as this RNA contains a 124-amino-acid ORF with homology to a proposed 180-amino-acid human protein of unknown function (Fig. 3A). However, the existence of neither protein has, as yet, been validated. Nevertheless, the existence of transcripts showing the characteristics of both an mRNA and a pri-miRNA prompted us to ask whether pri-miRNAs could reach the cytoplasm and function as mRNAs.
Analysis of both endogenous and overexpressed pri-miRNAs showed that very little full-length pri-miRNA reached the cytoplasm, probably because most pri-miRNA transcripts were processed by Drosha before they could be exported from the nucleus (Fig. 5). We were therefore surprised to find that insertion of a pri-miRNA expression cassette into the 3′ UTR of an mRNA encoding the luciferase indicator gene only moderately inhibited luciferase expression (Fig. 6B). This was not because processing of the luciferase-pri-miRNA fusion transcript was inefficient, as this precursor RNA gave rise to robust expression of the encoded miR-30as miRNA (Fig. 6A) and appeared to be efficiently cleaved, in the nucleus, at the site of the inserted miR-30 RNA hairpin (Fig. 6C). Surprisingly, however, the resultant 3′ truncated luc mRNA was able to reach the cytoplasm reasonably effectively, and we therefore hypothesize that the observed luc protein expression may, at least in part, result from translation of this 3′ truncated mRNA. If true, then this would suggest that a miRNA stem–loop precursor located in the 3′ UTR of an mRNA does not necessarily block expression of the encoded protein, even when miRNA processing is efficient. Of course, if processing of a miRNA stem–loop located in cis was inefficient, then expression of the linked ORF would likely be inhibited to an even lesser degree. In conclusion, our data suggest that pri-miRNAs are not only structurally similar to mRNAs but also have the potential to coordinately express both an encoded protein and a linked miRNA.
MATERIALS AND METHODS
Molecular clones
Plasmids pTRE2hyg, pTRE2hyg-Luc, pTet-Off and pEGFP-N3 were purchased from BD/Clontech. We have previously described the expression plasmid pCMV-Luc (Zeng and Cullen 2003). The full-length (FL) miR-21 gene, starting with the transcription initiation site and extending through the polyadenylation site to include 42 bp of flanking 3′ genomic sequences, was obtained by PCR from genomic human DNA after mapping the 5′ and 3′ end of the transcript using RACE. The pTRE-miR-21(FL) plasmid was generated by inserting this ~3.5-kb DNA fragment into 5′ Bam HI and 3′ SalI sites present in pTRE2hyg, after removal of the genomic β-globin poly(A) addition site. pTRE-miR-30 was generated by cloning a PCR-generated 414-bp genomic DNA fragment, centered on the miR-30 RNA stem–loop, into pTRE2hyg. Plasmids containing the miR-21 promoter element (−959 to +49 relative to the “T1” transcription start site) were generated by substitution of the CMV promoter in pEGFP-N3 with the miR-21 promoter, in either the sense or antisense orientation, using AseI and Bgl II sites. The gfp gene was then replaced with the firefly luciferase indicator gene, using EcoRI and Not I sites. pTRE-luc-miR-30 and pTRE-luc-03-Rim were derived from pTRE2hyg-Luc by insertion of a 414-bp genomic human DNA fragment, containing the entire miR-30 stem–loop, into a 3′ UTR SalI site in the sense or antisense orientation, respectively.
RNA preparation and RT PCR
Total RNA was isolated from HeLa cells using Trizol reagent (Invitrogen). Purification of capped RNA was performed as described by Choi and Hagedorn (2003). Briefly, 75 μg of heat-denatured total HeLa cell RNA was mixed with eIF4E beads for 1 h at room temperature. After extensive washing, the RNA-bound beads were recovered by centrifugation, extracted with acid phenol/chloroform, and the recovered RNA precipitated with ethanol.
Nuclear and cytoplasmic RNA fractions were prepared as previously described (Yi et al. 2003). For RT-PCR analysis, the reverse transcription reaction was performed using 4 μg of total HeLa cell RNA using SuperScript II Reverse Transcriptase and oligo(dT) primers (Invitrogen) or random primers (Stratagene), following the manufacturer’s protocols. All RNA samples were treated with RQ1 RNase-free DNase (Promega) prior to the reverse transcription step. For the polyadenylation analysis, high-purity oligo(dT)-primed HeLa cell QUICK-Clone cDNA (BD/Clontech) was used as the template. Two microliters of cDNA was used in each PCR reaction. Gene-specific primers were designed based on the genomic sequences known to flank each pre-miRNA stem–loop and were designed to give an ~410-bp PCR product with the exception of the miR-17 cluster, where the predicted PCR product was 1000 bp in length. Unique primers specific for the human histone 2A and gapdh genes were designed to give 400-bp and 528-bp PCR products, respectively. Thirty-five to 37 cycles of PCR were run at the following temperatures: 94°C, 60 s; 55°C, 60 s; 72°C, 60–90 s. To maintain linear amplification, only 25 cycles of PCR were run for GAPDH.
To determine the 5′ and 3′ end of the full-length pri-miR-21 transcript, 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR was performed using a SMART RACE cDNA Amplification kit (BD/Clontech). RACE experiments were carried out according to instructions included in the kit. PCR products were cloned into the TA cloning vector pCR2.1 (Invitrogen) and sequenced. Primer extension assays were performed as previously described (Zeng et al. 2002), except that 50 μg of total RNA, derived from transfected 293T cells, was used. The firefly luciferase gene-specific primer used for primer extension has the sequence 5′-GGGCCTTTCTTT ATGTTTTTGGCGTCTTCC-3′.
Transfections and luciferase assays
All transfections were performed using Fugene 6 (Roche Diagnostics) on human 293T cells cultured in 24-well tissue culture plates, according to the manufacturer’s instructions. For luciferase assays using pTRE-based plasmids, 10 ng of the indicator plasmid and an equal amount of the activator plasmid pTet-Off, where necessary, was cotransfected with 1 ng of a Renilla luciferase-based internal control plasmid. The total level of transfected DNA was maintained at 400 ng per transfection by supplementation with the pTRE2hyg parental vector. For miR-21 promoter-driven plasmids, 400 ng of the indicator plasmid and 1 ng of the Renilla luciferase control plasmid were cotransfected. Cells were lysed and assayed for both firefly and Renilla luciferase at 48 h after transfection.
Northern analyses
Transfections and preparation of nuclear/cytoplasmic RNA were performed as described above, except that 200 ng of the relevant indicator plasmids were cotransfected into 293T cells along with an equal amount of pTet-Off. Doxycycline (final concentration, 1 μg/ml, BD/Clontech) was used to suppress activation of the TRE-CMV promoter. Pri-miRNA expression was analyzed at 36 h after transfection, using formaldehyde-agarose gels, while pre-miRNA and mature miRNA expression was examined at 60 h after transfection, using 15% TBE-urea polyacrylamide gels. Blot transfer and signal detection were performed as previously described (Zeng and Cullen 2003).
Acknowledgments
This research was funded by the Howard Hughes Medical Institute and by NIH grants R01 GM71408 to B.R.C. and R01 CA063640 to C.H.H.
REFERENCES
- Aukerman, M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its _APETALA2_-like target genes. The Plant Cell 15**:** 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116**:** 281–297. [DOI] [PubMed] [Google Scholar]
- Bohnsack, M.T., Czaplinski, K., and Görlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export pre-miRNAs. RNA 10**:** 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht, J., Hunter, S., Eachus, R., Weeks, P., and Pasquinelli, A.E. 2004. _Trans_-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 10**:** 1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.-Z., Li, L., Lodish, H.F., and Bartel, D.P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303**:** 83–86. [DOI] [PubMed] [Google Scholar]
- Choi, Y.H. and Hagedorn, C.H. 2003. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl. Acad. Sci. 100**:** 7033–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Y., Yang, X.C., Kaygun, H., Dadlez, M., and Marzluff, W.F. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell 12**:** 295–305. [DOI] [PubMed] [Google Scholar]
- Dusetti, N.J., Jiang, Y., Vaccaro, M.I., Tomasini, R., Samir, A.A., Calvo, E.L., Ropolo, A., Fiedler, F., Mallo, G.V., Dagorn, J.-C., et al. 2002. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem. Biophys. Res. Comm. 290**:** 641–649. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106**:** 23–34. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404**:** 293–295. [DOI] [PubMed] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell specific microRNAs. Dev. Cell 5**:** 351–358. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., McLachlan, J., Pasquinelli, A.E., Bálint, É., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293**:** 834–838. [DOI] [PubMed] [Google Scholar]
- Johnson, S.M., Lin, S.-Y., and Slack, F.J. 2003. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 259**:** 364–379. [DOI] [PubMed] [Google Scholar]
- Johnston Jr., R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426**:** 845–849. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294**:** 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12**:** 735–739. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., and Tuschl, T. 2003. New microRNAs from mouse and human. RNA 9**:** 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75**:** 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.-T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21**:** 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Rådmark, O., Kim, S., et al. 2003. The nuclear RNase III drosha initiates microRNA processing. Nature 425**:** 415–419. [DOI] [PubMed] [Google Scholar]
- Lund, E., Güttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303**:** 95–98. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110**:** 563–574. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., Zavolan, M., Grasser, F.A., Chien, M., Russo, J.J., Ju, J., John, B., Enright, A.J., Marks, D., Sander, C., et al. 2004. Identification of virus-encoded microRNAs. Science 304**:** 734–736. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108**:** 501–512. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403**:** 901–906. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvágner, G., Haley, B., and Zamore, P.D. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10**:** 537–548. [DOI] [PubMed] [Google Scholar]
- Tam, W. 2001. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene 274**:** 157–167. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Ozaki, S., Osakada, F., Mori, K., Okubo, M., and Nakao, K. 1998. Cloning of follistatin-related protein as a novel autoantigen in systemic rheumatic diseases. International Immunol. 10**:** 1305–1314. [DOI] [PubMed] [Google Scholar]
- Weinstein, L.B. and Steitz, J.A. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11**:** 378–384. [DOI] [PubMed] [Google Scholar]
- Yi, R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17**:** 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y. and Cullen, B.R. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9**:** 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9**:** 1327–1333. [DOI] [PubMed] [Google Scholar]