A Network of Local and Redundant Gene Regulation Governs Arabidopsis Seed Maturation (original) (raw)
Abstract
In Arabidopsis thaliana, four major regulators (ABSCISIC ACID INSENSITIVE3 [ABI3], FUSCA3 [FUS3], LEAFY COTYLEDON1 [LEC1], and LEC2) control most aspects of seed maturation, such as accumulation of storage compounds, cotyledon identity, acquisition of desiccation tolerance, and dormancy. The molecular basis for complex genetic interactions among these regulators is poorly understood. By analyzing ABI3 and FUS3 expression in various single, double, and triple maturation mutants, we have identified multiple regulatory links among all four genes. We found that one of the major roles of LEC2 was to upregulate FUS3 and ABI3. The lec2 mutation is responsible for a dramatic decrease in ABI3 and FUS3 expression, and most lec2 phenotypes can be rescued by ABI3 or FUS3 constitutive expression. In addition, ABI3 and FUS3 positively regulate themselves and each other, thereby forming feedback loops essential for their sustained and uniform expression in the embryo. Finally, LEC1 also positively regulates ABI3 and FUS3 in the cotyledons. Most of the genetic controls discovered were found to be local and redundant, explaining why they had previously been overlooked. This works establishes a genetic framework for seed maturation, organizing the key regulators of this process into a hierarchical network. In addition, it offers a molecular explanation for the puzzling variable features of lec2 mutant embryos.
INTRODUCTION
The conquest of most terrestrial niches by land plants has been greatly facilitated by the appearance of seeds (Steeves, 1983). Seeds offer plants a unique opportunity to interrupt their life cycle by withstanding adverse environmental conditions in a desiccated state and then resuming growth using endogenous storage compounds. Seed-specific traits, such as desiccation tolerance, reserve accumulation, and entry into quiescence, are acquired during a developmental phase called seed maturation (Goldberg et al., 1994; Wobus and Weber, 1999; Vicente-Carbajosa and Carbonero, 2005). In Arabidopsis thaliana, this phase is genetically controlled by at least four genes, FUSCA3 (FUS3), ABSCISIC ACID INSENSITIVE3 (ABI3), LEAFY COTYLEDON1 (LEC1), and LEC2. ABI3, FUS3, and LEC2 encode related plant-specific transcription factors containing the conserved B3 DNA binding domain (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001), whereas LEC1 encodes a CBF transcription factor (Lotan et al., 1998). All four abi3, lec1, lec2, and fus3 mutants are severely affected in seed maturation and share some common phenotypes, such as reduced expression of seed storage proteins (SSPs) (Table 1). However, they also show some specific phenotypes, such as the absence of chlorophyll degradation in the dry seed (abi3), a reduced sensitivity to abscisic acid (ABA) (abi3 and, to a lesser extent, lec1), the accumulation of anthocyanins (fus3, lec1, and, to a lesser extent, lec2), an intolerance to desiccation (abi3, fus3, and lec1), or defects in cotyledon identity (lec1, fus3, and lec2) (Meinke, 1992; Bäumlein et al., 1994; Keith et al., 1994; Meinke et al., 1994; Parcy et al., 1994, 1997; West et al., 1994; Nambara et al., 1995; Lotan et al., 1998; Luerssen et al., 1998; Vicient et al., 2000a; Raz et al., 2001; Stone et al., 2001; Kroj et al., 2003). Despite numerous studies, the mechanisms through which these genes interact to control the various facets of seed maturation remain poorly understood (Bäumlein et al., 1994; Keith et al., 1994; Meinke et al., 1994; West et al., 1994; Parcy et al., 1997; Nambara et al., 2000; Vicient et al., 2000a; Raz et al., 2001). Since lec2, fus3, and abi3 single mutants have similar but distinct phenotypic traits, which are additive in double mutants, ABI3, LEC2, and FUS3 may work in parallel pathways (Keith et al., 1994; Meinke et al., 1994; West et al., 1994). Other genetic analyses have also suggested the existence of interactions between ABI3 and FUS3 but without elucidating their molecular nature (Parcy et al., 1997; Nambara et al., 2000; Vicient et al., 2000a). Recently, LEC1 was shown to regulate expression of ABI3 and FUS3 (Kagaya et al., 2005). Also, FUS3 and LEC2 were shown to act in a partially redundant manner to control SSP gene expression, and LEC2 was shown to locally regulate FUS3 expression in regions of the cotyledons (Kroj et al., 2003). These finding suggests that local and redundant regulation within this group of genes, which had been previously overlooked, might be a central requisite for the correct establishment of seed maturation and prompted us to systematically analyze their expression in various mutant combinations.
Table 1.
Main Phenotypes of Maturation Mutant Seeds
| Wild Type | abi3 | fus3 | lec2 | lec1 | |
|---|---|---|---|---|---|
| Chlorophyll accumulation in dry seed | No | Yes (f and g) | No (c) | Yes, in sectors (c and j) | Yes (g) |
| Anthocyanin in cotyledons | No | No (g) | Yes (b and g) | Yes, in sectors (i, c, and j) | Yes (a, g, and c) |
| Storage protein expression | Normal | Reduced (d, f, g, and j) | Reduced (b, c, g, h, and j) | Reduced (c and j) | Reduced (a, c, g, e, and h) |
| Ectopic trichomes on cotyledons | No | No (f) | Yes (b and c) | Yes (i and c) | Yes (a, e, j, and c) |
| Seed ABA sensitivity | Normal | Reduced (g) | Normal (g) | Normal (c) | Reduced in cotyledons (g and c) |
| Desiccation-tolerant seeds | Yes | No (d) | No (b) | Yes (c) | No (e) |
RESULTS
FUS3 Expression Is Regulated by LEC2, ABI3, and FUS3
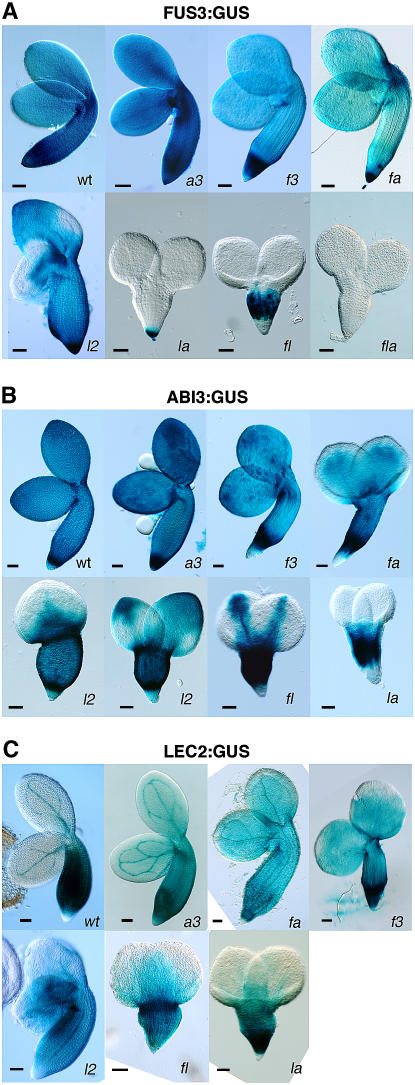
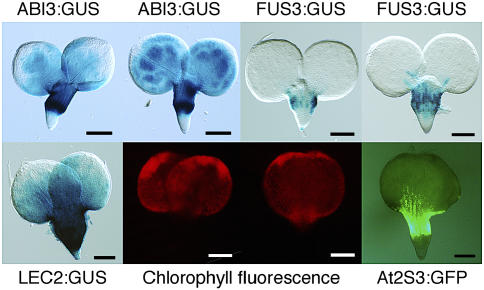
We tested the effect of the abi3, fus3, and lec2 mutations, alone and in combination, on the expression of ABI3, FUS3, and LEC2. We performed whole-mount β-glucuronidase (GUS) staining because this allowed direct visualization of the staining patterns in a large number of embryos. For the FUS3 gene, we used a FUS3:GUS reporter construct and analyzed its expression in mature embryos (10 d after pollination [DAP]) (Figure 1A). As demonstrated previously, FUS3 expression is detectable throughout the various parts of the wild-type embryo (root tip, embryo axis, and cotyledons) (Kroj et al., 2003; Tsuchiya et al., 2004). It was recently shown that this expression mainly originates from the protoderm of the embryo (Tsuchiya et al., 2004). Introduction of the FUS3:GUS reporter transgene in abi3, fus3, and abi3 fus3 mutants showed that the spatial expression pattern of FUS3 is not altered in these backgrounds (Figure 1A). As previously demonstrated, FUS3 expression is absent from sectors of lec2 mutant cotyledons (Figure 1A; Kroj et al., 2003). Strong alterations of FUS3 expression were revealed by crossing the lec2 mutant with abi3 or fus3. In the lec2 abi3 mutant, FUS3:GUS expression was restricted to the root meristem; in fus3 lec2, it was confined to the embryo axis; and in the fus3 lec2 abi3 triple mutant, FUS3:GUS expression was completely lost (Figure 1A). Comparison of fus3 lec2 abi3 with fus3 lec2 shows that ABI3 regulates FUS3 in the embryo axis. Comparison of lec2 abi3 with lec2 shows that ABI3 regulates FUS3 expression in the embryo axis and in the cotyledons. Similarly, the effect of the fus3 mutation in the lec2 mutant (by comparing lec2 with lec2 fus3) or in the lec2 abi3 background (by comparing lec2 abi3 with lec2 abi3 fus3) shows that FUS3 positively regulates its own expression in the root meristem and the cotyledons. Finally, the effect of the lec2 mutation in the fus3, abi3, or fus3 abi3 backgrounds shows that LEC2 regulates FUS3 expression throughout the embryo.
Figure 1.
ABI3:GUS, FUS3:GUS, and LEC2:GUS Activities in Wild-Type and Mutant Embryos.
Expression patterns of FUS3:GUS (A), ABI3:GUS (B), and LEC2:GUS (C) in wild-type, abi3 (a3), fus3 (f3), lec2 (l2), fus3 abi3 (fa), lec2 abi3 (la), fus3 lec2 (fl), and fus3 lec2 abi3 (fla) 10-DAP embryos and LEC2:GUS in 14-DAP embryos. The size and shape of the sectors with reduced ABI3 or FUS3 expression in lec2 mutant embryos are extremely variable: two examples of phenotypes are shown for ABI3:GUS and one for FUS3:GUS. LEC2:GUS persistent expression is also variable and not always as pronounced as shown here for lec2, fus3, or fus3 abi3. Bars = 50 μm.
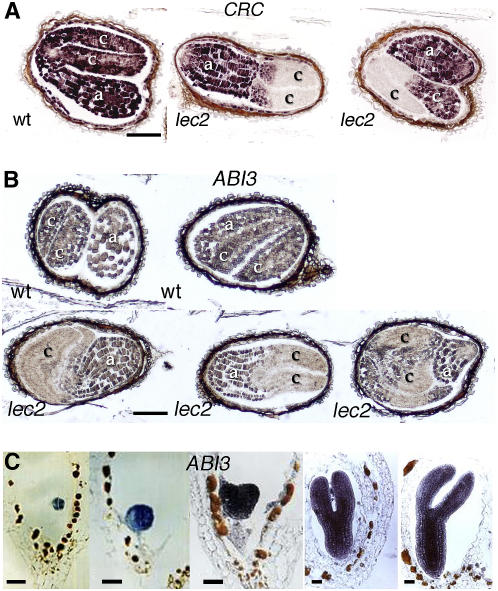
We tried to confirm some of these expression patterns using in situ hybridization. As opposed to the signal obtained with the highly expressed Cruciferin C (CRC) storage protein probe (Figure 2A), the FUS3 probe in the wild-type background gave a very weak and uniform signal (data not shown) and was not restricted to the epidermis as previously reported (Tsuchiya et al., 2004). We concluded that our experimental conditions were not sufficient to detect the weak level of FUS3 expression. However, based on in situ hybridization (Tsuchiya et al., 2004), mutant complementation (Gazzarrini et al., 2004), and functional evidence (Kroj et al., 2003), FUS3 expression appears to be faithfully reproduced by the sensitive FUS3:GUS reporter. Furthermore, this construct offers the unique opportunity to monitor FUS3 expression in the fus3 mutant backgrounds (fus3, fus3 abi3, fus3 lec2, and fus3 abi3 lec2) and to establish FUS3 autoregulation.
Figure 2.
Analysis of ABI3 and CRC Expression by in Situ Hybridization in Developing Seeds.
(A) CRC storage protein gene expression was analyzed in wild-type and lec2 backgrounds.
(B) ABI3 expression analyzed in wild-type and lec2 10-DAP embryos. Note the presence of variable sectors devoid of staining.
(C) Expression of ABI3 in early stages of embryo development.
a, embryo axis; c, cotyledons. Bars = 100 μm in (A) and (B) and 20 μm in (C).
ABI3 Expression Is Regulated by LEC2, ABI3, and FUS3
We also investigated ABI3 expression using the ABI3:GUS construct. As previously reported (Parcy et al., 1994; Lara et al., 2003), ABI3 expression was detected throughout the embryo axis and cotyledons in wild-type seeds (Figure 1B). The root meristem staining was usually weak. In the lec2 mutant, the ABI3 expression pattern was modified in the cotyledons where variable regions were devoid of staining (Figure 1B). These regions varied in shape and size as described for AT2S3, CRC, and FUS3 expression in lec2 (Figure 2A; Kroj et al., 2003). We concluded that LEC2 also controls ABI3 expression. ABI3:GUS expression was not reduced in abi3 or fus3 single mutants but appeared weaker in the periphery of the cotyledons of the abi3 fus3 double mutant, suggesting that ABI3 might also be regulated by FUS3 and by ABI3 itself. The effect of abi3 and fus3 mutations was very apparent in the lec2 mutant backgrounds. ABI3:GUS expression was absent from the lateral parts of the cotyledons of the fus3 lec2 mutants. The strong phenotypic variability observed in the lec2 mutant was abolished in fus3 lec2, and the pattern shown in Figure 1B was systematically observed, thereby demonstrating that FUS3 is involved in ABI3 regulation. In the lec2 abi3 double mutant, ABI3:GUS expression was completely absent from the cotyledons, confirming that ABI3 positively regulates its own expression in cotyledons. We complemented the analysis of ABI3:GUS transgenic plants by monitoring ABI3 mRNA expression by in situ hybridization (Figures 2B and 2C). In wild-type embryos, we confirmed that ABI3 expression was uniform throughout the cotyledons and embryo axis (Figures 2B and 2C). In lec2 embryos, ABI3 mRNA was absent from variable sectors of the cotyledons, exactly as shown by the ABI3:GUS constructs, thereby indicating that the ABI3:GUS construct faithfully reproduces the ABI3 mRNA pattern.
We also analyzed LEC2:GUS expression patterns in all single and double mutant backgrounds. In these experiments, we never observed any decrease in LEC2:GUS staining at any developmental stages examined (from the globular stage to desiccating embryos). At late developmental stages, when LEC2 expression disappears from the wild-type cotyledons (Kroj et al., 2003), we sometimes observed a sustained LEC2:GUS activity in the lec2, fus3, and lec2 fus3 mutant backgrounds, suggesting that LEC2 might be repressed by FUS3 and LEC2 (Figure 1C). Since we did not succeed in detecting LEC2 expression by in situ hybridization and since no obvious phenotypes are associated with the potentially extended LEC2 expression, we did not investigate this point further.
Confirmation of the Regulatory Links with Functional Tests
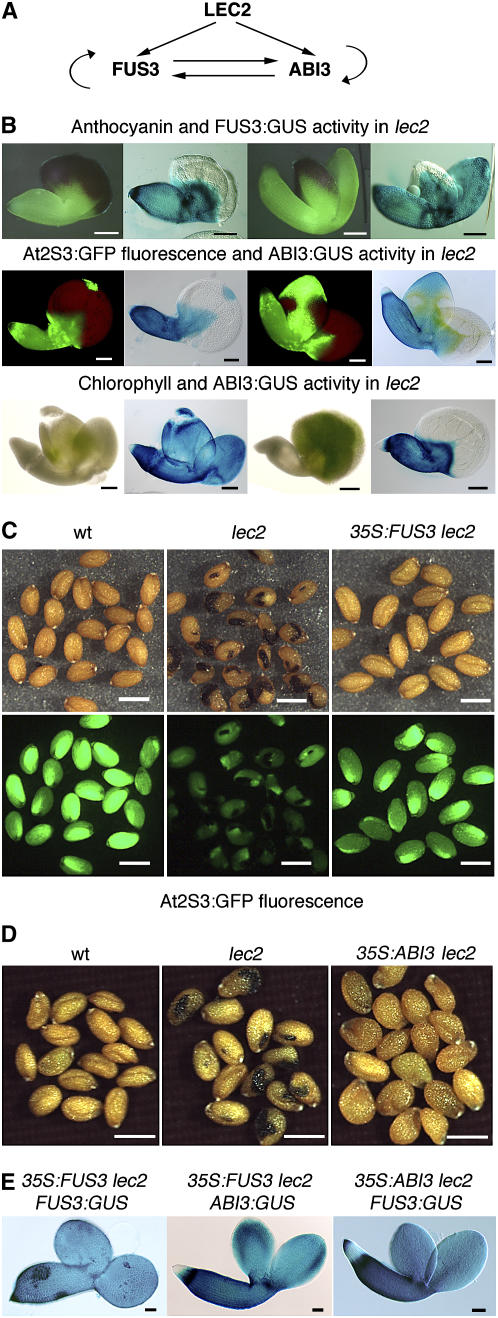
Cross-regulation between ABI3, FUS3, and LEC2, as established from expression analyses in wild-type and mutant backgrounds, is summarized in Figure 3A. These regulatory controls are largely corroborated by phenotypic analysis. Indeed, phenotypes characteristic of ABI3 (or FUS3) loss of function are obvious in several mutant backgrounds with a functional ABI3 (or FUS3) gene. For instance, the appearance of chlorophyll in dry embryos, which is one signature of ABI3 loss of function, was visible in sectors of lec2 (Figure 3B) and fus3 lec2 mutants (data not shown), consistent with the ABI3:GUS expression pattern. Similarly, anthocyanin accumulation in cotyledons, a characteristic of FUS3 loss of function, coincided with the loss of FUS3:GUS expression in lec2 and lec2 abi3 mutants (Figure 3B; data not shown; Meinke et al., 1994). Also, storage protein gene expression, monitored using the AT2S3:GFP (green fluorescent protein) reporter (Kroj et al., 2003), precisely coincided with the domains expressing both ABI3 and FUS3 in the lec2 mutant (Kroj et al., 2003; Figure 3B). In summary, phenotypic data provide evidence for the functional significance of mutual regulation within a network involving LEC2, ABI3, and FUS3.
Figure 3.
Network between B3 Transcription Factors (LEC2, ABI3, and FUS3).
(A) Positive interactions between ABI3, FUS3, and LEC2 as deduced from Figure 1.
(B) Spatial coincidence of the domains lacking ABI3 or FUS3 expression and the phenotypic traits in lec2, such as anthocyanin accumulation or reduced AT2S3:GFP fluorescence at 10 DAP or absence of chlorophyll degradation at 14 to 15 DAP.
(C) Rescue of the lec2 mutant phenotypes by constitutive expression of FUS3. The same groups of 15-DAP seeds are shown under white or blue illumination to observe the seed shape and color (top panels) and the AT2S3:GFP fluorescence (bottom panels). Dark spots on the lec2 seeds result from the combined presence of anthocyanin and chlorophyll.
(D) Rescue of the lec2 mutant phenotypes by constitutive expression of ABI3. Anthocyanin and chlorophyll accumulations in the lec2 mutant are suppressed by the 35S:ABI3 transgene.
(E) Rescue of ABI3:GUS and FUS3:GUS expression in 10-DAP lec2 embryos by ABI3 or FUS3 constitutive expression. The regions lacking ABI3 or FUS3 promoter activity observed in lec2 mutant (Figure 1) have disappeared when ABI3 or FUS3 are constitutively expressed.
Bars = 100 μm in (B), 500 μm in (C) and (D), and 50 μm in (E).
Our model suggests that part of LEC2 action might be indirect and that LEC2 controls several aspects of seed maturation, including the prevention of anthocyanin and chlorophyll accumulation, through the positive regulation of FUS3 and ABI3 (Figure 3A). If this model is correct, constitutive expression of FUS3 or ABI3 should complement some of the defects caused by the LEC2 loss of function. As shown in Figures 3C and 3E, the constitutive expression of FUS3 (directed by the 35S promoter) into lec2 AT2S3:GFP plants allowed a nearly complete phenotypic rescue of lec2, including the recovery of uniform AT2S3:GFP expression, the absence of anthocyanin and chlorophyll accumulation, and the suppression of ectopic trichomes (Table 2). 35S:FUS3 also restored a uniform FUS3:GUS expression (Figure 3E). Only the irregular shape of some of the lec2 embryo axes was unaffected by FUS3 expression (Figure 3E). Interestingly, constitutive expression of FUS3 also complemented defects associated with the lack of ABI3 (such as chlorophyll accumulation, which is not under FUS3 control), suggesting either that constitutively expressed FUS3 can fulfill ABI3 functions or that FUS3 might be able to restore uniform ABI3 expression in the lec2 mutant. To test the latter possibility, we introduced an ABI3:GUS reporter construct into the 35S:FUS3 lec2 background and observed that ABI3 expression indeed recovered a wild-type expression pattern in cotyledons (Figure 3E). These experiments suggest that many defects present in the lec2 mutant might indeed be indirectly due to the loss of FUS3 and ABI3 expression. They also show that FUS3 can positively regulate its own expression and that of ABI3, thereby confirming the conclusions previously inferred from loss-of-function analysis (Figure 3A).
Table 2.
Constitutive FUS3 or ABI3 Expression Suppresses Ectopic Trichome Formation in the lec2 Mutant
| Genotype | Wild Type | lec2 | 35S:FUS3 lec2 line 19 | |
|---|---|---|---|---|
| Plants with trichomes on cotyledons (%) | 0% (92) | 69% (202) | 0% (84) | |
| Genotype | Wild type | lec2 | 35S:ABI3 lec2 line 8 | 35S:ABI3 lec2 line 18 |
| Plants with trichomes on cotyledons (%) | 0% (45) | 37% (111) | 0% (57) | 0% (85) |
As for FUS3, constitutive expression of ABI3 in lec2 suppressed chlorophyll and anthocyanin accumulation and ectopic trichome formation (Table 2) and restored uniform FUS3:GUS expression in the lec2 mutant embryos (Figures 3D and 3E), confirming that ABI3 positively regulates FUS3 (Figure 3A). It is thus likely that ABI3 or FUS3 are sufficient to complement most of the lec2 mutant defects because constitutive expression of one of these two factors is sufficient to restore the uniform expression of the other.
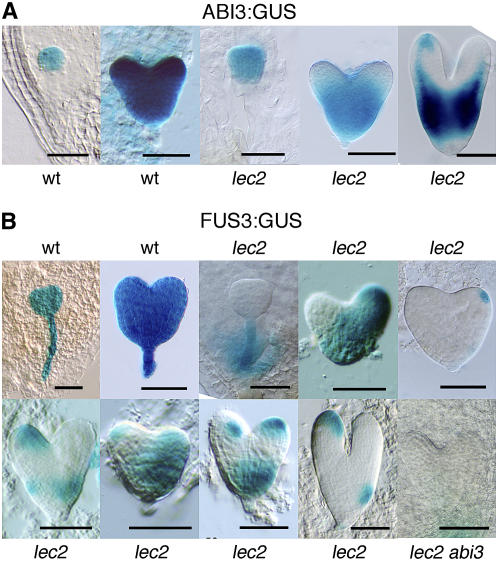
LEC2 Is Involved in FUS3 Initiation and ABI3 Maintenance
Cotyledons of lec2 exhibit variable sectors that are devoid of ABI3 and FUS3 expression. To explain this phenomenon, we reasoned that LEC2 might either play an essential role early on, during the initial induction phase of ABI3 and FUS3 expression, or alternatively, that it might be required only later to ensure uniform ABI3 and FUS3 expression patterns. To distinguish between these two possibilities, we analyzed the expression of ABI3 and FUS3 in early stages of embryo development. In wild-type embryos, ABI3 and FUS3 expression is first detected at the globular stage. ABI3 expression starts in the embryo proper at the globular stage (Figures 2C and 4A), while FUS3 expression initiates in the suspensor and, by the transition stage (between globular and heart stages), covers the whole embryo (Figure 4B; Parcy et al., 1994; Kroj et al., 2003; Tsuchiya et al., 2004). In the lec2 mutant, ABI3 expression was the same as that in the wild type in transition stage embryos (Figure 4A), whereas FUS3 expression was abolished in the embryo proper, though not in the suspensor (Figure 4B). This experiment indicates that initiation of FUS3 expression depends on LEC2, whereas that of ABI3 does not. Later on, in the heart and torpedo stages of embryo development, ABI3 expression started to fade from sectors of the cotyledons (Figure 4A), while FUS3 expression appeared in patches at variable locations (Figure 4B). By the bent cotyledon stage, ABI3 and FUS3 expression coincided, as deduced from the fact that they both shared the same pattern as the AT2S3:GFP marker (Figure 3B; Kroj et al., 2003). Therefore, LEC2 is required early for FUS3 initiation in the embryo proper and later to ensure ABI3 uniform expression (but not for ABI3 initiation). Since ABI3 regulates FUS3 in mature cotyledons, we wondered whether it was necessary for the appearance of FUS3 in patches in heart stage embryos of the lec2 mutant. We therefore analyzed FUS3:GUS expression in lec2 abi3 heart and torpedo stages and did not detect any FUS3:GUS activity in the embryo axis or cotyledons (Figure 4B), confirming that the late induction of FUS3 in lec2 mutants depends on ABI3.
Figure 4.
ABI3:GUS and FUS3:GUS Expression during Early Embryo Development.
(A) ABI3:GUS expression during wild-type and lec2 early embryo development. In the lec2 embryo, the ABI3:GUS expression pattern is the same as in wild-type embryos at globular stage but reduced in emerging cotyledons from heart stage on.
(B) FUS3:GUS expression during wild-type and lec2 early development. In the lec2 embryo, FUS3:GUS expression does not uniformly appear in the globular stage embryo (as in wild-type embryos) but in patches at heart stage. Patches are absent in the lec2 abi3 double mutant.
Bars = 50 μm.
LEC1 Controls ABI3 and FUS3 Expression in Cotyledons
LEC1 is the fourth master regulator of seed maturation in addition to LEC2, FUS3, and ABI3 and encodes the CBF-A subunit of the CCAAT binding trimeric transcription factor (Lotan et al., 1998). Based on single mutant phenotypes, LEC1 had been proposed to positively regulate LEC2 and FUS3 and to act independently of ABI3 (Meinke et al., 1994; Vicient et al., 2000a; Kagaya et al., 2005). Recently, LEC1 has been proposed to regulate ABI3 and FUS3 expression (Kagaya et al., 2005). However, double mutant analyses showed that ABI3, LEC2, and FUS3 were still active in the lec1 background (West et al., 1994; Parcy et al., 1997; Raz et al., 2001). We examined ABI3, LEC2, and FUS3 promoter activity in the lec1 mutant. We found a drastic local reduction of FUS3:GUS activity in cotyledons and root tips and a more moderate one for ABI3:GUS (Figure 5). LEC2 expression did not decrease but showed a moderate increase in late stages as observed in lec2 and fus3 mutants (Figure 1). Again, these modifications of reporter gene expression were consistent with local phenotypes. lec1 mutant embryos showed a marked chlorophyll accumulation at the periphery of the cotyledons, where ABI3 expression was most reduced (Figure 5). lec1 cotyledons have reduced ABA sensitivity, consistent with ABI3 reduced expression (Parcy et al., 1997). Finally, the lec1 mutant accumulated anthocyanin and showed reduced storage protein expression, consistent with the FUS3 expression pattern (Figure 5). This set of data indicates that LEC1 affects ABI3 and FUS3 expression locally (with a major effect on FUS3 expression in cotyledons) and suggests some of the _lec1-_associated phenotypes might actually be indirectly due to a reduction in the expression of ABI3 and FUS3.
Figure 5.
lec1 Mutation Affects the ABI3, FUS3, and LEC2 Expression Pattern.
All embryos are lec1 mutants and show reduced ABI3:GUS, FUS3:GUS, and AT2S3:GFP expression at 10 DAP and increased chlorophyll fluorescence at 14 to 15 DAP. Chlorophyll was present mainly in the regions where ABI3:GUS was reduced. AT2S3:GFP persisted in regions expressing FUS3:GUS. Bars = 100 μm.
DISCUSSION
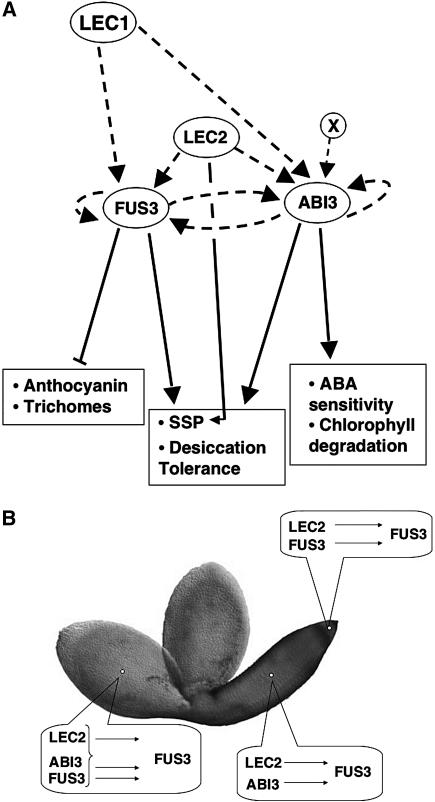
A Regulatory Network for Seed Maturation
In this study, we have unraveled the regulatory network linking ABI3, FUS3, LEC1, and LEC2, four major regulators of seed maturation (Figure 6A). In a previous study, we showed that LEC2 was necessary for uniform FUS3 expression in parts of the cotyledons (Kroj et al., 2003). We show here that FUS3 is controlled by a set of local and redundant regulations that vary spatially throughout the embryo and involve ABI3, LEC2, and FUS3 itself. In the root tip, FUS3 expression is redundantly controlled by LEC2 and by FUS3 itself; in the embryo axis, FUS3 expression is redundantly controlled by LEC2 and ABI3; in the cotyledons, FUS3 expression is under the control of all three regulators (Figure 6B). In addition to this set of regulatory controls, LEC1 also locally regulates FUS3 expression in cotyledons. Our data are consistent with recently published analyses showing that LEC2 and LEC1 can induce FUS3 expression in vegetative tissues (Kagaya et al., 2005; Santos Mendoza et al., 2005) and that FUS3 expression is reduced in whole-seed mRNA of the lec1 mutant (Brocard et al., 2002; Kagaya et al., 2005).
Figure 6.
Gene Network Architecture.
(A) Schematic description of the network. Relations between the regulators are depicted as dashed lines, whereas proposed downstream actions are show as solid lines. X represents the unknown factor inducing ABI3 expression at the globular stage.
(B) Summary of FUS3 spatial regulation by various combinations of the B3 transcription factors.
We also showed that ABI3 expression is controlled by LEC1, LEC2, FUS3, and by ABI3 itself in the developing cotyledons. The decreased ABI3 expression in fus3 lec2 and fus3 abi3 double mutants is consistent with previously obtained ABI3 protein level data (Parcy et al., 1997; Kroj et al., 2003). In lec1 mutants, a reduction of ABI3 expression was not systematically observed (Parcy et al., 1997; Kagaya et al., 2005). As suggested by Kagaya et al. (2005), this discrepancy very likely arose from differences in seed stages examined. In this study, we indeed observed that ABI3:GUS was weaker in lec1 embryos than in wild-type embryos at 10 DAP (Figure 5) but close to the wild-type expression in desiccating seeds (14 to 15 DAP; data not shown), which is the stage used by Parcy et al. (1997) for protein analysis. Interestingly, ABI3 regulation differs from that of FUS3, as it was not abolished in the embryo axis of the double mutants tested (abi3 fus3, abi3 lec2, and fus3 lec2). This expression therefore appears to be independent of FUS3 and LEC2. Consistent with this hypothesis, a 5′ deletion of the ABI3 promoter that removed all RY elements (likely binding sites for LEC2 and FUS3) resulted in an ABI3:GUS pattern confined to the embryo axis (F. Parcy, unpublished data).
Several of the features of the regulatory network studied here clearly rendered its analysis difficult. First, the gene regulatory controls in this network act locally, rather than on the whole embryo, thereby complicating genetic analyses that are often based on whole embryo characteristics (such as gene expression). Second, functional redundancies exist (for example, between ABI3 and FUS3 or between FUS3 and LEC2) that, again, are tissue dependent. FUS3 expression is regulated redundantly by ABI3 and LEC2 in the embryo axis but by LEC2 and FUS3 in the root tip (Figure 6B). Third, FUS3 and ABI3 autoregulate. Hints of such types of regulation were previously provided by the analysis of the weak abi3-1 allele (Parcy et al., 1997) or from the ectopic activation of seed regulators in vegetative tissues (Kagaya et al., 2005; Santos Mendoza et al., 2005). However, reporter constructs have been necessary to firmly establish these cases of autoregulation in strong mutant backgrounds. This network illustrates the intricate nature of regulatory circuitry and the difficulty of interpretation of global analyses of gene expression in whole seedlings or organs.
Functional Implications of the Network
Our results, together with other published data (Kroj et al., 2003; Kagaya et al., 2005), establish that LEC2 and LEC1 act on ABI3 and FUS3 expression. Moreover, we observed that FUS3 or ABI3 constitutive expression could rescue most aspects of the lec2 mutant phenotype, suggesting that ABI3 and FUS3 act downstream of LEC2. However, we should point out that interpretation of this result is not straightforward because ABI3, FUS3, and LEC2 encode homologous transcription factors, whose specificity might be altered by constitutive expression. For example, we found that 35S:FUS3 suppressed chlorophyll accumulation, a function known to be normally performed by ABI3. Since we also showed that 35S:FUS3 was capable of restoring ABI3 uniform expression in lec2 cotyledons, the simplest explanation is that FUS3 constitutive expression induces ABI3, which suppresses chlorophyll accumulation. However, we cannot exclude that constitutive expression of FUS3 acts on chlorophyll independently of ABI3. One way to test this hypothesis would be to introduce the 35S:FUS3 transgene in a lec2 abi3 double mutant and observe if chlorophyll accumulation remains. Similarly, it is conceivable that FUS3 or ABI3 constitutive expression complement the lec2 mutant because of the lack of specificity. We do not favor this hypothesis because FUS3 and ABI3 only complement phenotypic defects that are related to their endogenous function (such as chlorophyll breakdown, anthocyanin repression, or SSP gene expression). The abnormal shape of the lec2 mutant embryo axis is not complemented by FUS3 or ABI3 (Figure 3). Also, neither 35S:ABI3 nor _35S:FUS_3 induces the formation of ectopic embryos as LEC2 does (Stone et al., 2001). These results suggest that ABI3, FUS3, and LEC2 retain some specificity even when overexpressed. We concluded that LEC2 is upstream of ABI3 and FUS3 because (1) ABI3 and FUS3 expression are absent from sectors of lec2 embryos; (2) some mutant phenotypes, typical of ABI3 and FUS3 loss of function, appear precisely in these sectors; and (3) these defects can be complemented by constitutive expression of ABI3 or FUS3. However, it is clear that LEC2 does not act only via ABI3 or FUS3. Several complementary studies have shown that LEC2 is capable of activating directly 12S and 2S storage protein genes (Kroj et al., 2003; Santos Mendoza et al., 2005; Braybrook et al., 2006). It is thus likely that, in wild-type plants, LEC2 controls SSP gene expression through two different mechanisms: directly, by binding the SSP promoter, and indirectly, by activating FUS3 and ABI3 as indicated in Figure 6A.
Expression analyses of FUS3 and ABI3 in lec1 suggest that LEC1 also might indirectly control many aspects of seed maturation through regulation of ABI3 and FUS3 expression. Testing this hypothesis will require analyzing the extent to which the lec1 mutant phenotype can be complemented by ABI3 or FUS3 constitutive expression, as we did for the lec2 mutant. The question of whether LEC1 participates in ABA sensitivity has previously been a matter of debate (West et al., 1994; Parcy et al., 1997; Brocard et al., 2002). Based on observation of radicle growth, lec1 has been shown to be ABA sensitive, but lec1 cotyledon expansion is partially insensitive to ABA (West et al., 1994; Parcy et al., 1997). The observation that ABI3 expression is predominantly reduced in cotyledons offers a simple explanation for this apparent contradiction: LEC1 might affect ABA sensitivity only indirectly, through ABI3 regulation in the cotyledons. As recently suggested, LEC1 might also control storage protein expression through ABI3 and FUS3 control (Kagaya et al., 2005). Testing the direct and indirect target genes of LEC2 and LEC1 will require further experiments, possibly using versions of these transcription factors that can be posttranslationally activated (Kagaya et al., 2005; Santos Mendoza et al., 2005).
The Network Structure and Its Implications: A Confirmation of Theoretical Predictions?
The maturation network presented in Figure 6A resembles modules that are known to drive developmental progression or patterning in other higher eukaryotes (Davidson et al., 2002, 2003; Rudel and Sommer, 2003; Levine and Davidson, 2005). The network analyzed here appears to function to ensure a uniform expression of essential regulators, such as ABI3 and FUS3, during the entire phase of seed maturation in a manner that is independent of their initial activation. Our analysis at early developmental stages indicates that LEC2 induces FUS3 in the embryo at the globular stage (FUS3 is thus the earliest known LEC2 target gene), whereas ABI3 is induced independently of LEC2 (by an unknown factor X depicted in Figure 6A). However, as soon as the heart stage, mutual activation loops between ABI3 and FUS3 play a pivotal role in maintaining their uniform expression patterns. This maintenance might be of particular importance toward the end of seed maturation, when LEC2 expression decreases, and ABI3 and FUS3 are required to uniformly establish desiccation tolerance.
Interestingly, the structure of the network revealed here provides a possible explanation for the observed phenotypic variability of the lec2 mutant (Figure 3C; Meinke et al., 1994; Kroj et al., 2003). Our analyses at early stages of embryo development allowed us to trace back the origin of the observed variable lec2 defects to the heart stage, where ABI3 expression disappears and FUS3 expression appears in randomly positioned patches. These expression patterns seem surprising but are easily explained by the structure of the network. Modules with autoregulatory loops have been the focus of intense studies, and theoretical analyses have predicted that small variations (or noise) can be stabilized as a result of bistability generated by the network (Thattai and van Oudenaarden, 2001; Blake et al., 2003; Isaacs et al., 2003). Applying these principles to the situation analyzed here, we can propose the following explanation. In lec2 mutant embryo development, there might be a critical moment where ABI3 initial induction is fading. At this moment, in some localized parts of the lec2 embryo, ABI3 residual expression levels might be sufficient to trigger FUS3 expression, thereby feeding into the positive loop and leading to stabilization of the expression of both regulators (first stable state). In other parts of the lec2 embryo, ABI3 may not succeed in inducing FUS3, leading to the local loss of expression of both regulators (second stable state). The observed phenotype (embryo regions with both FUS3 and ABI3 expression, other regions lacking both) is the exact theoretical outcome of this bistable network (Thattai and van Oudenaarden, 2001; Blake et al., 2003; Isaacs et al., 2003). To our knowledge, our observations are one of the few experimental confirmations of such predictions. Our results therefore not only provide an explanation for a puzzling seed phenotype but also represent an experimental confirmation of theoretical studies of general interest.
Molecular Mechanism and Appearance in Evolution
What could be the molecular mechanisms underlying this network? Since ABI3, FUS3, and LEC2 are B3 transcription factors, and since RY elements are present in the promoters of FUS3 and ABI3, some of the regulatory controls might be direct and involve physical binding of the B3 transcription factor to the FUS3 and ABI3 promoters. However, FUS3 has been shown to act from the embryo epidermis and to regulate storage protein gene expression in internal cell layers using ABA as a mobile mediator (Gazzarrini et al., 2004). Thus, this indirect mode of action could also apply to ABI3 regulation by FUS3. This point will require further analysis.
Finally, it would be interesting to understand when and how this regulatory network was first established over the course of plant evolution. A likely mechanism (Wagner, 2001; Amoutzias et al., 2004; Teichmann and Babu, 2004) for the generation of this complex regulatory network of B3 transcription factors could be the repeated duplication of an ancestral B3 gene with autoregulatory properties, thereby generating three B3 transcription factors that regulate their own and each other's expression. The existence of ABI3 orthologs in monocotyledonous species (McCarty et al., 1991), together with the recent discovery of FUS3 and LEC2 homologs in cereals (F. Parcy and J. Vicente-Carbajosa, unpublished data), suggests that this network may also function in cereal grains and that it originated before the taxonomic split between the monocotyledons and eudicotyledons. Analysis of components of the network in basal angiosperms or gymnosperms might allow the identification of the ancestral autoregulatory gene.
METHODS
Plant Material
We used the following Arabidopsis thaliana strains: lec1-1 (Meinke, 1992) in Wassilewskija, AT2S3:GFP in Columbia-0 (Col-0) (line FP91.54.3) (Kroj et al., 2003), and lec2-1, abi3-6 gl1, and fus3-3 gl1 introduced in the AT2S3:GFP background (Kroj et al., 2003). Except for lec1-1 and abi3 fus3 lec2 triple mutants, all strains used in this study (including those mentioned as wild types in the figures) contained the AT2S3:GFP transgene. FUS3:GUS (line 3.2 in Col-0), LEC2:GUS (line 3.2 in Col-0), and ABI3:GUS (line LAG3-4 in Landsberg erecta) have been described (Parcy et al., 1994; Kroj et al., 2003).
Generation and Characterization of Transgenic Plants
A double enhanced 35S promoter–NOS terminator cassette (Parcy et al., 1994) and a Gateway cassette (Invitrogen) were successively introduced in the pZP312 vector (pZP derivative conferring Basta resistance in plants built by C. Fankhauser) to generate the pFP108-rfA plasmid in which FUS3 and ABI3 cDNAs were recombined according to Invitrogen's recommendations. Arabidopsis plants were grown and transformed as described (Kroj et al., 2003). The presence of mutations and transgenes was verified by PCR genotyping as described below.
Nineteen 35S:FUS3 lines in lec2 AT2S3:GFP were generated. Visual inspection of the T2 seeds (under white and blue illumination) showed lec2 phenotypic rescue in 15 of them. Four single-locus lines were followed until the homozygous stage and showed a degree of rescue similar to the line 35S:FUS3 #19, shown in Figure 3 and used for crosses to lec2 ABI3:GUS and lec2 FUS3:GUS. 35S:ABI3 (pFP111) was introduced directly into the lec2 FUS3:GUS background. Among 18 T1 lines generated, six showed apparent rescue of the lec2 phenotype, and five were followed until homozygous stage and showed a similar degree of rescue as line FP111 #3 shown in Figure 3.
Expression Analysis
The GUS staining and AT2S3:GFP fluorescence analyses were performed as described (Kroj et al., 2003). Individual embryos (at least 20 for each genotype) were observed and stained in rudimentary flow cells to directly compare AT2S3:GFP, GUS activity, and chlorophyll or anthocyanin accumulation as described by Kroj et al. (2003). When needed, anthocyanin was washed in 90% acetone (to allow observation of chlorophyll), and chlorophyll was washed with 95% ethanol (observation of anthocyanin). In situ hybridization was performed as described by Lara et al. (2003) using CRC, ABI3, or FUS3 as probes except for Figure 2C (performed as in Vicient et al., 2000b).
Counting of Trichomes on Cotyledons
To prevent ectopic trichomes on cotyledons to desiccate, siliques were harvested when still green (before the first sign of valve dehiscence) and were surface sterilized, and seeds were dissected out and sowed on germination medium (Parcy et al., 1994). Seedlings bearing trichomes were counted 5 to 7 d after stratification. Genotypes used were the following: AT2S3:GFP in Col-0 (indicated as wild type in Table 2), 35S:FUS3 lec2 AT2S3:GFP line 19, lec2 AT2S3:GFP, 35S:ABI3 FUS3:GUS lec2 line 8, and 35S:ABI3 FUS3:GUS line 18. The wild type and the lec2 mutant are presented twice because experiments were performed in two independent growth conditions.
PCR Genotyping of Mutations and the Transgene
Transgenes and mutations were followed by PCR amplification of genomic DNA using the oligonucleotides listed in Table 3.
Table 3.
Oligonucleotides Used for PCR Genotyping
| Mutation/Transgene | Oligonucleotides 5′–3′ | Comment |
|---|---|---|
| abi3-6 | gcttcttcatcaaaccaaa | 1.1-kb wild-type band |
| cgatgatggagaataacagtggt | 0.35-kb mutant band | |
| fus3-3 | gattcctcttccaaaaggaactca | _Rsa_I cuts the wild-type band twice and the mutant once |
| ggtcttaagatcgacatggataca | ||
| lec2-1 | acgtgcagatctccgacaagaa | Wild-type band 0.4 kb |
| ttaccaagtattggttcgagaaat | ||
| cgatggagatgcgatggtataatatgtggccgcaacgat | 0.6-kb mutant band | |
| lec1-1 | tgtgccgtttcgagttgccttgt | 0.75-kb wild-type band |
| atggcccggagaagacgattt | 0.52-kb mutant band | |
| tggaccccgttgagtagactgtt | ||
| ABI3:GUS transgene | cgatgatggagaataacagtggt | |
| tcacgggttggggtttctac | ||
| FUS3:GUS transgene | gaaacccaaagagatccacc | |
| tcacgggttggggtttctac | ||
| LEC2:GUS transgene | tgaatggctattaatggtgtttactct | |
| tcacgggttggggtttctac |
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4g27160 (AT2S3), At4g28520 (CRC), At3g24650 (ABI3), At3g26790 (FUS3), At1g28300 (LEC2), and At1g21970 (LEC1).
Acknowledgments
We thank H. Baumlein, P. McCourt, J. Harada, and the ABRC for providing seeds and cDNAs, C. Fankauser and Plant Genetic Systems for vectors, A. Marinière for generating FUS3:GUS fus3 lec2 abi3, M. Blazquez, J. Vicente, H. Meinhardt, and F. Berger for stimulating feedback, C. Scutt and R. Benlloch for proofreading, and M. Blazquez and C. Ferrandiz for hosting F.P. at the Instituto de Biologia Molecular y Celular de Plantas. F.P. was supported by the Centre National de la Recherche Scientifique and by a fellowship from the Spanish Ministry of Science, and G. Savino was supported by the REGIA FP5 European Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: François Parcy (francois.parcy@cea.fr).
References
- Amoutzias, G.D., Robertson, D.L., Oliver, S.G., and Bornberg-Bauer, E. (2004). Convergent networks by single-gene duplications in higher eukaryotes. EMBO Rep. 5 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumlein, H., Miséra, S., Luerssen, H., Kölle, K., Horstmann, C., Wobus, U., and Müller, A.J. (1994). The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J. 6 379–387. [Google Scholar]
- Blake, W.J., Kaern, M, Cantor, C.R., and Collins, J.J. (2003). Noise in eukaryotic gene expression. Nature 422 633–637. [DOI] [PubMed] [Google Scholar]
- Braybrook, S.A., Stone, S.L., Park, S., Bui, A.Q., Le, B.H., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 103 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard, I.M., Lynch, T.J., and Finkelstein, R.R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E.H., McClay, D.R., and Hood, L. (2003). Regulatory gene networks and the properties of the developmental process. Proc. Natl. Acad. Sci. USA 100 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E.H., et al. (2002). A genomic regulatory network for development. Science 295 1669–1678. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S., Tsuchiya, Y., Lumba, S., Okamoto, M., and McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7 373–385. [DOI] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B., de Paiva, G., and Yadegari, R. (1994). Plant embryogenesis: Zygote to seed. Science 266 605–614. [DOI] [PubMed] [Google Scholar]
- Isaacs, F.J., Hasty, J., Cantor, C.R., and Collins, J.J. (2003). Prediction and measurement of an autoregulatory genetic module. Proc. Natl. Acad. Sci. USA 100 7714–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya, Y., Toyoshima, R., Okuda, R., Usui, H., Yamamoto, A., and Hattori, T. (2005). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46 399–406. [DOI] [PubMed] [Google Scholar]
- Keith, K., Kraml, M., Dengler, N.G., and McCourt, P. (1994). fusca3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073. [DOI] [PubMed] [Google Scholar]
- Lara, P., Onate-Sanchez, L., Abraham, Z., Ferrandiz, C., Diaz, I., Carbonero, P., and Vicente-Carbajosa, J. (2003). Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 278 21003–21011. [DOI] [PubMed] [Google Scholar]
- Levine, M., and Davidson, E.H. (2005). Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102 4936–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. [DOI] [PubMed] [Google Scholar]
- Luerssen, H., Kirik, V., Herrmann, P., and Misera, S. (1998). FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15 755–764. [DOI] [PubMed] [Google Scholar]
- McCarty, D.R., Hattori, T., Carson, C.B., Vasil, V., Lazar, M., and Vasil, I.K. (1991). The viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66 895–905. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. (1992). A homeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258 1647–1650. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W., Franzmann, L.H., Nickle, T.C., and Yeung, E.C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., Hayama, R., Tsuchiya, Y., Nishimura, M., Kawaide, H., Kamiya, Y., and Naito, S. (2000). The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev. Biol. 220 412–423. [DOI] [PubMed] [Google Scholar]
- Nambara, E., Keith, K., McCourt, P., and Naito, S. (1995). A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121 629–636. [Google Scholar]
- Parcy, F., Valon, C., Kohara, A., Miséra, S., and Giraudat, J. (1997). The ABSCISIC ACID-INSENSITIVE 3 (ABI3), FUSCA 3 (FUS3) and LEAFY COTYLEDON 1 (LEC1) loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz, V., Bergervoet, J.H., and Koornneef, M. (2001). Sequential steps for developmental arrest in Arabidopsis seeds. Development 128 243–252. [DOI] [PubMed] [Google Scholar]
- Rudel, D., and Sommer, R.J. (2003). The evolution of developmental mechanisms. Dev. Biol. 264 15–37. [DOI] [PubMed] [Google Scholar]
- Santos Mendoza, M., Dubreucq, B., Miquel, M., Caboche, M., and Lepiniec, L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579 4666–4670. [DOI] [PubMed] [Google Scholar]
- Steeves, T.A. (1983). The evolution and biological significance of seeds. Can. J. Bot. 61 3550–3560. [Google Scholar]
- Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann, S.A., and Babu, M.M. (2004). Gene regulatory network growth by duplication. Nat. Genet. 36 492–496. [DOI] [PubMed] [Google Scholar]
- Thattai, M., and van Oudenaarden, A. (2001). Intrinsic noise in gene regulatory networks. Proc. Natl. Acad. Sci. USA 98 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, Y., Nambara, E., Naito, S., and McCourt, P. (2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37 73–81. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa, J., and Carbonero, P. (2005). Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49 645–651. [DOI] [PubMed] [Google Scholar]
- Vicient, C.M., Bies-Etheve, N., and Delseny, M. (2000. a). Changes in gene expression in the leafy cotyledon1 (lec1) and fusca3 (fus3) mutants of Arabidopsis thaliana L. J. Exp. Bot. 51 995–1003. [DOI] [PubMed] [Google Scholar]
- Vicient, C.M., Hull, G., Guilleminot, J., Devic, M., and Delseny, M. (2000. b). Differential expression of the Arabidopsis genes coding for Em-like proteins. J. Exp. Bot. 51 1211–1220. [PubMed] [Google Scholar]
- Wagner, A. (2001). Birth and death of duplicated genes in completely sequenced eukaryotes. Trends Genet. 17 237–239. [DOI] [PubMed] [Google Scholar]
- West, M.A.L., Matsuderai Yee, K., Danao, J., Zimmerman, J.L., Fisher, R.L., Goldberg, R.B., and Harada, J.J. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, U., and Weber, H. (1999). Seed maturation: Genetic programmes and control signals. Curr. Opin. Plant Biol. 2 33–38. [DOI] [PubMed] [Google Scholar]