An MMP liberates the Ninjurin A ectodomain to signal a lossof cell adhesion (original) (raw)
Abstract
Matrix metalloproteinases (MMPs) are important for developmental tissue remodeling and for the inflammatory response. Although the vertebrate MMP family is large and functionally redundant, the fruitfly Drosophila melanogaster has only two MMPs, both essential genes. Our previous work demonstrated that Mmp1 is required for growth of the tracheal system, and we suggested that the mutant phenotype resulted from aberrant persistence of cell adhesion to the extracellular matrix. Here we report the identification of NijA, a transmembrane protein whose vertebrate homologs regulate cell adhesion, as a two-hybrid binding partner for Mmp1. The binding of Mmp1 and NijA was confirmed by coimmunoprecipitation of endogenous proteins from flies, and the endogenous proteins were found to colocalize at the tracheal cell surface in larvae. When NijA is expressed in S2 cells, they lose adhesion to surfaces; this adhesion-loss phenotype is dependent on the expression and catalytic activity of Mmp1. Our data indicate that Mmp1 releases the N-terminal extracellular domain of NijA. This liberated ectodomain promotes the loss of cell adhesion in a cell-nonautonomous manner. We suggest that tracheal cell adhesion is regulated by a novel mechanism utilizing an MMP and a ninjurin family member.
Keywords: Adhesion, Drosophila, matrix metalloproteinase, tracheae, model organism
The matrix metalloproteinase (MMP) family of extracellular proteases contains 22 members in mice, and biochemically, this family is able to cleave dozens of substrates in vitro, including extracellular matrix and signaling molecules. MMPs are up-regulated in cancer and in inflammatory diseases and have been considered important therapeutic targets (for review, see Egeblad and Werb 2002). Clinical trials of MMP inhibitors have been stymied by side effects (Coussens et al. 2002), which suggests that in addition to their pathological functions, MMPs are important for normal physiological functions. Identifying these functions has been difficult because MMPs display genetic redundancy (Lambert et al. 2003; Stickens et al. 2004). Nevertheless, MMP mutant mouse phenotypes indicate that, in general, they appear to be required for tissue remodeling, both in development and for homeostasis. Mouse MMP-9 and MMP-13 are required to remodel bone during endochondrial ossification, when cartilage is replaced by bone (Vu et al. 1998; Inada et al. 2004; Stickens et al. 2004). MMP-14 is required to remodel connective tissue during development and adult life (Holmbeck et al. 1999). MMP-7 is required for several aspects of inflammation, a homeostatic response whose endpoints are pathogen clearance and tissue remodeling (Wilson et al. 1999; Li et al. 2002). Although these mouse phenotypes demonstrate that MMPs function in tissue remodeling, little is known about the molecular mechanisms underlying these phenotypes. In the clinic, these unknown pathways must be protected if MMPs are to be considered as therapeutic targets.
The fruitfly Drosophila melanogaster has only two MMPs in its genome, which simplifies problems of redundancy. The two MMP genes have been called Mmp1 and Mmp2, although they are not orthologous to any single vertebrate MMP and should not be considered the direct orthologs of vertebrate MMP-1 and MMP-2. The fly MMPs contain the stereotyped domain structure typical of the MMP family: a pro domain, a catalytic domain, and a hemopexin domain (Llano et al. 2000, 2002). The autoinhibitory pro domain is cleaved off at zymogen activation. The active site of the catalytic domain has a conserved HEXXHXXGXXH sequence; in many vertebrate MMPs, mutation of the E to A in the catalytic core causes the enzyme to become catalytically inactive (Crabbe et al. 1994). The hemopexin domain is believed to mediate substrate recognition and protein–protein interactions (Sternlicht and Werb 2001).
We previously made fly mutants for each MMP and have demonstrated that each is an essential gene with developmental phenotypes in tissue remodeling, specifically in tracheal growth during the larval period for Mmp1 and in histolysis during metamorphosis for Mmp2 (Page-McCaw et al. 2003). Others have demonstrated that they are required for remodeling the nervous system during metamorphosis (Kuo et al. 2005). Their expression patterns suggest, however, that there may be more requirements for the fly MMPs than have yet been reported (Page-McCaw et al. 2003). Thus, Drosophila is emerging as an excellent system for the functional analysis of MMPs.
The tracheal phenotype of Mmp1 mutants suggests that they are defective in their ability to remodel the attachment between cells and the extracellular matrix (Page-McCaw et al. 2003). The Drosophila tracheae are ramified epithelial tubes that enable oxygen to diffuse throughout the body. Tracheal morphogenesis takes place during the embryonic stages of development (Manning and Krasnow 1993). After its formation, the main tracheal trunk grows 14-fold in length and sevenfold in diameter during the larval period, all without cell division, requiring remodeling of the cells and the supporting extracellular matrix (Beitel and Krasnow 2000). Tracheal tubes are lined with cuticle, a specialized extracellular matrix, which is continuous with the exterior cuticle of the animal. This cuticle is shed twice during the interlarval molts, and this shedding of cuticle allows the tubes to dilate as the cells pull back from the cuticle at molts (Beitel and Krasnow 2000). In contrast to the smooth continuous tracheal tubes found in wild-type animals, Mmp1 mutant tracheal tubes have aberrant constrictions and frequent breaks. The constrictions suggest that cells have difficulty pulling back from the cuticle. Supporting this interpretation is the observation that some mutant larvae cannot fully shed their cuticles. Instead, the old cuticle remains stuck to the larva, either at the external epidermis or at a tracheal spiracle (Page-McCaw et al. 2003). In contrast to tube dilation, tube elongation occurs continually throughout the larval period (Beitel and Krasnow 2000), and the stretched and broken tracheal tubes of the Mmp1 mutant indicate that this process is also aberrant. These phenotypes suggest that Mmp1 is required for cells to release from the cuticle.
To identify substrates and binding partners that might be important for this MMP-mediated control of cell adhesion, we performed a yeast two-hybrid screen using the hemopexin domain of Mmp1 as bait. We identified seven potential partners, and here present the analysis of one interactor, a fly homolog of vertebrate ninjurin transmembrane proteins. Ninjurin A (NijA) was especially interesting to us because of studies showing that ninjurins act as adhesion molecules (Araki and Milbrandt 1996, 2000). Vertebrate Ninjurin-1 was first identified in a subtractive hybridization screen to identify genes up-regulated in response to nerve injury, and the family is named for this expression pattern: nerve injury induced (Araki and Milbrandt 1996). Vertebrate Ninjurin-2 was identified by its homology with Ninjurin-1, and it also is up-regulated in response to nerve injury (Araki and Milbrandt 2000). In addition to nervous system expression, Ninjurin-1 is widely expressed in uninjured epithelial tissues (Araki and Milbrandt 1996; Araki et al. 1997) and Ninjurin-2 is expressed in cells of the immune system (Araki and Milbrandt 2000). Ninjurins are considered to be cell-adhesion molecules because the vertebrate members each promote homophilic cell aggregation and neurite extension in culture (Araki et al. 1997; Araki and Milbrandt 2000).
Interestingly, microarray studies from flies have also determined that both NijA and Mmp1 are coregulated (De Gregorio et al. 2001; Boutros et al. 2002). These expression data suggest that NijA and Mmp1 function in the same processes. After confirming a physical interaction between Mmp1 and NijA, we determined that Mmp1 and NijA colocalize in vivo in tracheal cells. From cell-culture assays, we determined that NijA promotes a loss of cell adhesion. This _NijA_-induced loss of cell adhesion requires Mmp1, which liberates an extracellular domain of NijA. The liberated NijA domain then acts cell nonautonomously to cause cells to detach. Our data identify a novel mechanism for how an MMP alters cellular adhesion. Mmp1 liberates the NijA ectodomain, which regulates cell adhesion in a cell-nonautonomous fashion.
Results
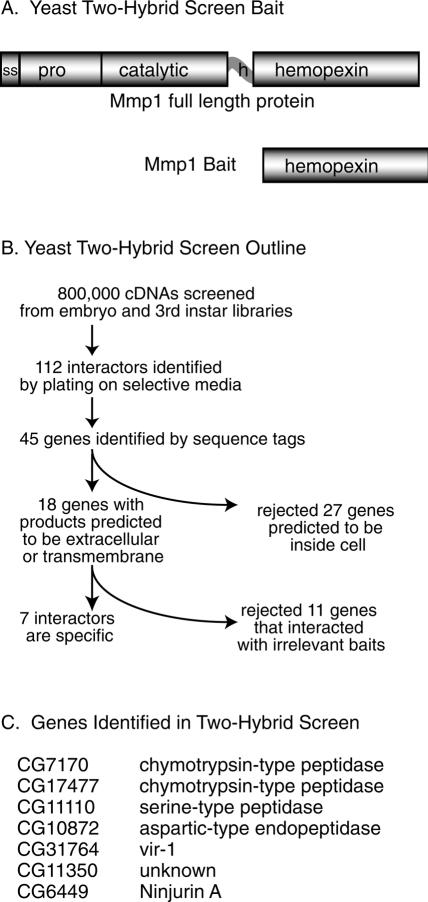
To identify binding partners and substrates of Drosophila Mmp1, we performed a yeast two-hybrid screen (James et al. 1996). Although a Drosophila large-scale systematic yeast two-hybrid screen has been reported (Giot et al. 2003), some genes were absent from that screen, including both Mmp1 and the binding partner we describe here, Ninjurin A. We reasoned that a protease might cleave rather than stably bind potential partners, and so we used as bait the isolated hemopexin domain, which mediates substrate recognition and has no proteolytic activity (Fig. 1A). We screened ∼106 cDNA prey constructs and identified 50 colonies from an embryo library and 62 colonies from a larval library by plating on selective medium. We PCR-amplified and sequenced target DNA from 89 colonies, identifying 45 independent genes. For each interacting gene, the predicted full-length sequence (Stapleton et al. 2002) was analyzed for its likely cellular location. All intracellular genes were rejected, as these were likely to be irrelevant partners for an extracellular protease. Finally, all target genes predicted to be extracellular or transmembrane were experimentally checked for Mmp1-binding specificity by testing their ability to interact with two irrelevant baits (Drosophila genes Argonaute 1 and arrow); any gene that interacted with either of these was rejected as potentially nonspecific (see Fig. 1B for an overview).
Figure 1.
A yeast two-hybrid screen to identify proteins that bind to the Mmp1 hemopexin domain. (A) Schematic of Mmp1 domain structure in the wild-type protein and in the bait construct. The hemopexin domain of splice form 1 was used as two-hybrid bait. (ss) Signal sequence; (h) hinge domain. (B) Outline of the screen and secondary screens. (C) The seven genes identified in this screen.
Seven genes interacted specifically with Mmp1 (see Fig. 1C). Of these candidate interactors, CG6449, or Ninjurin A (NijA), was especially interesting because of its homology with vertebrate ninjurin genes and its expression data. NijA encodes a 245-amino-acid protein containing a ninjurin domain (Fig. 2A), which is highly conserved between flies and humans (Fig. 2B). Like vertebrate ninjurins, NijA is predicted to have two transmembrane domains oriented such that the protein termini are extracellular (Fig. 2C).
Figure 2.
NijA protein sequence, homology, and topology. (A) Drosophila NijA amino acid sequence, from CG6449. Gray boxes indicate predicted transmembrane domains. The black box indicates the region corresponding to the antigenic peptide. NijA splice form B is shown; isoform A removes VNRT (amino acids 23–26); isoform C removes VPETDDDDNDDRPFV (amino acids 52–66). The cDNA fragment identified by two-hybrid screening encodes amino acids 43–end, consistent with either isoform A or B. (B) ClustalW alignment of human Ninjurin-1 and Ninjurin-2 and Drosophila Ninjurin A. Black boxes indicate identical residues; gray boxes indicate similar residues. Isoform B of NijA is shown, and the human splice forms are the ones reported by Araki and Milbrandt (1996, 2000). (C) Predicted NijA topology, with the termini outside the cell. (D) Phylogenetic tree relating all ninjurin domain-containing proteins identified to date. Non-insect proteins shown are rat Ninjurin-1 and Ninjurin-2, human Ninjurin-1 and Ninjurin-2, BAE34827.1 (Mus musculus), XP 853215.1 (Canis familiaris), XP 528716.1 (Pan troglodytes), XP 586318.2 (Bos taurus), XP 414328.1 (Gallus gallus), NP 001008021.1 (Xenopus tropicalis), CAG06884.1 (Tetraodon nigroviridis), XP 416382.1 (Gallus gallus), XP 854702.1 (Canis familiaris), CAF97213.1 (Tetraodon nigroviridis), XP 522310.1 (Pan troglodytes), XP 586544.2 (Bos taurus), and XP 783902.1 (Strongylocentrotus purpuratus).
Our two-hybrid screen identified Ninjurin A (NijA), and subsequent database searching identified two other Drosophila genes containing ninjurin domains: CG11637, which we call Ninjurin B (NijB), and CG14394, which we call Ninjurin C (NijC). To determine the evolutionary relationships between the different ninjurin family members, we built a phylogenetic tree of all the ninjurin domain-containing proteins that are reported in GenBank (Fig. 2D). This analysis suggests that insects and vertebrates independently duplicated a primordial ninjurin; consequently, NijA appears equally related to vertebrate Ninjurin-1 and Ninjurin-2.
Drosophila genome-wide expression studies have shown that macrophage-like S2 cells immediately up-regulate NijA and Mmp1 in response to lipopolysaccharide (Boutros et al. 2002); and both NijA and Mmp1 are up-regulated in wounded Drosophila adult whole animals (De Gregorio et al. 2001). These expression data suggest that NijA participates in the inflammatory response, which is interesting because MMPs are already known to play important roles in inflammation (Parks et al. 2004). Because of its coregulation with Mmp1 and its role as an adhesion molecule, we chose to pursue NijA as a potential interactor with Mmp1.
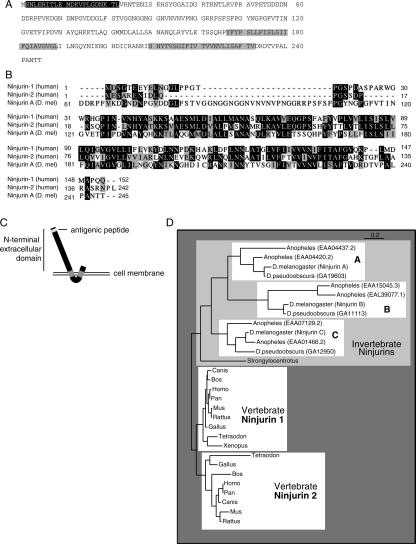
Vertebrate MMPs are known to have overlapping substrate specificities. To determine the specificity of NijA protein binding to the Drosophila MMP family, we tested several constructs by yeast two-hybrid assay (Fig. 3). Mmp1 has two splice forms that differ by only a few amino acids within the hemopexin domain at the C terminus of the protein (Fig. 3B), and the functional significance of these splice forms is unknown. The original bait that identified NijA was the hemopexin domain of splice form 1 (Mmp1.f1). The full-length Mmp1.f1, containing a mutation at the proteolytic core designed to mimic catalytically dead vertebrate MMP mutants (Crabbe et al. 1994), bound NijA equally well by the yeast two-hybrid assay. In contrast, the Mmp1.f2 hemopexin domain did not interact with NijA; a full-length, catalytically dead Mmp1.f2 interacted only weakly with NijA. This indicates that the hemopexin domain of Mmp1.f1 is a major site of NijA interaction, and the isoform 1-specific last 40 amino acids are absolutely required for this binding. It also suggests that there is a secondary weak interaction at the catalytic domain, which is the same in splice forms 1 and 2. Interestingly, for Mmp2, neither the hemopexin domain nor the catalytically dead full-length Mmp2 interacted with NijA in the two-hybrid screen, although other two-hybrid targets were shared by the two fly MMPs (data not shown). These experiments demonstrated that the last 40 amino acids of Mmp1.f1 are necessary for a strong interaction with NijA.
Figure 3.
Specificity of the two-hybrid interaction between NijA and Mmp1. (A) The indicated bait constructs were tested for their ability to interact with a NijA prey construct. Interaction was measured by the growth of 18 replicate colonies on −leu −trp medium. (B) Comparison of the form 1 and form 2 splice forms of Mmp1.
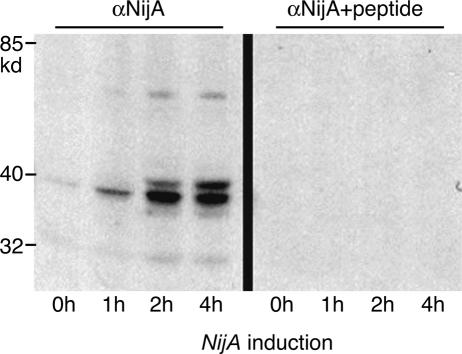
To further investigate the interaction between Mmp1 and NijA proteins, we raised guinea pig antibodies against an N-terminal peptide of NijA protein (Fig. 2A). We determined the specificity of these antibodies by Western blots of S2 cells stably transformed to express NijA. Uninduced cells showed a faint NijA immunoreactive band that was strongly intensified after 4-h NijA induction. Induced NijA protein appeared as a doublet ∼40 kd in molecular weight, with fainter 70- and 32-kd bands appearing as well (Fig. 4). Each of these bands was competed away by preabsorbing the antibodies with the peptide to which they were raised (Fig. 4). NijA protein is predicted to be a 27-kd protein with multiple potential glycosylation sites, and thus, the observations that these higher molecular weight bands are induced with a NijA transgene and are competed away by the immunogenic peptide indicate that NijA protein is subject to post-translational modifications.
Figure 4.
NijA antibody. S2 cells stably transformed with a copper-inducible NijA plasmid expressed a protein recognized by anti-NijA antibodies when induced. (Right side) Cell extracts probed with anti-NijA whole serum. (Left side) Cell extracts probed with anti-NijA antibodies preabsorbed against the antigenic peptide, demonstrating the specificity of the antibodies. Length of NijA induction is shown for each lane.
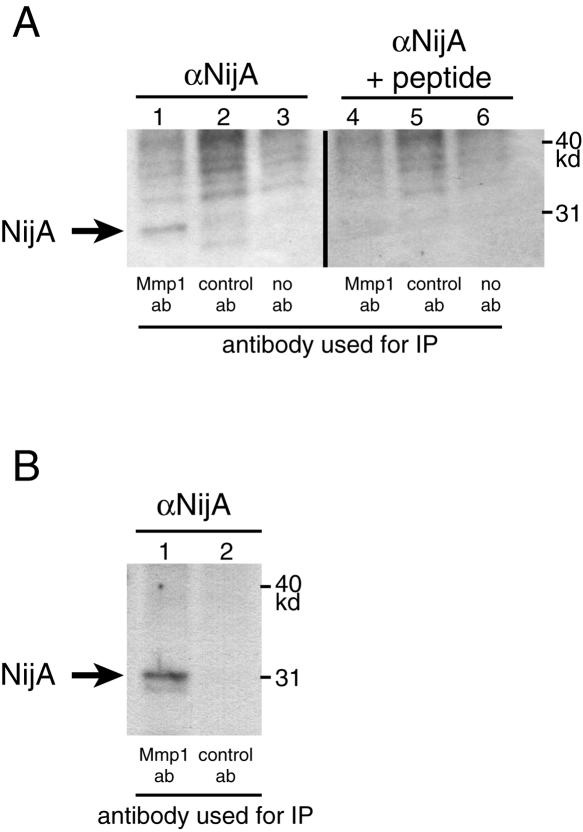
We used these antibodies to examine whether NijA protein is pulled down by anti-Mmp1 antibodies in an immunoprecipitation. Because of concern that a substrate might be cleaved rather than precipitated, we used extracts of whole-transgenic flies expressing a catalytically dead Mmp1. As shown in Figure 5, the 32-kd form of NijA protein was coprecipitated with anti-Mmp1 antibodies. The band was eliminated when competitor peptide was added to the hybridizing anti-NijA antibody, and the band was not evident when a control monoclonal antibody was used to probe the precipitate; these control experiments confirm that NijA protein coimmunoprecipitates with Mmp1 protein. To determine whether NijA protein was able to coimmunoprecipitate with wild-type Mmp1 protein in addition to catalytically dead Mmp1 protein, we first affinity purified the anti-NijA antibody to reduce background. We used these purified antibodies to probe blots of the precipitates of wild-type animals pulled down with anti-Mmp1 antibodies. Importantly, a band of the expected size was identified in immunoprecipitations with anti-Mmp1 antibodies and not with control antibodies (Fig. 5B). These results indicate that NijA and wild-type Mmp1 proteins can form a stable complex.
Figure 5.
NijA coimmunoprecipitates with Mmp1 in vivo. (A) NijA coimmunoprecipitates with catalytically inactive Mmp1. Protein extracts were made from transgenic animals expressing catalytically dead Mmp1.f1 ubiquitously (genotype tubP-GAL4/+; UAS-Mmp1.f1E225A/+). Extracts were precipitated with mixed monoclonal anti-Mmp1 antibodies (lanes 1, 4), control monoclonal antibodies (lanes 2, 5), or no primary antibodies (lanes 3, 6). Two identical blots were probed: one with anti-NijA antibodies (lanes 1–3), and one with anti-NijA antibodies preabsorbed with the competing immunogenic peptide (lanes 4–6). (B) NijA coimmunoprecipitates with endogenous wild-type Mmp1 in vivo. Protein extracts were made from wild-type animals and precipitated with mixed monoclonal anti-Mmp1 antibodies (lane 1) or control antibodies (lane 2). The blot was probed with affinity-purified anti-NijA antibodies.
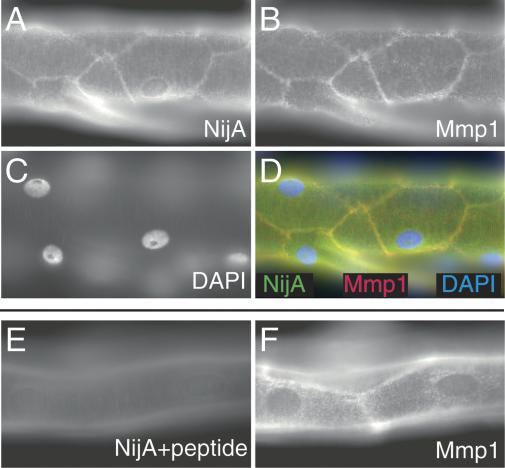
To determine whether this physical interaction might be relevant in vivo, we examined the expression of Mmp1 and NijA proteins in wild-type third instar tracheal trunks, a tissue affected by mutations in Mmp1 (Page-McCaw et al. 2003). The dorsal trunks are multicellular tubes consisting of large polyploid cells. We identified NijA at cell surfaces within the tracheal tubes, a pattern consistent with it being a transmembrane protein (Fig. 6A). Significantly, in double-labeling experiments, we identified Mmp1 in the same pattern as NijA (Fig. 6B,D). When the NijA antibodies were prehybridized with the antigenic peptide, the membrane localization was not observed (Fig. 6E), although this did not interfere with the Mmp1 signal (Fig. 6F). This colocalization, in concert with the coimmunoprecipitation, strongly suggests that Mmp1 and NijA interact in the tracheal epithelium.
Figure 6.
NijA and Mmp1 colocalize to cell surfaces in trachea. A–D are images of one sample; E–F are images of another sample. (A) NijA whole serum recognizes cell surfaces in tracheal dorsal trunks. (B) Mmp1 antibodies also recognize cell surfaces in dorsal trunks. (C) DAPI staining of nuclei in the same sample. (D) A merged image of the images in A–C showing the colocalization of NijA and Mmp1. (E) The cell surface staining is blocked by preabsorbing the NijA antibody with the antigenic peptide, demonstrating the specificity of the antibody in fixed tissue. (F) Mmp1 staining is unaffected by antibody preabsorption with the NijA antigenic peptide.
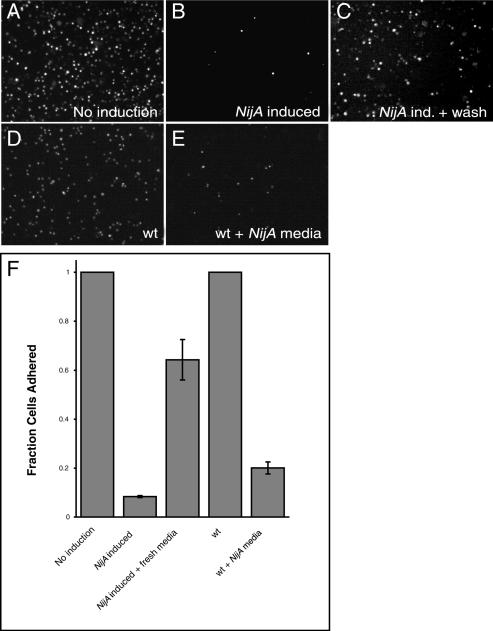
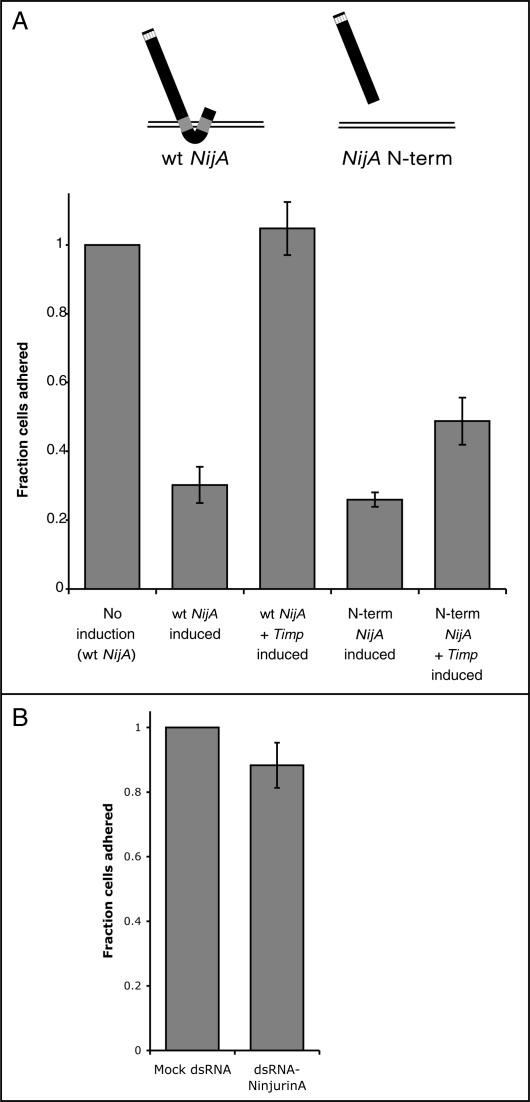
To determine the functional relationship between NijA and Mmp1, NijA was overexpressed in stably transformed S2 cells under a copper-inducible promoter. S2 cells are weakly adherent Drosophila cells that can be grown either in suspension or on surfaces, and they express both NijA (weakly) and Mmp1 endogenously (Fig. 4; Page-McCaw et al. 2003). When we examined fixed S2 cells overexpressing NijA, we found that few cells remained adhered to the glass slide (Fig. 7A,B). This absence of cells adhered to the slide was not caused by death of the S2 cells, as trypan blue exclusion indicated that the cells expressing NijA were not dying (data not shown). Because the expression of either vertebrate Ninjurin-1 or Ninjurin-2 in Jurkat cells causes cells to aggregate (Araki and Milbrandt 1996, 2000), we considered the possibility that cells were not adhering to the slide because they were aggregating with each other. We examined S2 cells expressing NijA for their ability to aggregate into clumps of four or more cells when grown in suspension (Berndorff et al. 1994), but we found that aggregated cells accounted for only ∼5%–10% of the total, whether or not cells were expressing NijA, even when we inhibited Mmp1 activity (data not shown). This data was consistent with our visual inspection of NijA-expressing cells, and indicated that NijA was not causing cells to aggregate. Rather, NijA expression interfered with the ability of cells to adhere to both glass slides and plastic tissue culture dishes. We developed an assay to measure their adhesive capacity; we induced NijA for 3 h by the addition of copper to the medium, then put the cells into a depression slide well and let them settle for 2 h, after which we removed the medium, fixed the cells, and stained them with DAPI. NijA induction caused a striking failure of adherence compared with uninduced cells (Fig. 7A,B,F). Because NijA induced a loss of cell- surface adhesion, it appeared to function oppositely from vertebrate Ninjurin-1 and Ninjurin-2, which increase cell–cell adhesion.
Figure 7.
NijA conditions the medium to inhibit cell adhesion. (A–F) _NijA_-expressing cells were induced for 3 h; half were resuspended in fresh medium, and both samples were allowed to settle for 2 h on glass slides (B, C; columns 2,3 in F). Wild-type cells plated at the same density as the NijA cells were split; half were resuspended in the medium conditioned by the NijA cells, and both samples were allowed to settle on glass slides (D, E; columns 4,5 in F). (A–E) Photographs of DAPI-stained cells of the indicated genotypes, treated as described: (A) _NijA_-transformed cells, not induced to express NijA. (B) _NijA_-transformed cells induced to express NijA for 3 h. (C) _NijA_-transformed cells induced to express NijA for 3 h, then resuspended in fresh medium. (D) Wild-type cells. (E) Wild-type cells removed to _NijA_-conditioned medium. (F) Quantification of the fraction of adherent cells. The fraction of adherent cells in columns 1–3 was determined by dividing the number of adherent cells counted in each sample by the number of adherent _NijA_-uninduced cells counted in the sample shown in column 1. The fraction adhering in columns 4 and 5 was determined by dividing the number of adherent cells by the number of wild-type adherent cells in the sample shown in column 4. Thus, columns 1 and 4 are defined as 1. Error bars indicate SEM; data from four experiments are shown.
Importantly, the ability of _NijA_-expressing cells to adhere was significantly restored if they were resuspended in fresh medium after the 3-h induction and then allowed to settle for 2 h (Fig. 7B,C,F). Inversely, when wild-type cells were allowed to settle in medium that had been conditioned for 3 h by _NijA_-expressing cells, the wild-type cells were unable to adhere (Fig. 7D–F). These reciprocal experiments demonstrated that the activity causing cells to lose adhesion was secreted or shed into the medium by the _NijA_-expressing cells, and this activity was sufficient to make wild-type cells fail to adhere. Thus, the NijA activity works in trans, or cell nonautonomously.
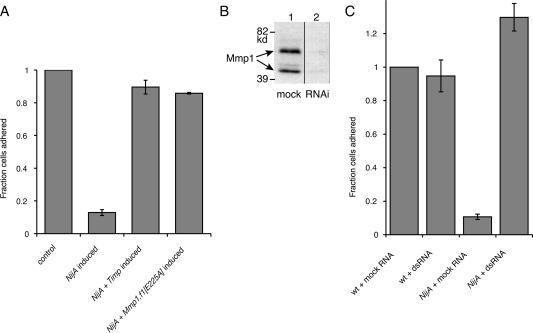
We previously demonstrated that S2 cells express both fly MMPs endogenously (Page-McCaw et al. 2003). Because we identified NijA as an Mmp1 interactor, we tested whether the presence of the _NijA_-induced activity in the medium was Mmp1 dependent. We coexpressed the Drosophila MMP inhibitor Timp (tissue inhibitor of metalloproteinases) (Godenschwege et al. 2000; Page-McCaw et al. 2003) with NijA. When NijA was expressed in the presence of the Timp inhibitor, cells maintained their attachment to the surface, similar to uninduced cells and in contrast to cells where NijA was induced without the Timp inhibitor (Fig. 8A). Because fly Timp can also inhibit other metalloproteinases as well as MMPs (Wei et al. 2003), we determined whether expression of the catalytically inactive Mmp1, carrying the E225A mutation, was able to block the _NijA_-induced loss of cell adhesion. We transfected the NijA cell lines with a copper-inducible Mmp1.f1E225A transgene. When induced, these cells expressing NijA and the catalytically dead Mmp1 were able to adhere at a rate similar to that of control-transfected cells (Fig. 8A). The restoration of cellular adhesion indicated that the catalytic activity of Mmp1 is required for the _NijA_-induced loss of cell adhesion, and the catalytically inactive Mmp1 was acting in a dominant negative fashion.
Figure 8.
Mmp1 activity is required for the _NijA_-mediated loss of adhesion. (A) S2 cells stably transformed with inducible NijA were transiently transformed with MT-GAL4 and UAS-Timp, so that the addition of copper induced the expression of Timp (Klueg et al. 2002); or the plasmid pRmHa-3.Mmp1.f1E225A, which expresses a catalytically inactive mutant of Mmp1 upon induction; or MT-GAL4 as a control. Column 1 shows the baseline adherence of uninduced control-transfected cells. Column 2 shows the adherence of control-transfected cells induced to express NijA. Column 3 shows the adherence of cells transfected and induced to express NijA and Timp. Column 4 shows the adherence of cells transfected and induced to express NijA and Mmp1.f1E225A. Adherent fractions were determined by normalizing to the number of adherent cells in the control transformed, uninduced cells (shown in column 1). Error bars indicate SEM; data from three experiments are shown. (B) Western blot with anti-Mmp1 antibodies to determine the level of Mmp1 in dsRNA experiments. (Lane 1) Untreated cells. (Lane 2) Cells treated with dsRNA against Mmp1. (C) S2 cells stably transformed with inducible NijA were treated with dsRNA against Mmp1, or mock-treated with no RNA. Column 1 shows the fraction of adherent wild-type cells mock treated and put under induction conditions. Column 2 shows the fraction of adherent wild-type cells treated with dsRNA against Mmp1. Column 3 shows the fraction of adherent mock-treated _NijA_-expressing cells. Column 4 shows the fraction of adherent _NijA_-expressing cells, treated with dsRNA against Mmp1. Adherent fractions were determined by normalizing to the number of wild-type mock-treated adherent cells (column 1). Error bars indicate SEM; data from four experiments are shown.
To independently confirm the requirement of Mmp1 for the NijA phenotype, we used an RNA interference (RNAi) strategy to knockdown levels of Mmp1 specifically. The addition of Mmp1 double-stranded RNA (dsRNA) reduced Mmp1 expression in S2 cells substantially, as observed by Western blot (Fig. 8B). _NijA_-expressing cells treated with dsRNA against Mmp1 retained their surface adhesion (Fig. 8C). Thus, Mmp1 expression is required for the _NijA_-induced loss of adhesion. These three experiments, coexpression of Timp, coexpression of catalytically inactive Mmp1, and RNAi knock-down of Mmp1, demonstrate that Mmp1 activity is required for cells to lose adhesion in response to NijA expression.
As medium conditioned by NijA-expressing cells causes wild-type cells to lose adhesion, the simplest explanation for the role of Mmp1 in this process is that the Mmp1 protease cleaves off, or sheds, an extracellular domain of NijA protein into the medium, where it acts on cells in trans. NijA protein has two extracellular domains at the N and C termini; because the N-terminal domain is the most highly conserved (Fig. 2B), we hypothesized that the Mmp1 protease was releasing the N-terminal extracellular domain (ectodomain) of NijA into the medium. Although our anti-NijA antibodies recognize the N-terminal domain, we were unable to demonstrate the presence of this domain in the conditioned medium, possibly because the N terminus was further processed, removing the epitope. As an alternate means of testing our hypothesis that the Mmp1 protease liberates the N-terminal domain of NijA protein, we transiently transfected cells with a copper-inducible expression construct containing a secreted NijA N-terminal domain (Fig. 9A). A full-length NijA control plasmid demonstrated that the loss of cell adhesion was observed under transient transfection conditions (Fig. 9A). The expression of a secreted N-terminal domain of NijA, engineered with an exogenous signal sequence to ensure that it was secreted from the cell, completely replicated the _NijA_-induced failure of adherence. These experiments demonstrate that the release of the NijA N terminus from the cells into the medium is sufficient to cause the _NijA_-induced loss of cell adhesion.
Figure 9.
The secreted N-terminal fragment of NijA signals the loss of cell adhesion in an MMP-independent manner. (A) Wild-type S2 cells were transiently transfected with the NijA constructs shown, either with or without the MT-GAL4 and UAS-Timp plasmids that confer Timp inducibility. Fraction of adherent cells is shown for each of the genotypes. (Column 1) Adherent fractions were normalized to the number of adherent cells in uninduced cells transformed with wild-type NijA. Error bars indicate SEM. Data from three experiments are shown in columns 2 and 3; six experiments are shown in columns 4 and 5. (B) Wild-type S2 cells were treated with dsRNA against NijA to knock down the endogenous protein levels or mock treated with no RNA. Column 2 shows the fraction of adherent cells after dsRNA treatment, normalized to the number of adherent cells in column 1 (mock-treated). Error bars indicate SEM; data from three experiments are shown.
To address the role of Mmp1 in the release of NijA N terminus into the medium, we coexpressed these NijA constructs with Timp. As expected, transiently transfected full-length NijA was not able to induce the loss of cell adhesion in the presence of the MMP inhibitor Timp (Fig. 9A), as Mmp1 activity is required for the induction of this phenotype. In contrast, however, when Timp was coexpressed with the secreted N-terminal fragment of NijA, the fragment was still able to promote cells to lose adhesion (Fig. 9A). The full-length NijA is completely unable to cause the loss-of-adhesion phenotype in the presence of Timp inhibitor, whereas the secreted N terminus is still able to induce ∼50% of the cells to lose adhesion in the presence of the inhibitor. This relaxation of the requirement for Mmp1 indicates that MMP proteolysis is no longer an essential step for the induction of the NijA phenotype once the N terminus of NijA is released from the cell surface. These results indicate that Mmp1 is responsible for the generation of a NijA N-terminal fragment, which acts nonautonomously on cells to cause them to lose adhesion to surfaces.
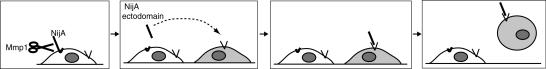
The Mmp1-liberated NijA fragment could, in principle, act as a signaling molecule, inducing cells to alter their adhesive properties. Alternatively, if intact endogenous NijA is responsible for maintaining cell adhesion directly, the liberated ectodomain could act as a dominant negative, inhibiting the intact NijA from contacting surfaces. To test the role of NijA directly, we knocked down the expression of endogenous NijA by RNAi and examined whether the adhesion of cells was affected. We determined that NijA is not required to adhere cells to surfaces (Fig. 9B). Thus, we conclude that the NijA ectodomain acts as a signaling molecule, carrying information to surrounding cells instructing them to release adhesion (Fig. 10).
Discussion
We report here the physical interaction between a Drosophila matrix metalloproteinase and a fly homolog of the ninjurin family of cellular adhesion modulators. The interaction between Mmp1 and NijA proteins has been demonstrated by yeast two-hybrid and by coimmunoprecipitation of the endogenous wild-type proteins from flies, and the two proteins colocalize in vivo. Yeast two-hybrid assays suggest that a small motif of Mmp1 is required for this physical interaction in yeast. When NijA is expressed in S2 cells, it causes the cells to lose adhesion to glass or plastic by secreting an activity into the medium. This activity can act nonautonomously on nonexpressing wild-type cells, and the phenotype can be rescued by replacement of the medium with fresh, nonconditioned medium. This activity is not observed when Mmp1 is inhibited by any of three methods: coexpression of the Timp inhibitor, coexpression of a dominant-negative catalytically dead Mmp1, or RNAi knock-down of Mmp1 protein levels. When the N terminus of NijA is secreted directly into the medium, MMP activity is not absolutely required for the cells to lose adhesion. These data, in combination, demonstrate that the Mmp1 protease is responsible for shedding the N terminus of NijA protein from cells, and that this N terminus acts nonautonomously on cells to cause them to lose adhesion (Fig. 10). The shedding of adhesion molecule ectodomains by metalloproteases has many precedents, including the MMP-7-mediated shedding of E-cadherin (McGuire et al. 2003). However, this is the first report of shedding in the ninjurin family.
Ninjurin-1 was originally identified as a message up-regulated in injured rat neurons and subsequently discovered to be widely expressed in many organs and tissue types. Both vertebrate Ninjurin-1 and Ninjurin-2 are two-pass transmembrane proteins, and expression of either causes cultured Jurkat cells to aggregate in a homotypic manner and causes cultured neurons to extend longer processes. These cellular phenotypes suggest that they function as cell-adhesion molecules. Drosophila NijA shares a conserved ninjurin domain with the vertebrate family members and is predicted to have the same topology as in vertebrates. The conserved domain encompasses roughly the last half of the protein, and it includes a region important for homotypic adhesion, identified in Ninjurin-2 as amino acids 15–45 (Araki and Milbrandt 1996). Our data suggest an opposite function for Drosophila NijA (loss of adhesion) compared with the vertebrate ninjurins (promotion of adhesion), although it is possible that these phenotypes can be reconciled. Our RNAi data suggests that in the absence of Mmp1, high levels of NijA may indeed promote increased adhesion to a substrate in our system. If this were the case, NijA would control both functions, acting as a switch upon cleavage to change from promotion of adhesion to loss of adhesion. NijA is not solely responsible for the adhesion of S2 cells, however, because when NijA is knocked down with RNAi, the cells remain adherent. Although a cell–cell adhesion assay did not detect any homotypic adhesion in _NijA-_expressing S2 cells, it is possible that S2 cells are not a cell type that can support _NijA_-mediated cell–cell homotypic adhesion, but that other cell types might be capable of binding to each other via _NijA_-mediated homotypic adhesion.
The simplest explanation for our data is that the Mmp1 protease cleaves NijA. We have demonstrated a physical interaction between Mmp1 and NijA proteins. Using an S2 cell-adhesion assay, we have also demonstrated that NijA’s activity in S2 cells is dependent on Mmp1 expression and catalytic activity, and that this proteolytic function is no longer required once the NijA ectodomain is shed by another mechanism. However, we cannot completely exclude the possibility that Mmp1 and NijA are two components of a larger proteolytic complex, and that another member of this complex is responsible for the direct cleavage of NijA.
Once NijA is liberated from the cell surface by Mmp1, it acts nonautonomously to change the adhesive character of cells. This is evident because medium conditioned by _NijA_-expressing cells changes the adhesiveness of wild-type cells. The simplest explanation is that the NijA N-terminal fragment itself binds to the receiving cell, to a receptor or docking molecule that is constitutively expressed. Our data are consistent with the NijA-derived signal either directly blocking cell surface adhesion receptors, or initiating a fast-acting signaling cascade whose endpoint is changing the adhesive properties of the cell.
Significance for tracheal development
The Mmp1 mutant tracheal phenotype had previously indicated that Mmp1 was required for regulation of adhesion in the tracheal system. Our findings that Mmp1 and NijA physically interact in vivo, colocalize to the tracheae, and regulate cell adhesion in culture strongly suggest that Mmp1 and NijA regulate cell adhesion in the tracheal system through a similar mechanism. Our model is that the Mmp1-generated ectodomain of NijA is required for tracheal cell release from the cuticle, both at molting for tube dilation and throughout larval growth to allow the tubes to elongate. Because it is cell nonautonomous, the NijA ectodomain signal may allow the tracheal epithelial cells to act in concert as they detach from the ECM. Without Mmp1, the extracellular NijA signal is not produced, and the cells cannot coordinate their detachment from the cuticle. This model predicts that NijA mutants would also have defective tracheae. A preliminary analysis of recently generated NijA mutants indicates that these larvae have tracheal phenotypes that are similar to Mmp1 mutant tracheae, including stuck cuticles, hypoxia-induced behavior, and tracheal breaks, although they appear to have highly variable penetrance (N. Simms, A. Kabra, and A. Page-McCaw, unpubl.).
Significance for inflammation
Although there are no reports of the study or analysis of fly NijA, microarray analysis has identified it as a message immediately up-regulated in response to the bacterial endotoxin LPS. Similarly, microarrays have shown that NijA is up-regulated 12-fold in whole animals within 1.5 h of septic wounding (De Gregorio et al. 2001). The up-regulation of NijA in response to wounding and to the bacterial product LPS implicates it in inflammation.
The assay we used to analyze the interaction between NijA and Mmp1, that of cell adhesion to glass, is an in vitro model for some aspects of insect immunity. In vivo, wild-type hemocytes in a functional insect immune system adhere to and encapsulate or engulf foreign invaders (Evans et al. 2003). However, insect hemocytes lose the ability to adhere to foreign substances when infected with the Microplitis demolitor bracovirus in vivo, because the virus suppresses the immune system (Schmidt et al. 2001). Similarly, insect hemocyte cell lines in culture also lose the ability to adhere to foreign substances, including glass, when infected with this virus. This loss-of-adhesion phenotype has been reproduced by the transgenic expression of a single viral gene product, Glc1.8, in both cultured S2 cells and another insect hemocyte cell line, High Five cells (Beck and Strand 2005). Thus, for induction of the Glc1.8 viral protein, the S2 loss-of-adhesion to glass is an assay for immunosuppression. Because NijA expression in S2 cells phenocopies Glc1.8 expression, one interpretation is that Mmp1-processed NijA acts as an endogenous negative regulator of the immune/inflammatory response. We report here a novel mechanism by which a Drosophila MMP can modify cell adhesion through the generation of a liberated ninjurin ectodomain. It will be interesting to determine whether MMPs and ninjurins function together in innate immunity.
Materials and methods
Two-hybrid screening
The bait plasmid pGBD-C1.Mmp1pexf1 was constructed by inserting the hemopexin domain (beginning at isoleucine 299) amplified from an Mmp1.f1 cDNA (Page-McCaw et al. 2003) into pGBD-C1 (James et al. 1996). We screened 3.6 × 105 cDNAs from a Drosophila embryo library and 4.4 × 105 cDNAs from a third instar larval library (Durfee et al. 1993) using the methods of James et al. (1996). Prey sequence tags were amplified directly out of yeast: Colonies were lysed by microwaving for 1 min and PCR mix was added directly to the lysate. Sequences were BLASTed to retrieve predicted full-length genes, and assigned cellular locations with SignalP (Nielsen and Krogh 1998), PSORT (Nakai and Horton 1999), and TMHMM (Krogh et al. 2001). Two-hybrid binding specificity of prey constructs was tested against pGBD.dLRPC (arrow cytoplasmic domain) and pGBD.ZAPS1 (full-length Ago1) generously provided by Brian Avery and Bobby Williams. MMP specificity of the NijA prey was tested against the following MMP bait constructs: Mmp1.f1 starting at Ser 106 (pro domain removed) carrying the E225A mutation at the active site; Mmp1 splice form 2 hemopexin domain starting at Ser 298; Mmp1.f2 starting at Ser 106 (pro domain removed) carrying the _E225A_-inactivating mutation; Mmp2 hemopexin domain starting at Asp 514; and Mmp2 beginning at Ser 130 (pro domain removed) carrying the mutation E258A. Point mutations were engineered by splice overlap extension (Vallejo et al. 1994). Specificity was tested by transforming yeast with bait and prey plasmids and testing 18 replicate patches for growth on selective medium.
Phylogenetic analysis
The Web-based program Blockmaker (Henikoff et al. 1995) was used to identify stretches of homology in rat Ninjurin-1 and Ninjurin-2 and the three Drosophila ninjurin family members. The Cobbler consensus sequence (Henikoff and Henikoff 1997) of the Motif blocks identified were used to search the GenBank database (Henikoff and Henikoff 1994). To generate a tree, all sequences identified (those in Fig. 2D and the bacterial CAD14037.1 from Ralstonia solanacearum as an outgroup) were aligned with Blockmaker and drawn with njplot. Danio rerio sequences were excluded, as they appeared incomplete or anomalous.
Antibodies, Western blotting,and immunoprecipitations
Guinea pig anti-NijA antibodies were generated against a synthesized peptide corresponding to residues 2–22 of NijA. For Western blots, whole serum was used at 1:5000; preabsorbed antibody was preincubated 30 min with the peptide (1 mg/mL), both diluted 1:5000. NijA antibodies were affinity-purified against the peptide using the Sulfo-Link kit (Pierce) and were diluted 1:200 for probing blots. HRP-labeled goat anti-guinea pig (Santa Cruz) was diluted 1:5000. Western protein samples were prepared by boiling cells in Laemmli sample buffer.
Immunoprecipitations were performed on extracts of 10 white prepupae dounce-homogenized in 10 mL lysis buffer (150 mM NaCl, 1% NP40, 50 mM Tris 8.0, protease inhibitors [Roche Mini-Complete]). Supernatant (1.5 mL) was precleared by addition of 75 μL of normal rabbit serum (Calbiochem) and 150 μL of fixed Staphylococcus aureus cowan (Calbiochem). Complexes were precipitated with Protein G Plus/Protein A beads (Calbiochem). Blots were probed with monoclonal anti-Mmp1 antibodies 5H7, 3B8, and 3A6 mixed 1:1:1 (Page-McCaw et al. 2003), or control monoclonal anti-Cyclin B F2F4 (donated by Pat O’Farrell to the Developmental Studies Hybridoma Bank).
NijA and Mmp1 colocalization experiments were performed by fixing dissected third instar larvae in 4% formaldehyde for 20 min, hybridizing with an anti-Mmp1 monoclonal cocktail (a 1:1:1 mixture of 3B8, 5H7, and 23G1, diluted 1:100) and anti-NijA whole serum (1:500) simultaneously. Secondary antibodies were Cy3 goat anti-mouse (1:1500, Jackson) and FITC donkey anti-guinea pig (1:500, Jackson Laboratory). Competing peptide (1 mg/mL stock) was added to primary antibodies at 1:500 1 h before antibody hybridization to samples. Tracheal trunks were dissected out before mounting and photographing with a Zeiss AxioImager Z1.
S2 cell culture
Drosophila S2 cells were maintained at 25°C in Schneider’s Drosophila medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 U/mL Pen-strep (Cellgro), and 2 mM L-glutamine (Gibco). Gene expression from the pRmHa-3 metallothionine promoter was induced with 0.7 M copper sulfate diluted 1:1000. Trypan blue was 0.4% in PBS.
Cells were transiently transfected with 5 μg of total DNA using Superfect (Qiagen). Plasmids transfected were pMT-GAL4 (Klueg et al. 2002) and UAS-Timp (Page-McCaw et al. 2003); pRmHa-3.Mmp1f1E225A, where the mutation E225A renders the protease catalytically inactive; pRmHa-3.NijAA, made by PCR-amplifying the NijA (isoform A) cDNA RE57443; and the _NijA_-secreted N terminus plasmid, pRMHa-3.SS-NijAA-EC1, made by splice overlap extension of the signal sequence from Mmp1 with the 5′ end of the NijA sequence encoding a protein ending with His161. Adhesion assays were performed 24 h after transfection.
Stably transformed NijA cell lines were made by transfecting wild-type S2 cells with pRmHa-3.NijAA-polyo, containing the NijA cDNA fused to a 3′ polyoma epitope and cotransfecting with the α-amanitin-resistant plasmid pPC4. After 48 h, transformed cells were selected with 5 μg/mL α-amanitin (Sigma) in 50% conditioned medium for three to four passages, checked for NijA expression, and cloned by limiting dilution.
Adhesion assay
Cells were seeded in duplicate at 3 × 106 cells/well in 3 mL of complete medium in a six-well plate and grown overnight at 25°C. One well was induced for 3 h. Cells were resuspended, and 100 μL of each sample was placed in one well of a 12-well Multitest slide (MP Biomedicals) and allowed to settle for 2 h. The medium was removed and cells were fixed with 2% formaldehyde (Ted Pella) in PBS. After washing in PBS, cells were stained with 0.1 μg/mL DAPI. A researcher blinded to sample identity photographed 10 randomly chosen fields with a 20× objective mounted on a Zeiss AxioImager Z1 epifluorescence microscope. The threshold level tool of the program ImageJ was used to count DAPI-stained nuclei in photographs, and the sum from the 10 fields was divided by a control sum where NijA was not induced (as indicated in the figure legends) to determine the fraction cells adhered.
For medium-swapping experiments, after the 3-h NijA induction, cells were pelleted and half were resuspended in fresh medium. Simultaneously, wild-type cells plated at the same density as the NijA cells were pelleted, and half were resuspended in the _NijA_-conditioned medium. One-hundred microliters of each of the four samples was added to single wells of a 12-well slide and allowed to settle for 2 h, then processed as above.
DsRNA was made with the Megascipt T7 kit (Ambion) using 5 μL of a 1.6-kb Mmp1 PCR product or 1.2-kb NijA PCR product generated with primers carrying exogenous T7 sites. Thirty micrograms of dsRNA was added to 3 × 106 S2 cells in 1 mL of serum-free medium in a six-well plate. After gentle rocking for 30 min at room temperature, 2 mL of complete medium was added. Forty-eight hours later, cells were assayed for adhesion.
Figure 10.
Proposed mechanism of Mmp1 and NijA promoting the loss of cell adhesion. Our data indicate that Mmp1, shown as scissors, is responsible for shedding the NijA extracellular domain. The white cell expresses NijA and the gray cell does not. After cleavage, the liberated ectodomain of NijA binds to an unknown receptor (shown as a V) on the gray cell in a nonautonomous fashion. The gray cell then loses cell adhesion and releases from the substrate.
Acknowledgments
The early phases of this work were initiated in the laboratory of Gerald M. Rubin when A.P-M. was a post-doctoral fellow, and we gratefully acknowledge his support, resources, and generosity. We thank David King at HHMI/UCBerkeley for peptide synthesis, Jessica McGovern for initiating the construction of the secreted N-terminal NijA plasmid, and Patrick Page-McCaw for helpful ideas and discussion. Bobby Williams and Brian Avery kindly provided unpublished two-hybrid bait constructs. We acknowledge the Developmental Studies Hybrodoma Bank for antibodies. We are grateful to Heather Broihier, Fern Finger, and Patrick Page-McCaw for critically reading the manuscript.
Footnotes
References
- Araki T., Milbrandt J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron. 1996;17:353–361. doi: 10.1016/s0896-6273(00)80166-x. [DOI] [PubMed] [Google Scholar]
- Araki T., Milbrandt J. Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J. Neurosci. 2000;20:187–195. doi: 10.1523/JNEUROSCI.20-01-00187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T., Zimonjic D.B., Popescu N.C., Milbrandt J. Mechanism of homophilic binding mediated by ninjurin, a novel widely expressed adhesion molecule. J. Biol. Chem. 1997;272:21373–21380. doi: 10.1074/jbc.272.34.21373. [DOI] [PubMed] [Google Scholar]
- Beck M., Strand M.R. Glc1.8 from Microplitis demolitor bracovirus induces a loss of adhesion and phagocytosis in insect high five and S2 cells. J. Virol. 2005;79:1861–1870. doi: 10.1128/JVI.79.3.1861-1870.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel G.J., Krasnow M.A. Genetic control of epithelial tube size in the Drosophila tracheal system. Development. 2000;127:3271–3282. doi: 10.1242/dev.127.15.3271. [DOI] [PubMed] [Google Scholar]
- Berndorff D., Gessner R., Kreft B., Schnoy N., Lajous-Petter A.M., Loch N., Reutter W., Hortsch M., Tauber R. Liver–intestine cadherin: Molecular cloning and characterization of a novel Ca2+-dependent cell adhesion molecule expressed in liver and intestine. J. Cell Biol. 1994;125:1353–1369. doi: 10.1083/jcb.125.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., Agaisse H., Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- Coussens L.M., Fingleton B., Matrisian L.M. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Crabbe T., Zucker S., Cockett M.I., Willenbrock F., Tickle S., O’Connell J.P., Scothern J.M., Murphy G., Docherty A.J. Mutation of the active site glutamic acid of human gelatinase A: Effects on latency, catalysis, and the binding of tissue inhibitor of metalloproteinases-1. Biochemistry. 1994;33:6684–6690. doi: 10.1021/bi00187a039. [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P.T., Rubin G.M., Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Egeblad M., Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Evans C.J., Hartenstein V., Banerjee U. Thicker than blood: Conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell. 2003;5:673–690. doi: 10.1016/s1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Godenschwege T.A., Pohar N., Buchner S., Buchner E. Inflated wings, tissue autolysis and early death in tissue inhibitor of metalloproteinases mutants of Drosophila. Eur. J. Cell Biol. 2000;79:495–501. doi: 10.1078/0171-9335-00072. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J.G. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J.G. Embedding strategies for effective use of information from multiple sequence alignments. Protein Sci. 1997;6:698–705. doi: 10.1002/pro.5560060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J.G., Alford W.J., Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S.A., Mankani M., Robey P.G., Poole A.R., Pidoux I., et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- Inada M., Wang Y., Byrne M.H., Rahman M.U., Miyaura C., Lopez-Otin C., Krane S.M. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl. Acad. Sci. 2004;101:17192–17197. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klueg K.M., Alvarado D., Muskavitch M.A., Duffy J.B. Creation of a GAL4/UAS-coupled inducible gene expression system for use in Drosophila cultured cell lines. Genesis. 2002;34:119–122. doi: 10.1002/gene.10148. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuo C.T., Jan L.Y., Jan Y.N. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin–proteasome, and ecdysone signaling. Proc. Natl. Acad. Sci. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert V., Wielockx B., Munaut C., Galopin C., Jost M., Itoh T., Werb Z., Baker A., Libert C., Krell H.W., et al. MMP-2 and MMP-9 synergize in promoting choroidal neovascularization. FASEB J. 2003;17:2290–2292. doi: 10.1096/fj.03-0113fje. [DOI] [PubMed] [Google Scholar]
- Li Q., Park P.W., Wilson C.L., Parks W.C. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111:635–646. doi: 10.1016/s0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- Llano E., Pendas A.M., Aza-Blanc P., Kornberg T.B., Lopez-Otin C. Dm1-MMP, a matrix metalloproteinase from Drosophila with a potential role in extracellular matrix remodeling during neural development. J. Biol. Chem. 2000;275:35978–35985. doi: 10.1074/jbc.M006045200. [DOI] [PubMed] [Google Scholar]
- Llano E., Adam G., Pendas A.M., Quesada V., Sanchez L.M., Santamaria I., Noselli S., Lopez-Otin C. Structural and enzymatic characterization of Drosophila Dm2-MMP, a membrane-bound matrix metalloproteinase with tissue-specific expression. J. Biol. Chem. 2002;277:23321–23329. doi: 10.1074/jbc.M200121200. [DOI] [PubMed] [Google Scholar]
- Manning G., Krasnow M.A. Development of the Drosophila tracheal system. In: Bate M., Martinez Arias A., editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1993. pp. 609–685. [Google Scholar]
- McGuire J.K., Li Q., Parks W.C. Matrilysin (matrix metalloproteinase-7) mediates E-cadherin ectodomain shedding in injured lung epithelium. Am. J. Pathol. 2003;162:1831–1843. doi: 10.1016/S0002-9440(10)64318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Horton P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:122–130. [PubMed] [Google Scholar]
- Page-McCaw A., Serano J., Sante J.M., Rubin G.M. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev. Cell. 2003;4:95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- Parks W.C., Wilson C.L., Lopez-Boado Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- Schmidt O., Theopold U., Strand M. Innate immunity and its evasion and suppression by hymenopteran endoparasitoids. Bioessays. 2001;23:344–351. doi: 10.1002/bies.1049. [DOI] [PubMed] [Google Scholar]
- Stapleton M., Carlson J., Brokstein P., Yu C., Champe M., George R., Guarin H., Kronmiller B., Pacleb J., Park S., et al. A Drosophila full-length cDNA resource. Genome Biol. 2002;3:RESEARCH0080. doi: 10.1186/gb-2002-3-12-research0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht M.D., Werb Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickens D., Behonick D.J., Ortega N., Heyer B., Hartenstein B., Yu Y., Fosang A.J., Schorpp-Kistner M., Angel P., Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo A.N., Pogulis R.J., Pease L.R. In vitro synthesis of novel genes: Mutagenesis and recombination by PCR. PCR Methods Appl. 1994;4:S123–S130. doi: 10.1101/gr.4.3.s123. [DOI] [PubMed] [Google Scholar]
- Vu T.H., Shipley J.M., Bergers G., Berger J.E., Helms J.A., Hanahan D., Shapiro S.D., Senior R.M., Werb Z. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93:411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Xie Z., Filenova E., Brew K. Drosophila TIMP is a potent inhibitor of MMPs and TACE: Similarities in structure and function to TIMP-3. Biochemistry. 2003;42:12200–12207. doi: 10.1021/bi035358x. [DOI] [PubMed] [Google Scholar]
- Wilson C.L., Ouellette A.J., Satchell D.P., Ayabe T., López-Boado Y.S., Stratman J.L., Hultgren S.J., Matrisian L.M., Parks W.C. Regulation of intestinal-defensin activation by the metalloproteinase matrilysin in innate host edefense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]