ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform (original) (raw)
Abstract
DNA microarray technology has led to an explosion of oncogenomic analyses, generating a wealth of data and uncovering the complex gene expression patterns of cancer. Unfortunately, due to the lack of a unifying bioinformatic resource, the majority of these data sit stagnant and disjointed following publication, massively underutilized by the cancer research community. Here, we present ONCOMINE, a cancer microarray database and web-based data-mining platform aimed at facilitating discovery from genome-wide expression analyses. To date, ONCOMINE contains 65 gene expression datasets comprising nearly 48 million gene expression measurements form over 4700 microarray experiments. Differential expression analyses comparing most major types of cancer with respective normal tissues as well as a variety of cancer subtypes and clinical-based and pathology-based analyses are available for exploration. Data can be queried and visualized for a selected gene across all analyses or for multiple genes in a selected analysis. Furthermore, gene sets can be limited to clinically important annotations including secreted, kinase, membrane, and known gene-drug target pairs to facilitate the discovery of novel biomarkers and therapeutic targets.
Keywords: Cancer, transcriptome, gene expression, microarray, ONCOMINE
Introduction
Gene expression profiling with DNA microarrays has emerged as a powerful approach to study the cancer transcriptome. More than 100 published studies have presented analyses of human cancer samples, identifying gene expression signatures for most major cancer types and subtypes, and uncovering gene expression patterns that correlate with various characteristics of tumors including tumor grade or differentiation state, metastatic potential, and patient survival [1–24]. Also, novel tissue [25,26] and serum [27,28] biomarkers as well as potential therapeutic targets [29,30] have been identified using these genome-wide screens. These discoveries highlight the remarkable impact that DNA microarrays have had on cancer research; however, we argue that due to limitations of data availability and integration, the full potential of gene expression profiling with microarrays has not been realized. For most published microarray studies, which may comprise thousands of gene measurements across tens or hundreds of cancer specimens, the authors have presented one interpretation of their data and have reported on only a subset of genes that demonstrate their particular hypothesis. The complete microarray datasets are sometimes made available as supplementary data, but even if that is the case, the datasets often sit as cryptic text files, stored and processed in an unsystematic manner, and thus only useful to those with computational expertise. Although standards have now been set for recording and exchanging microarray data [31], and authors have been urged to provide their complete datasets upon publication [32], the full potential of cancer microarray data will only be reached when it is unified, logically analyzed, and made easily accessible to the cancer research community.
Here we describe our ongoing effort to systematically curate, analyze, and make available all public cancer microarray data via a web-based database and data-mining platform, designated ONCOMINE (www.oncomine.org). Our effort also includes centralizing gene annotation data from various genome resources to facilitate rapid interpretation of a gene's potential role in cancer. Furthermore, we are integrating microarray data analysis with other resources including gene ontology annotations and a Therapeutic Target Database. In this report, we describe microarray data collection and analysis, and data retrieval and visualization methods available at ONCOMINE, and demonstrate the potential for important discoveries.
Data Collection and Analysis
As the goal of this ongoing effort is to compile, analyze, and serve all public cancer microarray data, we identified all potential studies by literature searching, focusing on those that have generated gene expression profiles of human cancer tissue samples. We retrieved the complete datasets if available and, if not, we contacted the authors to request for the dataset. As of May 1, 2003, we cataloged information on 152 cancer microarray studies (catalog available at ONCOMINE), of which 40 studies were available and compiled—in total, 37,901,459 gene measurements from 3,762 microarray experiments. We processed and normalized each dataset independently by a single method (see Methods section) and mapped each microarray feature to Unigene build 159.
Although many analytical methods have been applied to microarray data, we chose differential expression analysis using _t_-statistics as a measure of differential expression, and false discovery rates [33] as a corrected measure of significance. To define potential differential expression analyses, we reviewed the samples in each dataset. Thirty-four datasets had samples corresponding to both classes of at least one comparison of interest including cancer versus respective normal tissue, high-grade (undifferentiated) cancer versus low-grade (differentiated cancer) cancer, poor outcome (metastases, recurrence, or cancer-specific death) cancer versus good outcome (long-term or recurrence-free survival) cancer, metastatic cancer versus primary cancer, and cancer subtype 1 (e.g., estrogen receptor-positive) versus subtype 2 (e.g., estrogen-receptor negative). We conducted a total of 81 differential expression analyses, encompassing 939,117 gene/cancer hypotheses. The genes most differentially expressed in these analyses can be explored at ONCOMINE (see below).
GENE Module
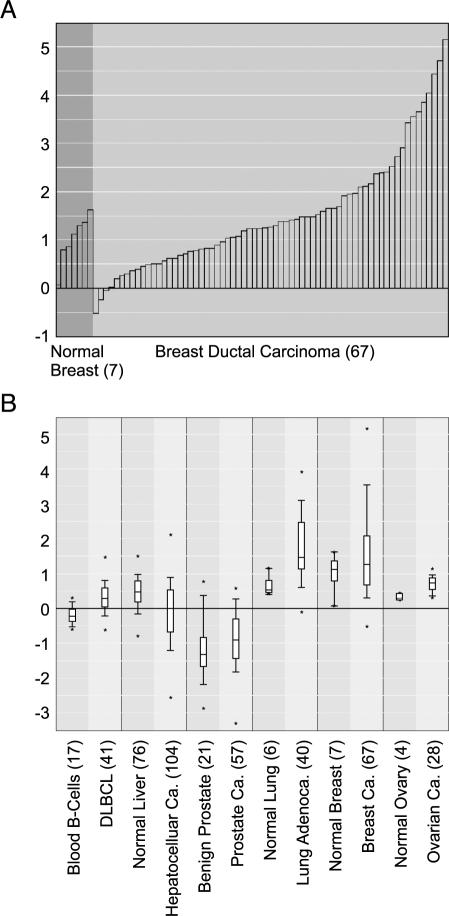
Unifying cancer microarray data and then processing, normalizing, and analyzing all datasets by a single method allow for gene centric analysis. Typically, researchers use a single microarray dataset to identify a set of genes that are associated with a particular cancer type or subtype. With ONCOMINE, users can now assess and visualize the differential expression of a selected gene across all available datasets and differential expression analyses. After searching for a gene of interest, ONCOMINE lists all differential expression analyses in which the gene was included, and allows the user to select analyses of interest. For the selected analyses, the statistical results are provided and linked to graphical representations of the microarray data. To illustrate the value of gene centric analysis with ONCOMINE, we performed a search for ERBB2 (i.e., HER2/neu), an oncogene known to be amplified in a subset of breast tumors and targeted by the antibody therapeutic, Herceptin [34]. We first looked at the expression of ERBB2 in breast cancer as per the study of Sorlie et al. [21]. We found that, as expected, ERBB2 is highly overexpressed in a fraction of breast cancer samples relative to normal breast samples (P = .057; Figure 1_A_). Next, we looked at ERBB2 expression in all “cancer versus normal” analyses. Interestingly, ERBB2 was significantly overexpressed in diffuse large B-cell lymphoma (DLBCL) relative to normal blood B-cells (P = 1.2e–6), in non small cell lung cancer (NSCLC) relative to normal lung (P = 1.7e–5 and P = 1.1e–5), and in ovarian carcinoma relative to normal ovary (P = 1.0e–5), but not in the majority of other cancer types. Figure 1_B_ depicts these analyses, along with selected others that were not significant, as a multidataset box plot for ERBB2. It is notable that the associations of Her2/neu with NSCLC and ovarian cancer as revealed by ONCOMINE have been documented by other independent studies [35], and clinical trials of Herceptin use for NSCLC are underway [36].
Figure 1.
ERBB2 (Her2/neu) gene centric expression analysis as revealed by ONCOMINE. (A) ERBB2 is overexpressed in a subset of breast cancers relative to normal breast tissue (P = .0567). (B) ERBB2 is significantly overexpressed in DLBCL relative to normal blood B-cells (P = 1.2e-6), in non small cell lung cancer relative to normal lung (P = 1.1e-5), and in ovarian carcinoma relative to normal ovary (P = 1.0e-5), but not in hepatocellular carcinoma or prostate cancer relative to their respective normal tissue. Y-axis units are normalized expression values (standard deviations above or below the median per array). The number of samples in each class is given in parentheses. Adenoca. indicates adenocarcinoma; Ca. indicates carcinoma; DLBCL indicates diffuse large B-cell lymphoma.
STUDY Module
The STUDY module provides a standard gene expression color map to visualize genes most differentially expressed in a selected analysis. Many of the differential expression analyses are analogous to those performed in the original publications; however, with ONCOMINE, they are centralized and apply a single, robust statistical method. Furthermore, some analyses available at ONCOMINE were not performed in the original publications, thus increasing the value of these microarray datasets. For example, Ramaswamy et al. published a report on multicancer type classification highlighting a focused gene set that can accurately classify tumor types of different origin [16]. Because the dataset also included respective normal tissue samples for many of the cancer types, we performed multiple “cancer versus normal” differential expression analyses, including pancreatic cancer versus normal pancreas—a hypothesis that was not testable from any of the other available datasets. A final point about the STUDY module: direct links are provided to the GENE module, so that if the gene of interest is identified by exploring a differential expression analysis, the user can quickly evaluate the gene's expression in other differential expression analyses (as demonstrated below with prostasin).
Gene Ontology Integration
The focus of many cancer microarray studies is to identify potential therapeutic targets or diagnostic markers. Genes are usually considered as potential targets or markers if they are highly overexpressed in a particular cancer, and their molecular function or localization suggests that they might be amenable to pharmacologic inhibition or detection in serum or tissue. To provide a platform for the discovery of potential targets or markers that are overexpressed in cancer, we annotated genes with relevant gene ontology descriptors. Three ontology categories were created by combining gene ontology annotations from GO ontology consortium [37]: 1) membrane-bound, which could be targeted by antibody therapies; 2) kinase, which could be inhibited by small molecule kinase inhibitors; and 3) secreted, which could serve as serum biomarkers. Significantly overexpressed genes from each ontology category were present in nearly all analyses. The genes in a particular ontology category (e.g., membrane) that are most differentially expressed in a specific analysis (e.g., lung adenocarcinoma versus normal lung) can be explored at ONCOMINE. Furthermore, specific GO annotations (e.g., DNA binding) can also be used to filter differential expression analyses.
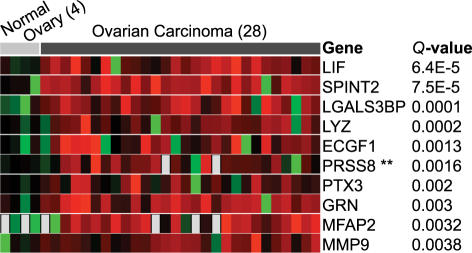
To demonstrate the utility of this approach, we will highlight an analysis using ONCOMINE to identify serum biomarkers for ovarian cancer. Ovarian cancer is in particular need of improved serum biomarkers to aid in early detection as it often presents late in the course of disease when treatment options are limited. Recently, a study was published suggesting prostasin as a potential serum biomarker for ovarian cancer [28]. The authors profiled a small number of ovarian cancer cell lines and found that prostasin was overexpressed relative to normal ovary cell lines and then used enzyme-linked immunosorbent assay to show that prostasin protein is found at high levels in the serum of ovarian cancer patients. Using the “secreted” filter in ONCOMINE, we looked for genes overexpressed in ovarian cancer based on a study by Welsh et al. [23], which had profiled 27 primary ovarian carcinomas. This search independently confirmed prostasin as one of the most highly overexpressed genes with a secreted annotation in ovarian cancer (Figure 2). Had this resource been available to the authors of the prostasin study [28], they could have avoided their microarray analysis of cell lines moving straight from ONCOMINE to validation studies. Of note, genes encoding five other secreted proteins were found to be more significantly overexpressed than prostasin (LIF, SPINT2, LGALS3BP, LYZ, and ECGF1), suggesting that more accurate biomarkers may exist. A gene centric analysis of prostasin revealed that this gene is also highly expressed in prostate cancer, as defined by two independent datasets, and a subset of lung cancers, suggesting a broadened role for this marker.
Figure 2.
Genes encoding secreted proteins most significantly overexpressed in ovarian carcinoma relative to normal ovary samples as revealed by ONCOMINE. PRSS8, the sixth most significant gene, was previously shown to be an accurate serum biomarker for ovarian carcinoma [28]. Red signifies overexpressed relative to the mean normal value, black equally expressed, and green underexpressed. The number of samples in each class is given in parentheses.
Known Therapeutic Target Integration
Based on the hypothesis that therapeutic agents are most effective in cancer types in which their targets are highly expressed (e.g., ERRB2 overexpression in breast cancer leads to Herceptin susceptibility), we sought to provide a platform to explore the expression of all known therapeutic targets in cancer, even those that are targeted in diseases other than cancer. We hypothesized that this platform may lead to novel drug target-cancer type associations, suggesting novel applications of therapeutic agents currently in use. We compiled a set of 148 known drug targets and their respective drugs by querying the Therapeutic Target Database [38] and by automated PubMed searches (see Methods section). Sixty-five of these targets were found to be significantly overexpressed in at least one differential expression analysis (data not shown).
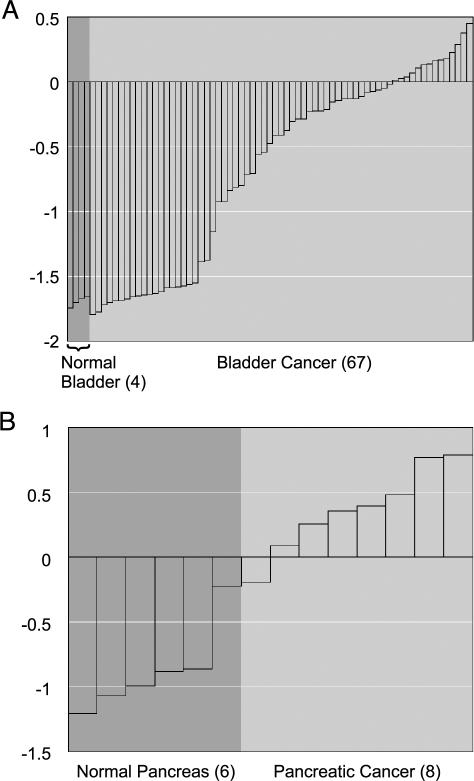
Within the STUDY module, the user can apply the therapeutic target filter to identify the targets most overexpressed in a particular differential expression analysis. For example, we found that PTGS2, otherwise known as COX-2, is the most significant overexpressed drug target in bladder cancer relative to normal bladder tissue (Q = 3.1e–15; Figure 3_A_). COX-2 is the key enzyme in prostaglandin biosynthesis and is targeted by nonsteroidal anti-inflammatory medications such as aspirin. Unknown to us, COX-2 had previously been shown to be overexpressed in bladder cancer, and a COX-2 inhibitor, Celcoxib, was shown to inhibit bladder tumor formation in rats [39] and is currently in phase III clinical trials for the prevention of bladder cancer in humans [40]. Although this association was previously made, our coincidental finding supports the value of this approach.
Figure 3.
Therapeutic targets overexpressed in cancer as revealed by ONCOMINE. (A) PTGS2 (COX-2) is significantly overexpressed in bladder cancer relative to normal bladder samples (Q = 3.1e-15), confirming previous work that COX-2 is a potential target for bladder cancer. (B) ABL1 is significantly overexpressed in pancreatic cancer relative to normal pancreas samples (Q = 0.0097), suggesting that the Abl tyrosine kinase inhibitor, Gleevec, should be investigated for use. The number of samples in each class is given in parentheses.
The majority of hypotheses generated by this approach remain to be explored. For example, effective treatment strategies are desperately needed for pancreatic cancer, as current treatments have limited efficacy with survival rates less than 5% [41]. By applying the drug target filter, we found that ABL1 (Abl tyrosine kinase) is the most significant overexpressed drug target in pancreatic cancer relative to normal pancreas (Q = 0.0097; Figure 3_B_). Abl kinase is targeted by Gleevec, a small molecule inhibitor that has recently been approved for first-line therapy in chronic myelogenous leukemia [42]. Although the number of pancreatic samples in which ABL1 was overexpressed is small (n = 8), the association is novel and worth exploring. If further studies confirmed ABL1 overexpression and demonstrated its role in pancreatic carcinogenesis, perhaps Gleevec could be useful in its management. A gene centric analysis of ABL1 further revealed that it is overexpressed in glioblastoma (P = .0012) and medulloblastoma (P = .0005).
ONCOMINE Extras and Future Directions
To facilitate the rapid interpretation of a gene's potential role in cancer, ONCOMINE provides a centralized gene annotation resource, integrating information from other bioinformatics resources including Swiss-Prot, LocusLink [43], and Unigene, and providing direct links to Human Protein Reference Database (HPRD) [44] and SOURCE [45], and the pathway resources Kyoto Encyclopedia of Genes and Genomes (KEGG) [46] and Biocarta. An online tutorial is provided at the ONCOMINE website to demonstrate its functionality through a series of sample analyses. Future work will include the collection of additional microarray datasets as they become available, increased integration with other genome resources, and correlation-based analysis. ONCOMINE also serves as a platform to explore the “metasignatures” identified identified from the cancer microarray compendium, as described in our companion report (Submitted for publication).
In summary, ONCOMINE is a powerful platform for bioinformatic discovery that brings cancer microarray data and analysis capabilities to the fingertips of the cancer research community. We hope that this work and the continued support and development of ONCOMINE will stimulate further research and maximum access to and hypothesis generation from cancer microarray data, ultimately leading to the improved understanding of cancer and the development of novel diagnostic and therapeutic strategies.
Methods
Data Collection, Processing, and Storage
Microarray datasets were downloaded from public websites or provided by the authors upon request. The web addresses to download particular datasets are listed at ONCOMINE (www.oncomine.org). All data that were available from the authors were included in processing and analysis, except that negative values were not included. All data were log-transformed, median centered per array, and standard deviation normalized to one per array. Studies were named by the following convention: FirstAuthor_TissueTypeProfiled (e.g., Dhanasekaran_Prostate). To facilitate multistudy analysis, microarray features were mapped to Unigene Build 159. Data were stored in an Oracle 8.1 relational database.
Data Analysis
For each of the 40 microarray studies present in the database, we reviewed the samples profiled. Thirty-four studies had at least four samples corresponding to both classes of one analysis of interest and were further analyzed. Analyses of interest included cancer versus respective normal tissue, high-grade (undifferentiated) cancer versus low-grade (differentiated cancer) cancer, poor outcome (metastases, recurrence, or cancer-specific death) cancer versus good outcome (long-term or recurrence-free survival) cancer, primary cancer versus metastatic disease, and subtype 1 versus subtype 2. Following the assignment of samples to classes, each gene was assessed for differential expression with _t_-statistics using Total Access Statistics 2002 (FMS Inc., Vienna, VA). _t_-Tests were conducted both as two-sided for differential expression analysis and one-sided for specific overexpression analysis. For the purpose of whole study analysis, P values were corrected for multiple comparisons by the method of false discovery rates. Corrected P values are designated as Q values [33], where Q = P* n / i (n = total number of genes; i = sorted rank of P value).
Drug Target
Drug targets were defined by two methods. First, the Therapeutic Target Database [38] was queried for all targets that had a defined antagonist, inhibitor, or antibody. One hundred nine unique drug targets were identified. The targets were mapped to Unigene build 159 using gene names, symbols, and aliases as provided by SOURCE [45]. Second, all drug names present in the National Cancer Institute (NCI) clinical trials database (http://www.nci.nih.gov/clinicaltrials/) were subjected to automated PubMed searches, identifying articles with the drug name and the word “inhibitor” or “antibody” in the title. This list of titles was manually investigated for drugs and their specific targets (e.g., rituximab, CD20). Fifty-three unique targets were identified by this method. In total, 148 unique gene targets with specific drug inhibitors or antibodies were identified.
Gene Ontology
GO gene ontology [37] annotations linked to Unigene Cluster IDs were downloaded from SOURCE [45]. Three ontology categories were created by combining multiple annotations. The following annotations were part of the membrane-bound category: cell adhesion receptor, G-protein protein coupled receptor, plasma membrane, peripheral plasma membrane protein, transmembrane receptor, and transmembrane receptor protein tyrosine kinase. The following were in the kinase category: 1-phosphatidylinositol 3-kinase, cyclin-dependent protein kinase, diacylglycerol kinase, guanylate kinase, mitogen-activated protein (MAP) kinase, MAP kinase kinase, MAP kinase kinase kinase, nonmembrane-spanning protein tyrosine kinase, protein kinase, protein kinase C, protein serine/threonine kinase, protein tyrosine kinase, receptor signaling protein tyrosine kinase, transmembrane receptor protein serine/threonine kinase, and transmembrane receptor protein tyrosine kinase. Lastly, the following annotations were part of the secreted category: extracellular, extracellular matrix, and extracellular space.
ONCOMINE
ONCOMINE was developed using three-tier architecture. The back end consists of an Oracle 8i database for storing microarray data and statistics, and a series of key-indexed flat files for various biological databases. The middle tier, which handles application logic and core functionality, was developed with Python (www.python.org). The front-end client was implemented using ZOPE (www.zope.org). ONCOMINE is available at www.oncomine.org.
Acknowledgements
We thank Vasudeva Mahavisno for graphics and Douglas Gibbs for hardware support. D.R.R. is a fellow of the Medical Scientist Training Program and A.M.C. is a Pew Scholar.
Footnotes
1
This was funded by pilot funds from the Dean's Office, Department of Pathology, DOD grant PC02322, and the Bioinformatics Program.
2
The authors contributed equally to this work.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 7.Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoff TF. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 8.Frierson HF, Jr., El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol. 2002;161:1315–1323. doi: 10.1016/S0002-9440(10)64408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengerbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 11.Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 12.Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, Catalona WJ, Watson MA, Milbrandt J. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res. 2001;61:5692–5696. [PubMed] [Google Scholar]
- 13.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 14.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 15.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 16.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai W, Darrah DM, Reed H, Ellenson LH, Giordano TJ, Fearon ER, Hanash SM, Cho KR. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- 18.Rickman DS, Bobek MP, Misek DE, Kuick R, Bliavas M, Kurnit DM, Taylor J, Hanash SM. Distinctive molecular profiles of highgrade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001;61:6885–6891. [PubMed] [Google Scholar]
- 19.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 20.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 21.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 23.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriquez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 25.van de Rijn M, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, Torhorst J, Sauter G, Zuber M, Kuchli OR, Mross F, Dieterich H, Seitz R, Ross D, Botstein D, Brown P. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–1996. doi: 10.1016/S0002-9440(10)64476-8. Erratum in: Am J Pathol 2003; 163: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. Alphamethylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 27.Tanwar MK, Gilbert MR, Holland EC. Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res. 2002;62:4364–4368. [PubMed] [Google Scholar]
- 28.Mok SC, Chao J, Skates S, Wong K, Yiu GK, Muto MG, Berkowitz RS, Cramer DW. Prostasin, a potential serum marker for ovarian cancer: identification through microarray technology. J Natl Cancer Inst. 2001;93:1458–1464. doi: 10.1093/jnci/93.19.1458. [DOI] [PubMed] [Google Scholar]
- 29.Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, Ma ZC, Wu ZO, Ye SL, Liu YK, Tang ZY, Wang XW. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, Golub TR, Griffin JD, Korsmeyer SJ. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 31.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causten HC, Gaasterland T, Glennison P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 32.Perou CM. Show me the data! Nat Genet. 2001;29:373. doi: 10.1038/ng1201-373. [DOI] [PubMed] [Google Scholar]
- 33.Storey J. A direct approach to false discovery rates. R Stat Soc. 2002;64:479–498. [Google Scholar]
- 34.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 35.Fujimura M, Katsumata N, Tsuda H, Uchi N, Miyazaki H, Hidaka T, Sakai M, Saito S. HER2 is frequently over-expressed in ovarian clear cell adenocarcinoma: possible novel treatment modality using recombinant monoclonal antibody against HER2, trastuzumab. Jpn J Cancer Res. 2002;93:1250–1257. doi: 10.1111/j.1349-7006.2002.tb01231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinner RG, Kim J, Herbst RS. Non-small cell lung cancer clinical trials with trastuzumab: their foundation and preliminary results. Lung Cancer. 2002;37:17–27. doi: 10.1016/s0169-5002(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 37.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grubbs CJ, Lubet RA, Koki AT, Leahy KM, Masferrer JL, Steele VE, Kelloff GJ, Hill DL, Seibert K. Celecoxib inhibits N-butyl-N-(4-hydroxybutyl)-nitrosamine-induced urinary bladder cancers in male B6D2F1 mice and female Fischer-344 rats. Cancer Res. 2000;60:5599–5602. [PubMed] [Google Scholar]
- 40.Gee J, Sabichi AL, Grossman HB. Chemoprevention of superficial bladder cancer. Crit Rev Oncol Hematol. 2002;43:277–286. doi: 10.1016/s1040-8428(01)00190-1. [DOI] [PubMed] [Google Scholar]
- 41.Konner J, O'Reilly E. Pancreatic cancer: epidemiology, genetics, and approaches to screening. Oncology (Huntington) 2002;16:1615–1622. (1631 – 1612; discussion 1632 – 1613, 1637 – 1618) [PubMed] [Google Scholar]
- 42.Gleevec approved for first-line treatment of CML. FDA Consum. 2003;37:5. [PubMed] [Google Scholar]
- 43.Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI genecentered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro JD, Niranjan V, Peri S, Jonnalagadda CK, Pandey A. From biological databases to platforms for biomedical discovery. Trends Biotechnol. 2003 doi: 10.1016/s0167-7799(03)00108-2. (in press) [DOI] [PubMed] [Google Scholar]
- 45.Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, Alizadeh AA. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]