AraCyc: A Biochemical Pathway Database for Arabidopsis (original) (raw)
Abstract
AraCyc is a database containing biochemical pathways of Arabidopsis, developed at The Arabidopsis Information Resource (http://www.arabidopsis.org). The aim of AraCyc is to represent Arabidopsis metabolism as completely as possible with a user-friendly Web-based interface. It presently features more than 170 pathways that include information on compounds, intermediates, cofactors, reactions, genes, proteins, and protein subcellular locations. The database uses Pathway Tools software, which allows the users to visualize a bird's eye view of all pathways in the database down to the individual chemical structures of the compounds. The database was built using Pathway Tools' Pathologic module with MetaCyc, a collection of pathways from more than 150 species, as a reference database. This initial build was manually refined and annotated. More than 20 plant-specific pathways, including carotenoid, brassinosteroid, and gibberellin biosyntheses have been added from the literature. A list of more than 40 plant pathways will be added in the coming months. The quality of the initial, automatic build of the database was compared with the manually improved version, and with EcoCyc, an Escherichia coli database using the same software system that has been manually annotated for many years. In addition, a Perl interface, PerlCyc, was developed that allows programmers to access Pathway Tools databases from the popular Perl language. AraCyc is available at the tools section of The Arabidopsis Information Resource Web site (http://www.arabidopsis.org/tools/aracyc).
The genome of the flowering plant Arabidopsis was the first plant genome to be fully sequenced (Arabidopsis Genome Initiative, 2000). Initially, approximately 26,000 genes were identified in the genomic sequence, based on different computational methods, and were assigned to functional categories (Arabidopsis Genome Initiative, 2000). About 9% of these genes have been studied experimentally (Arabidopsis Genome Initiative, 2000), and about 32% of all genes in Arabidopsis could not yet be assigned to any functional category (Reiser et al., 2002). From the initial annotation of the genome, it has been estimated that about 4,000 genes may be involved in metabolism (Arabidopsis Genome Initiative, 2000).
In this work, we used Pathway Tools software (Karp et al., 2002a) to build a database for Arabidopsis metabolism. The software allows automatic generation of pathway databases using functional assignment of genes and also allows manual editing of pathways through a graphical user interface. Although most of the functional annotations were derived computationally, we hypothesized that there was enough information to build an initial metabolism database, which could be used to facilitate manual literature curation of genes involved in metabolism.
The Pathway Tools software suite is a comprehensive system to identify, curate, store, and publish biochemical pathways on the Web in the form of pathway genome databases (PGDBs; Karp et al., 2002a). PGDBs contain the entire genomic information of an organism, including its metabolic compounds, reactions, biochemical pathways, enzymes, and enzyme complexes. There are three components in the Pathway Tools: (a) Pathologic, which allows a new PGDB to be built from data sets consisting essentially of gene annotations; (b) Pathway/Genome Editor, which allows pathways to be edited and new pathways to be added; and (c) Pathway/Genome Navigator, which allows users to query and browse the database, both locally and on the Web. The Pathologic analysis predicts the pathways of an organism using a reference PGDB from which pathways are extracted using a pathway-scoring algorithm (Paley and Karp, 2002). The reference PGDB used in this work is MetaCyc (http://metacyc.org; Karp et al., 2002b), a metabolic-pathway database that describes 449 curated pathways and 1,115 enzymes occurring in 158 organisms.
The Pathway Tools system has been applied extensively to annotate microbial genomes and has been optimized to a point where it exceeds expert analyses in comprehensiveness and matches expert analyses in accuracy (Paley and Karp, 2002). However, it was unknown how well it would handle a eukaryotic genome. The software had been applied previously to only one eukaryote, yeast. Because of this limited exposure to eukaryotic organisms, we expected a lower accuracy of the initial database build as compared with prokaryotic databases. A eukaryotic genome not only is more complex, but also has an enormous difference in scale. A typical bacterium, such as Escherichia coli, contains 4.6 million bp of DNA and has on the order of 4,392 genes; the E. coli pathway/genome database, EcoCyc, lists 164 different pathways and 914 enzymes. In comparison, Arabidopsis has a genome of 125 million bases, and comprises more than 26,000 genes, which corresponds to 20 times the amount of DNA and almost five times the number of genes in E. coli. The resulting pathway/genome database can therefore be expected to be many-fold more complex than EcoCyc. In addition, eukaryotes have subcellular compartments, many different cell-types, and elaborated life cycles with a complex series of developmental stages, which qualitatively increases the complexity of biochemical processes.
In this paper, we describe how AraCyc was initially built, we compare the quality of the resulting database to the version of AraCyc that has been improved through manual verification and annotation, and we compare the overall quality of AraCyc to EcoCyc. We also describe what adaptations to the Pathway Tools software were necessary to better accommodate a eukaryotic organism.
RESULTS
Pathologic Analysis
The Pathologic module of Pathway Tools was run using Arabidopsis enzyme annotations that were obtained from the Arabidopsis sequencing project (Arabidopsis Genome Initiative, 2000), which were edited manually to remove extraneous words and characters that could interfere with the enzyme name-matching software. A total of about 6,000 genes were retained and formatted for input into Pathologic according to the Pathway Tools documentation (P. Karp and S. Paley, unpublished data). Pathologic recognizes enzyme functions using an enzyme name-matching program and a database of enzyme names and synonyms, and extracts corresponding pathways from the MetaCyc database using a pathway scoring algorithm (see “Materials and Methods”; Paley and Karp, 2002).
Overall Statistics of the Initial Build
Pathologic recognized 1,858 enzymes for which it knew a defined function (roughly 7% of the total number of genes in the genome), and another 1,650 gene products (6.3% of the genome) were identified as putative enzymes (Table I). The putative enzymes comprised both enzymes annotated with generic names such as “kinase,” for which the precise function was unknown, as well as enzymes that were specific to plants that were not in MetaCyc, such as “gibberellin oxidase.”
Table I.
Summary data for the AraCyc data sets and comparison with EcoCyc
The number of pathways, reactions, genes, and missing enzyme annotations in pathways are given for AraCyc, the initial build of AraCyc, and EcoCyc for comparison. AraCyc has approximately 42% of reactions with missing annotations, down from 45% for the initial build. EcoCyc has only approximately 7% missing annotations. Twenty-two pathways were deleted from the intial build, and 23 new ones were added. More pathways will be added in the future.
| AraCyc | AraCyc Initial Build | EcoCyc | |
|---|---|---|---|
| Pathways (excluding superpathways) | 174 | 173 | 164 |
| Reactions, total | 1,096 | 1,132 | 845 |
| Reactions, unique | 833 | 750 | 706 |
| Unique genes associated with pathways | 958 | 767 | 695 |
| Missing enzyme annotations, total | 469 (42%) | 521 (45%) | 60 (7%) |
| Missing enzyme annotations, unique | 403 (48%) | 408 (54%) | 52 (7%) |
| Genes per annotated reaction | 2.2 | 2.2 | 1.06 |
| Overlapping pathways with EcoCyc | 76 | 79 | 164 |
| Reactions in overlapping set | 458 (403 unique) | 467 (434 unique) | 462 |
| Missing enzyme annotations in overlapping set | 164 (38%; 151 unique) | 204 (43%; 186 unique) | 11 (2.3%) |
| Pathways added manually since initial build | 23 | — | — |
| Pathways deleted from initial build | 22 | — | — |
In total, AraCyc contained 173 pathways after the initial build (Table I), containing 767 enzymes and 1,132 reactions (or 750 unique reactions if same reactions in different pathways are counted once). One or more enzymes were annotated to 611 (342 unique) reactions, whereas 521 reactions, or 45% (408 unique, or 54%), lacked enzyme annotations. There were thus 883 enzymes with a defined function that were not attributed to any pathway; this was the case for many generic enzymes such as cytochrome P450s, where the reaction is not specific enough to place it in a pathway.
Statistics of Manual Editing of AraCyc
AraCyc has been manually edited since the automatic build. Curation includes deleting inappropriate pathways, adding missing pathways, or updating existing pathways (for more details, see “Analysis of Pathways”). Twenty-two pathways (or 12.7% of the original 173) were manually deleted from the original Pathway Tools analysis. Among these were lowscoring pathways with few enzymes annotated to them (6 pathways), pathways that were thought not to occur in plants (12 pathways), and close variants of other pathways in the database (4 pathways), which were merged. The complete list of deleted pathways is available on-line (http://www.Arabidopsis.org/tools/aracyc/aracyc.deleted.pathways.html). Five pathways in the database are questionable and are “on hold,” meaning that they may be deleted in the future. Deleting them would bring the total pathways deleted to 27 or 15% of the initial set. Twenty-three new pathways comprising 194 (185 unique) reactions with 212 gene annotations were added, containing 90 (86 unique) missing enzyme annotations (46%, 46% unique). Some pathways that were retrieved from MetaCyc were incomplete. Most notably, the pathway “isopentenyl diphosphate biosynthesis, mevalonate-independent” consisted of only two reactions. The pathway has been completed with four additional reactions (not all of the reactions in the pathway are currently known).
In total, the AraCyc database presently contains 174 pathways containing 1,096 reactions (833 unique). Of the reactions in the pathways, 469 (43%) are missing enzyme annotations (403 or 48% unique). A total of 958 genes were annotated to one or more reactions. The automatic building process missed mainly annotations of reactions catalyzed by large enzyme complexes with complex subunit compositions such as pyruvate dehydrogenase and ketoglutarate dehydrogenase. The reactions were missed not because Pathologic could not handle them, but rather because the enzyme names in the input files were not always accurately specified.
AraCyc contains an average of 2.2 genes per annotated reaction. This may seem to be a low number, just roughly twice the number of E. coli genes per reaction. However, there were big differences in the number of genes annotated per reaction among the different pathway categories. In Energy Metabolism, the average number was 3.3 genes/reaction, in Degradation 2.5, in Intermediary Metabolism 2.4, and in Biosynthesis 2.07. Between these categories, there were also large differences in the number of reactions that were lacking annotations, indicating that not all of the pathway categories are equally well understood: In the Energy Metabolism category, only 16.5% reactions lacked annotations, compared with 29% in Biosynthesis, 41% in Intermediary Metabolism, and 58% in Degradation. At any rate, the glycolysis pathway itself has no reactions lacking annotations and has an average of 5.1 genes annotated to a reaction. Some reactions in that pathway have more than a dozen annotated genes. In E. coli, glycolysis has an average of 1.6 reactions and a maximum of three genes per reaction. This suggests that certain pathway categories, such as the Energy Metabolism category, have a higher potential degree of regulation than the other pathway categories.
Comparison with EcoCyc
To compare these benchmarks with a database that has been manually curated over a long period of time, we compared AraCyc with the EcoCyc database (Karp et al., 2002c). EcoCyc is a database specific for the metabolism of E. coli and has been manually curated since the mid-1990s. It contains 164 pathways (not counting super-pathways) comprising 845 reactions (706 unique). EcoCyc contains only 60 missing enzyme annotations (54 unique), which means that more than 93% of all reactions have at least one enzyme annotation.
Pathways conserved between AraCyc and EcoCyc have a higher percentage of annotated reactions. We analyzed the 76 pathways (excluding super-pathways) that AraCyc shares with EcoCyc, and we found that they contain a total of 458 reactions (403 unique reactions), 164 of which were missing enzyme annotations (151 unique) in AraCyc. The percentage of missing enzyme annotations in these pathways is therefore only 36% in AraCyc, compared with 43% when all of the pathways in AraCyc are considered. In EcoCyc, these pathways contain only 11 missing annotations (10 unique) or 2%! The pathways that occur in both AraCyc and EcoCyc are therefore a subset that is much better described than the other pathways. A look at the pathways shows that they are mostly central, conserved metabolism and includes pathways such as glycolysis and biosynthesis of amino acids. The pathways that occur in AraCyc but not in EcoCyc (98 pathways) had 636 reactions (518 unique) and 315 missing annotations (278 unique), or 49.5% (53% unique) missing enzyme annotations.
Analysis of Experimental Evidence for Genes in AraCyc
To estimate how many of the annotations that were used to build the pathways were solely based on sequence similarity-based predicted information, we counted how many genes had a gene symbol synonym. A list of gene symbol aliases for each locus were obtained from The Arabidopsis Information Resource (TAIR) FTP site (ftp://ftp.arabidopsis.org/Genes/). Of the 958 unique genes currently annotated to pathways in AraCyc, only 155 had synonyms (16%). A large fraction of the gene annotations used in the Pathologic analysis are likely to be based on sequence similarity alone; the accuracy of the functional annotation based on sequence is difficult to estimate and can be verified only with future experimentation.
Analysis of Pathways in AraCyc
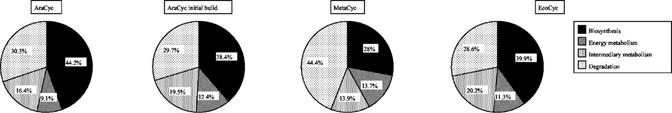
The Pathway Tools classification hierarchy defines four categories of pathways at its top level: Biosynthesis, Intermediary Metabolism, Degradation, and Energy Metabolism. In AraCyc, these categories contain 73, 27, 50, and 15 pathways, respectively (Fig. 1). The Biosynthesis class contains the largest number of pathways, largely due to pathways that we added manually since the initial build. In comparison, the largest class in MetaCyc is Degradation. This is probably due to the many bacterial degradation pathways that have been characterized. Overall, however, the distribution of pathways between these classes are very similar between the curated version of AraCyc and EcoCyc.
Figure 1.
Comparison of pathway distribution between AraCyc, AraCyc initial build, MetaCyc, and EcoCyc. The number of pathways in the different top-level classifications (Biosynthesis, Energy Metabolism, Intermediary Metabolism, and Degradation) are shown as pie charts. The major class in AraCyc is the biosynthesis class with 73 or 44.2% of pathways, up from 38.4% in the initial build. The major class in MetaCyc is the degradation class, probably reflecting the many bacterial degradation pathways that are known. The distribution within these classes in EcoCyc is, however, very similar to AraCyc.
In the Biosynthesis category of AraCyc, all amino acid biosyntheses have been inferred from Pathologic except the biosynthesis for Glu. The biosyntheses of Tyr and Phe that were inferred did not correspond to the plant versions; in plants, the biosynthesis of these amino acids is assumed to go through arogenate, instead of prephenate (Jung et al., 1986). Noticeably missing were biosynthesis of phospholipids and the mevalonate pathway. The latter is important as a precursor for terpenoid biosynthesis and was copied from MetaCyc to AraCyc manually. MetaCyc also classifies phytoalexin, flavonoid, and mevalonate metabolism under the “Fatty Acid and Lipids” class. These three pathways were also inferred in AraCyc, but subsequently moved to the newly created “Secondary Metabolite Biosynthesis” class, which was added under Biosynthesis. Apart from the secondary metabolites (flavonoids, phytoalexins) inferred under the Fatty Acid and Lipids class, no plant secondary metabolite pathways were in MetaCyc and therefore could not be identified by Pathologic. Therefore, the following pathways have been added manually: carotenoid biosynthesis, camalexin biosynthesis, and phenylpropanoid ester biosynthesis.
The Flavonoid biosynthetic pathway had to be modified extensively; the original pathway contained errors and was not very comprehensive. The phytoalexin pathway is almost an exact copy of that initial flavonoid pathway and will probably be deleted in the future. Chlorophyll biosynthesis was newly created under “heme biosynthesis.” Conspicuously, “NAD biosynthesis” was not inferred and added manually from MetaCyc. Polyisoprenoid metabolism was moved to the “Terpenoid Biosynthesis” under the “Secondary Metabolites” class. Both pyrimidine and purine biosyntheses were inferred correctly. In addition, under the newly created class “Plant Hormone Biosynthesis,” we added cytokinin, brassinosteroid, jasmonic acid, gibberellin, and abscisic acid biosynthesis pathways. The biosynthesis of ethylene was inferred correctly by Pathologic and moved to the Plant Hormones class.
The Energy Metabolism class contained 15 pathways, including glycolysis (2 instances, of which one [glycolysis 2] was deleted as a duplicate variant), the tricarboxylic acid cycle, and the Calvin cycle. In plants, glycolysis can use pyrophosphate instead of ATP for the phosphorylation of Fru. These plant-specific features will be added to AraCyc manually in the future. Several fermentation pathways were also inferred, most of which have been deleted due to insufficient evidence; the fermentation pathways usually contained some of the preceding glycolysis reactions that obviously had good evidence, but the actual fermentation reactions had no enzyme matches. The two fermentation pathways—“Glc fermentation” and “anaerobic fermentation”—that were not deleted from the database had good evidence for the fermentation-specific reactions. In general, fermentation reactions seem to be less well studied in Arabidopsis than other metabolic processes; some of the fermentation pathways we deleted due to insufficient evidence may have to be restored in the future as we learn more about them.
Intermediary Metabolism contained 27 pathways, including carnitine metabolism. Interestingly, although there is a carnitine metabolism pathway in MetaCyc, there is no carnitine biosynthetic pathway. Carnitine accumulates in many plants (Panter and Mudd, 1969), although its presence in Arabidopsis is uncertain. The 50 pathways in the Degradation class did not include the amino acid degradation pathways for Gln, His, Phe, and Pro. Pathologic found evidence for several pathways for xenobiotics degradation such as “pentachlorophenol degradation pathway.” Most of these pathways are known to exist in certain bacteria but are unlikely in plants. Not all have been deleted from the database yet, because some contain a large number of enzyme annotations. These pathways could potentially be present in Arabidopsis but remain to be characterized; they could therefore represent pathways discovered by Pathologic.
Modification of the Controlled Vocabularies of Pathway Tools for AraCyc
Because the Pathway Tools software has been used primarily to describe metabolism of prokaryotic organisms, the descriptions of intracellular structures in the database were limited and had to be extended for the use with Arabidopsis. The cellular compartment ontology consisted of only five different keywords: periplasm, membrane, inner-membrane, outer-membrane, and mitochondria. We extended this vocabulary to represent eukaryotic structures and plant organelles; it now comprises 35 terms, including chloroplast, the inner structures of the chloroplast, endoplasmatic reticulum, nucleus, etc. The complete listcanbefoundon-line(http://www.arabidopsis.org/tools/aracyc/intracellular.html). These modifications were also adopted by the MetaCyc database. In the future, it may be desirable to integrate the Gene Ontology (The Gene Ontology Consortium, 2001; http://www.geneontology.org) system into the Pathway Tools.
Another limitation of the Pathway Tools software is the lack of support for different tissues and developmental stages, for which TAIR has developed the necessary ontologies. For example, a pathway may be active only in a subset of tissues and/or at certain developmental stages, but this information cannot yet be captured in the database.
Modification of the Classification Hierarchies in Pathway Tools
We modified the chemical compound hierarchy to better accommodate plant metabolism, adding Plant Hormones and Secondary Metabolites as new classes.
PerlCyc
Pathway Tools is written in Lisp, a powerful language that is popular in the artificial intelligence community. The most popular language in biology is probably Perl, due to its simplicity and built-in string handling features such as regular expressions. To facilitate the access to the internal Pathway Tools functions, such as automated queries and batch-loading of data, we implemented a Perl module called perlcyc.pm. The module is available for download at http://www.arabidopsis.org/tools/aracyc/perlcyc. PerlCyc allows the user to write small programs in Perl that formulate more complex queries, such as: How many reactions have multiple enzyme annotations that include enzymes located in both the cytoplasm or in the chloroplast?
DISCUSSION
We have built a database for Arabidopsis metabolic pathways using the Pathway Tools software. The automatic build was edited by manual curation and addition of Arabidopsis-specific pathways. The database contains 174 pathways (excluding super-pathways) comprising more than 1,000 reactions and 958 different enzyme annotations. The database contains 822 metabolic compounds. Our aim is to represent the metabolism of Arabidopsis in AraCyc to the extent that it is known through ongoing manual curation efforts. AraCyc will allow to pinpoint the gaps in our understanding of Arabidopsis metabolism, and to facilitate researchers to fill in the gaps. The database is also a tool for the annotation of Arabidopsis biochemical enzymes, a resource for researchers who want to explore Arabidopsis metabolism, and a tool for teaching plant metabolism.
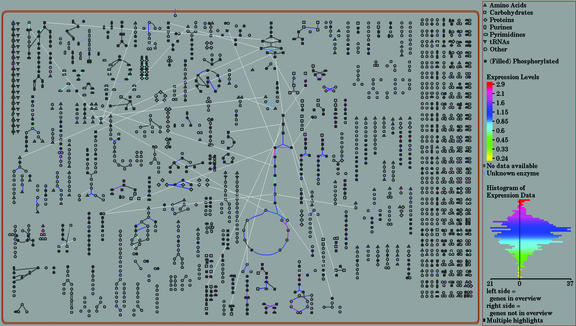
A noteworthy feature of Pathway Tools is the integrated expression viewer that allows expression data from microarray or DNA chip experiments to be visualized on the metabolic overview diagram (Fig. 2). In this example, we took data from a previously published microarray experiment (Arabidopsis Functional Genomics Consortium experiment no. 10615; Ramonell et al., 2002). A number of differentially expressed enzymes can clearly be distinguished. The expression viewer is also available through the TAIR Web site.
Figure 2.
Overlaying expression data on the overview diagram. The overview diagram gives a bird's eye view of all of the pathways in the database. The pathways are shown as glyphs consisting of nodes, which represent the metabolites, and lines, which represent the reactions. Expression data can be uploaded as a simple tab-delimited file. The lines representing the reactions are painted in a color relative to the expression level, with a dynamically generated scale depicted on the right side of the screen. For this example, data from a published data set (Ramonell et al., 2002), downloaded from the Arabidopsis Functional Genomics Consortium site, was used.
A comparison with EcoCyc shows that the AraCyc database has many more reactions lacking annotations than EcoCyc. EcoCyc has only 7% reactions lacking annotations as compared with 43% for AraCyc. This may also reflect the research priority in the Arabidopsis community to some extent. Other areas of research, such as development and disease resistance, seem to be studied more extensively in this organism than metabolism.
For primary plant metabolism, Arabidopsis should be an excellent model system for other dicots. For secondary metabolites, Arabidopsis can only be a model for the 36 secondary metabolites it has been shown to produce (Chapple et al., 1994). They fall into four classes: flavonoids, hydroxycinnamic acid esters, glucosinolates, and indole phyoalexins. Important classes of plant secondary metabolites such as alkaloids and terpene secondary metabolites have not yet been identified in Arabidopsis. Whether these compounds are not produced by Arabidopsis or have not yet been detected is an open question. The Arabidopsis genomic sequence reveals sequences homologous to enzymes that are involved in the biosynthesis of terpenes and alkaloids (Arabidopsis Genome Initiative, 2000). Recently, two myrcene/(E)-beta ocimene synthases have been cloned from Arabidopsis that had enzymatic activity when expressed in E. coli (Bohlmann et al., 2000). For the biosynthesis of alkaloids, there are 10 enzymes annotated as berberine bridge enzyme in Arabidopsis, although it is not known if they are expressed or can form active enzymes. Conversely, not all pathways that are known to operate in Arabidopsis are well characterized. In the camalexin biosynthetic pathway, the major indole phytoalexin in Arabidopsis, only one gene has been cloned, and even the precursor molecule is uncertain. Clearly, more research is needed to define the metabolic complement of Arabidopsis.
How complete is AraCyc now and when will it be finished? One way to estimate the completeness is to compare the number of estimated metabolic enzymes to the number of enzymes stored in AraCyc. It has been estimated that approximately 4,000 enzymes are involved in metabolism in Arabidopsis (Arabidopsis Genome Initiative, 2000). However, this number should be considered as an upper limit, because it is likely to include kinases, phosphatases, etc., that are specific for proteins and not for small metabolite metabolism. In AraCyc, Pathologic identified 1,850 enzymes with a defined biochemical function and a further 1,650 probable enzymes (again, most of which were annotated to imprecise functions such as “kinase,” which may not be specific to small molecule metabolism), for a total of 3,500 enzymes. Presently, there are 958 different enzymes annotated to one or more pathways. Hence, AraCyc could presently be considered one-fourth complete to the extent of what is known. Considering that roughly one-half of the reactions do not have annotations, just filling in the missing reactions should bring completeness to one-half (assuming that the average number of enzymes per reaction is similar for the missing annotations). The rest of the enzymes would probably be in pathways that are not yet in AraCyc. Again assuming that these additional pathways have a distribution of reactions and annotations similar to the present ones, the complete AraCyc database reflecting the current knowledge would have an upper limit of just more than 300 pathways.
In another attempt to estimate completeness, we compared the compounds in AraCyc with compounds identified in a metabolic profiling experiment. In the experiment, which analyzed the metabolites found in leaves, hundreds of compounds were resolved, of which 94 could be identified (Fiehn et al., 2000). Of these 94 compounds, 24 were not found in AraCyc. This seems like a large fraction considering that the metabolic profiling experiment identified relatively simple low _M_r compounds and did not detect the many complex molecules that are present in plant cells. However, 15 of the 94 compounds were classified by the authors as “uncommon plant metabolites” that had never before been seen in plants. These 15 compounds were all part of the 24 missing compounds; the nine remaining compounds, which included mostly sugars that are likely involved in cell wall biosynthesis, point us to pathways that we will have to add to AraCyc in the near future. Additional profiling experiments will be a great help in verifying AraCyc completeness in the future, when the technology will allow more compounds to be identified.
In the coming months, we will add approximately 30 pathways (refer to http://www.Arabidopsis.org/tools/aracyc), with a focus on carbohydrate and lipid biosynthesis, bringing the total of pathways to more than 200. Of course, many pathways are presently in a canonical form and will have to be extended to reflect the peculiarities of Arabidopsis metabolism. For example, the genes that are known to be involved in the biosynthesis of anthocyanin pigments, which is relatively well-studied in Arabidopsis, account for the biosynthesis of cyanidin 3-glucoside. The major anthocyanin in Arabidopsis, however, has been shown to be cyanidin (3-_O_-[2-O(2-_O_-(sinapoyl-β-d-xylopyranosyl)-6-_O_-(4-_O_-(β-d-glucopyranosyl)-_p_-coumaroyl-β-d-glucopyranoside] 5-_O_-[6-_O_-(malonyl) β-d-glucopyranoside]; Bloor and Abrahams, 2002), which is a long way from cyanidin 3-glucoside.
CONCLUSIONS
AraCyc still has a way to go to be on a par with databases such as EcoCyc in annotation quality. At least partially, this may be due to the fact that the metabolism of Arabidopsis is not as well described as the metabolism of E. coli. The Pathway Tools system has, however, permitted us to construct a relatively high-quality, comprehensive database of Arabidopsis metabolism in a short time, forming an excellent basis for further refinement through manual corrections, curation, and experimentation. Pathways that are added to AraCyc are also added to the MetaCyc database, so that these pathways will be available for future database builds for other plant species. AraCyc is available on the TAIR Web site (Huala et al., 2001).
MATERIALS AND METHODS
Pathway Tools Installation
The Pathway Tools software was downloaded from the Web by a link provided by SRI International (Menlo Park, CA). For information on obtaining Pathway Tools, contact ptools-info@ai.sri.com. The installation was performed according to instructions provided. The hardware used consisted of a SunBlade 100 workstation from Sun Microsystems (Palo Alto, CA), running Solaris 8. Pathway Tools can be run with an Oracle database backend or using flatfiles for data persistence. In this work, the flat file-based version was used. The two modes of operation are completely transparent to the user. The flat file version is easier to install and is cheaper because it does not require the purchase of an Oracle license.
Initial Build of AraCyc
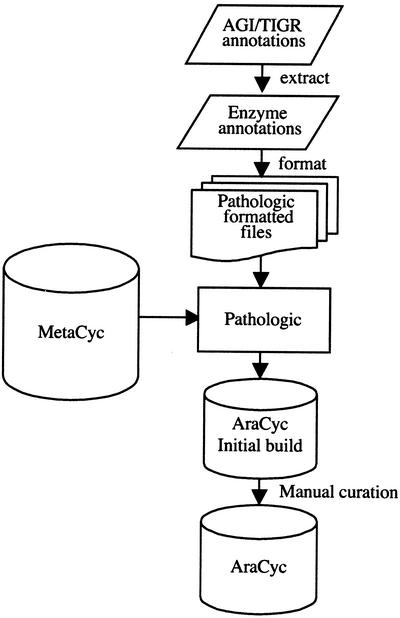
The flowchart outlining the steps in generating AraCyc is described below and is shown in Figure 3.
Figure 3.
Building AraCyc. AraCyc was built using a selection of The Institute for Genomic Research gene models that were annotated as enzymes or putative enzymes. These annotations were formatted into a Pathologic-specific format according to the documentation for Pathway Tools (P. Karp and S. Paley, unpublished data) and then analyzed with Pathologic, using MetaCyc as a reference database. The resulting database, AraCyc initial build, was then manually curated, resulting in AraCyc.
Input Files
The Institute for Genomic Research's Arabidopsis genome annotation data (http://www.tigr.org) were manually edited to include only enzyme names. Enzymes labeled as “putative” or “similar to” were also included in the data set. Any string that might interfere with the enzyme name-matching algorithm of Pathologic was removed. These strings included descriptions of subcellular locations or gene names following the enzyme name. The edited list was then formatted into a Pathologic-specific file format, which requires one file per chromosome describing their genes and one file describing the number and nature of the chromosomes (such as whether the chromosome is circular or linear etc.; P. Karp and S. Paley, unpublished data). Only nuclear-encoded genes were included in the data set.
Running Pathologic
Pathologic imports the genes and proteins described by the input files into a new database that is structured using the Pathway Tools schema and then matches the enzymes listed in the annotated genome against the enzymes required by every pathway in a reference pathway database MetaCyc (http://metacyc.org; Karp et al., 2002b). The program assesses the pathways using a pathway-scoring algorithm and only those pathways with significant scores are imported into the new PGDB. The scoring and pathway import algorithm have been described elsewhere (Paley and Karp, 2002).
Pathologic generates reports that summarize the amount of evidence supporting each pathway predicted to be present in the new PGDB and that list the “pathway holes,” i.e. the enzymes missing from each predicted pathway. This information can help the curator decide on which of the pathways imported by Pathologic should be kept in the database.
Modifying the Object Class Structures
The Pathway Tools classification hierarchy for biosynthetic pathways and chemical compound was modified to accommodate plant pathways using the built-in editing tool in Pathway Tools, GKB-Editor. In addition, some attributes, such as the subcellular location attribute of enzymes, were modified for use with eukaryotic and plant cells, using the GKB-Editor.
Manual Annotation
The manual curation process includes both editing existing pathways and adding new pathways. Information from the literature is collected and added to the pathway, reaction, compound, enzyme, and gene frames. For a pathway, we add a summary of what it does and a short description of its significance. Regarding reactions, we add, if known, EC number, free energy, whether the reaction is novel or hypothetical, and whether it is spontaneous in vitro or in vivo. For compounds, chemical structures are added if they are not already in the database. For enzymes, subcellular location, native _M_r, subunit composition, subunit _M_r, known cofactors, activators, and inhibitors are added. The _K_m, _K_i, optimum pH, and optimum temperature of an enzyme are added if known. If an enzyme is a complex of multiple subunits, comments on the role of each subunit are added. For enzyme isoforms, we capture the substrate specificity, tissue/cell type, and developmental stage specificity. Genes are linked to TAIR locus detail pages by their locus identification. Finally, synonyms of pathways, reactions, enzymes, genes, and compounds are added, and literature citations are provided by entering PubMed identification.
Acknowledgments
We thank Peter Karp, Suzanne Paley, John Pick, Cindy Krieger, and Pepe Romero from SRI for their help in carrying out this work and Peter Karp, Leonore Reiser, and Margarita Garcia-Hernandez for critically reading the manuscript.
This is Carnegie Institution of Washington, DPB, publication no. 1623.
1
This work was supported by the National Science Foundation (grant no. DBI–9978564) and by the National Institutes of Health (grant no. R01–GM65466–01).
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408**:** 796–815 [DOI] [PubMed] [Google Scholar]
- Bloor SJ, Abrahams S (2002) The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59**:** 343–346 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Martin D, Oldham NJ, Gershenzon J (2000) Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-beta-ocimene synthase. Arch Biochem Biophys 375**:** 261–269 [DOI] [PubMed] [Google Scholar]
- Chapple C, Shirley B, Zook M, Hammerschmidt R, Somerville S (1994) Secondary metabolism in Arabidopsis. In E Meyerowitz, C Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 989–1030
- Fiehn O, Kopka J, Trethewey RN, Willmitzer L (2000) Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal Chem 72**:** 3573–3580 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res 11**:** 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman A, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang J, Huang W et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and Web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29**:** 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E, Zamir LO, Jensen RA (1986) Chloroplasts of higher plants synthesize l-phenylalanine via l-arogenate. Proc Natl Acad Sci USA 83**:** 7231–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P, Paley S, Romero P (2002a) The Pathway Tools software. Bioinformatics Suppl 1 18**:** S225–S232 [DOI] [PubMed] [Google Scholar]
- Karp PD, Riley M, Paley SM, Pellegrini-Toole A (2002b) The MetaCyc Database. Nucleic Acids Res 30**:** 59–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Riley M, Saier M, Paulsen IT, Collado-Vides J, Paley SM, Pellegrini-Toole A, Bonavides C, Gama-Castro S (2002c) The EcoCyc Database. Nucleic Acids Res 30**:** 56–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley SM, Karp PD (2002) Evaluation of computational metabolic-pathway predictions for Helicobacter pylori. Bioinformatics 18**:** 715–724 [DOI] [PubMed] [Google Scholar]
- Panter RA, Mudd JB (1969) Carnitine levels in some higher plants. FEBS Lett 5**:** 169–170 [DOI] [PubMed] [Google Scholar]
- Ramonell K, Zhang B, Ewing R, Chen Y, Xu D, Stacey G, Somerville S (2002) Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol Plant Pathol 3**:** 301–311 [DOI] [PubMed] [Google Scholar]
- Reiser L, Mueller LA, Rhee SY (2002) Surviving in a sea of data: a survey of plant genome data resource and issues in building data management systems. Plant Mol Biol 48**:** 59–74 [PubMed] [Google Scholar]