Mouse Tumor Biology Database (MTB): status update and future directions (original) (raw)
Abstract
The Mouse Tumor Biology (MTB) database provides access to data about endogenously arising tumors (both spontaneous and induced) in genetically defined mice (inbred, hybrid, mutant and genetically engineered mice). Data include information on the frequency and latency of mouse tumors, pathology reports and images, genomic changes occurring in the tumors, genetic (strain) background and literature or contributor citations. Data are curated from the primary literature or submitted directly from researchers. MTB is accessed via the Mouse Genome Informatics web site (http://www.informatics.jax.org). Integrated searches of MTB are enabled through use of multiple controlled vocabularies and by adherence to standardized nomenclature, when available. Recently MTB has been redesigned and its database infrastructure replaced with a robust relational database management system (RDMS). Web interface improvements include a new advanced query form and enhancements to already existing search capabilities. The Tumor Frequency Grid has been revised to enhance interactivity, providing an overview of reported tumor incidence across mouse strains and an entrée into the database. A new pathology data submission tool allows users to submit, edit and release data to the MTB system.
INTRODUCTION
The laboratory mouse is one of the most important animal model systems used to study human disease. The well-developed genetic tools (e.g. targeted and conditional mutations), similar genetics and physiology to humans, a sequenced genome and a large number of established inbred strains provide the underpinnings for analyzing the cause of many disorders. Numerous mouse models have been developed to examine the genetics and progress of different syndromes, including many types of cancer. A search of PubMed for the terms ‘mouse model’ and ‘cancer’ returns over 2000 publications since 2000, emphasizing the impact of mouse in current cancer research. MTB was established to integrate tumor data obtained from these cancer models and make them available to the scientific community in a robust database that can inform users of existing models, support hypothesis generation, and enable the development of new cancer models.
The Mouse Tumor Biology (MTB) database was first released on the World Wide Web in 1998 (1). Data stored in MTB includes incidence and latency of mouse tumors, pathology reports and images, genomic changes occurring in the tumors, genetic (strain) background, and literature or contributor citations. Integrated searches of MTB are enabled through use of multiple controlled vocabularies and by adherence to standardized nomenclature. Data can be queried using several web-based query forms centered on the primary data types in MTB: tumor type, mouse strain, genetics, pathology and reference. Users specify one or more parameters to retrieve a Results Summary Page listing all MTB entries that satisfy the query parameters. An advanced search form combines features of the strain, genetics, pathology and tumor search forms, allowing users to ask complex questions. MTB is updated weekly with data obtained from curation of the primary literature and from direct database submissions from researchers. MTB is accessible from the Mouse Genome Informatics website (MGI) and shares database infrastructure and standard gene nomenclature with the Mouse Genome Database (MGD) (2) and the Gene Expression Database (GXD) (3). MTB also provides links to other related onlineresources such as the Mouse Phenome Database (MPD) (4), the Biology of the Mammary Gland website (http://mammary.nih.gov/), Festing's Listing of Inbred Strains of Mice (5), the JAX® Mice website (http://jaxmice.jax.org/), the Mammary Cancer in Humans and Mice: A Tutorial for Comparative Pathology: The CD-ROM Web site (6), and the Mouse Models of Human Cancers Consortium's (MMHCC's) Mouse Repository (7). Links to additional resources, such as Ensembl, are accessed using reference links to MGI and the associated gene detail pages. Direct submission of mouse tumor data and pathology images from the cancer research community is encouraged and MTB has developed a web-based system to facilitate entry of these data.
ENHANCEMENTS TO MTB
Since the last reports (8,9), the MTB data model and database schema have been redesigned and the system implemented in a Sybase relational database management system (RDMS) to enhance performance and reliability. Web-based search capabilities and on-line support have been improved, data content has dramatically increased, and the pathology data and image submission capabilities enhanced. This article describes these changes and the current status of MTB. Screen shots and additional data illustrating these enhancements are available in the online Supplementary Data.
MTB INFRASTRUCTURE
Version 1.0 of the redesigned MTB database was released in May 2005. The new MTB uses Sybase ASE 12.5 as its underlying database management system, which provides a scalable foundation for future growth. In addition, MTB's web interface has been completely revamped to take advantage of the new schema design and modernize its appearance and functionality.
MTB applications utilize the latest Java technologies along with the latest industry standard design patterns. Java (version 1.5) is the foundation upon which each component of the MTB system is based upon. Database access is achieved through a module structured after the Data Access Object Design Pattern and utilizing JDBC (Java Database Connectivity). The web interface relies on an Apache Tomcat 5.5 and Apache Struts 1.2, which is designed upon the Model-View-Controller (MVC) Design Pattern. Axis 2.0 is used to allow programmatic access to MTB through Web Services.
SEARCH FORMS
Search capabilities of MTB's web-based forms have been improved. MTB can be queried using multiple data type specific search forms (Tumor, Strain, Genetics, Pathology Images, Reference and an Advanced Search Form) or by using a quick keyword search box accessed from the left side of every MTB web page. In addition the MTB homepage offers a Quick Organ/Tissue Search. The search forms are all similar in nature enabling queries on one or more of multiple data type specific terms. For example, the Pathology Image Search allows one to search using the following parameters: Organ or Tissue of Tumor Origin; Tumor Classification, Organ or Tissue Affected, Stain or Histological Method used and Probe (i.e. antibody). A list of over 200 mouse-specific antibodies is available, in both HTML and Microsoft® Excel format (10,11).
ADVANCED SEARCH
A new advanced search form combines the functions of MTB's basic search forms. Query terms from the strain, genetics, pathology and tumor forms are utilized in the advanced search form to enable complex searches not possible from a single basic search form. For example, using the advanced search form one can query for lymphomas from mice carrying the Trp53tm1Tyj knockout. Ninety-one tumor records with 100 tumor frequencies are returned by this query. This search could be further restricted to only those records from congenic strains carrying this gene knockout where pathology data are available. This refined search returns 2 tumor records and 3 tumor frequencies. Examples of Advanced Search Form queries and results are available in Supplementary Figure S1.
ENHANCEMENT TO THE TUMOR FREQUENCY GRID
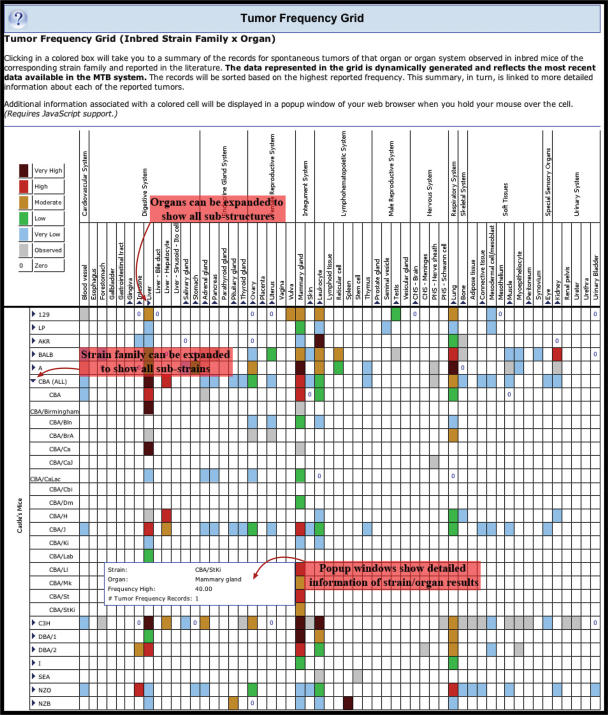
The Tumor Frequency Grid graphically displays published tumor frequency data for inbred strains of mice in a grid format as a function of the strain and organ of tumor origin (7) (Figure 1). A Tumor Frequency is the percentage of a population of mice that develop a specific lesion (defined by background, alleles, type of tumor and organ affected) as reported in a specific reference. The highest recorded tumor frequency for a given strain is represented by color coding of the grid cells. Tumor frequency is represented by six colors corresponding to Very High, High, Moderate, Low, Very Low and Observed; in addition to a zero frequency. Each frequency cell links to the detailed tumor results for that inbred strain/organ combination. We have added several enhancements to the tumor frequency grid. A rollover pop-up window has been added for each grid cell that shows strain of origin, organ of origin, highest reported frequency and number of tumor frequency records. Thus users can more easily screen for datasets that match their interests. The strain family and organ axes have been made expandable to show frequency results for sub-strains of a strain family and sub-structures of an organ. Using expansion toggles, one can access data for 307 sub-strains of the 23 basic strains and 115 sub-structures of the 33 base organs. This provides significantly more flexibility to users. In addition the Grid is now generated dynamically from the database rather than being updated manually. The Tumor Frequency Grid can be accessed from any page in MTB by using the ‘Tumor Frequency Grid’ link in the Menu Bar on the left side of every web page.
Figure 1.
The Tumor Frequency Grid graphically displays a summary of inbred strain spontaneous tumor data in MTB. Each cell is color coded to represent tumor frequency for an inbred strain and organ of origin combination and is hyperlinked to detailed frequency data.
SUBMITTING PATHOLOGY IMAGES/DATA
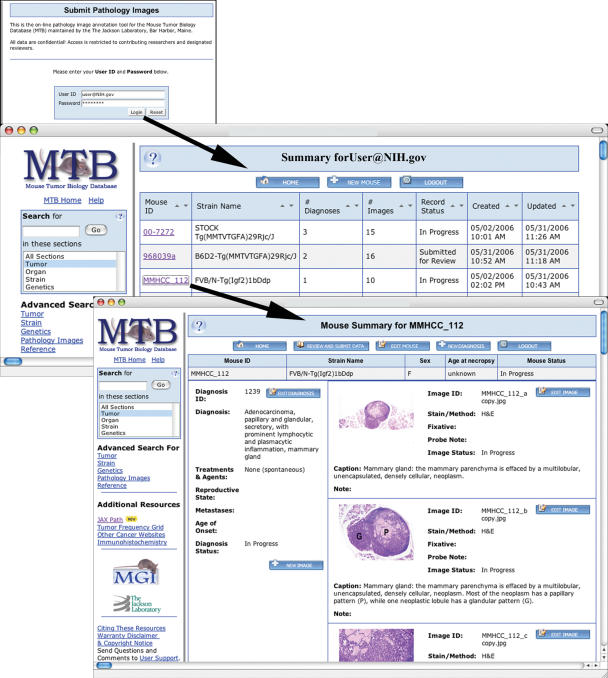
Most of the data in MTB is acquired through direct review of published scientific literature by members of the MTB curatorial staff who have expertise in biology of cancer and mouse genetics. To increase the data available to the scientific community MTB also encourages direct submission of pathology data and images, including unpublished and Supplementary Data from researchers. MTB currently displays 1668 histopathology images submitted by 45 investigators from 31 research institutions. The previous JaxPath submission tool has been completely rewritten to streamline the submission process, enhance utility (allow uploading of images) and make the system compatible with our new database structure (Figure 2). The new web-based Pathology Submission mechanism allows authors to submit data directly to MTB and view how the data will appear when it is released to the public MTB website. Users can partially complete a submission and return to finish it at a later date or update or alter previously submitted information. Once approved by the author, submitted data are reviewed by MTB staff and promptly released. The new Pathology Submission form enables researchers to create mouse records including strain and genetics information, generate tumor diagnoses specific for a mouse record, include detailed pathology and treatment descriptions in each diagnosis, attach images to a diagnosis to complement the pathology descriptions and have editorial access to any data they have submitted to MTB. Authors are assigned an ID and password that allows access to all of their in-progress and submitted records. Further examples of the new Pathology Submission form are available in Supplementary Figure S1.
Figure 2.
Examples of the Pathology Data Submission pages. The User Summary page displays all mouse records entered by a user and the Mouse Summary page shows details for a specific mouse including diagnoses and images.
To improve the quality and utility of pathology images in MTB the pathology image module recently has been enhanced to use the Zoomify program (http://www.zoomify.com/). Zoomify makes fast and interactive high-quality images on the web using HTML, JPEGs and Flash. All of these programs are freely available. With Zoomify, users can change the perspective and magnification of an image dynamically. Users whose systems do not support Flash technology will still see pathology images in a static view. MTB has only recently added zoomify capability and many existing MTB images are not yet zoomified. The number of zoomify images will increase rapidly as new images are added and existing data updated.
CURRENT STATUS OF MTB
Data in MTB are updated weekly and new software features are released periodically. Data release date and software release version are shown on the bottom of the left hand Menu Bar. Software releases are announced in the ‘What's new in MTB’ section. As of September 2006 over 1150 references have been curated. From these references and direct researcher submissions, over 25 000 tumor frequency records have been obtained. The current data content of MTB is listed in Table 1.
Table 1.
Current data content for the Mouse Tumor Biology Database
| References | 1167 |
|---|---|
| Tumor Frequency Records | 25 796 |
| Genetically Defined Strains | 2762 |
| Tumor Records | 14 193 |
| Tumors With Specific Gene Associations (germ line mutations) | 13 024 |
| Tumors with Specific Gene Associations (somatic mutations) | 1258 |
| Tumor Pathology Images | 1668 |
| Tumor Pathology Reports | 2618 |
For June 2006 MTB had 129 605 web hits and 3565 unique visitor IP addresses. This represents a 219% increase in web hits and an 85% increase in unique IP addresses from June 2005.
FUTURE DIRECTIONS
Tumor data from mouse strains will continue to be added to MTB from the scientific literature and researcher data submission. Further planned development of MTB will expand data coverage to include measures of genomic/gene expression changes in tumors using techniques such as Fluorescence in situ hybridization (FISH), spectral karotyping (SKY), gene expression arrays and cancer quantitative trait loci (QTL) data. Collection of more detailed tumor genetic change data, such as comparative genome hybridization (CGH), and links to specific data resources will also be a future emphasis of MTB. To provide relevant graphical user interfaces for these data we will develop methods for visualization of large amounts of data and genome wide patterns.
ADDRESSES AND USER SUPPORT
The MTB Database can be accessed at the MTB Home Page (http://tumor.informatics.jax.org) which is part of the MGI group web pages (http://www.informatics.jax.org). User support for MTB is available in the form of online documentation, email, fax and phone: http://www.informatics.jax.org/mgihome/support/support.shtml, Email: mgi-help@informatics.jax.org; Tel: +1 207 288 6445; Fax: +1 207 288 6132.
Enhanced online user support is available on the MTB pages via pop-up window definitions for each field displayed. This feature is a shortcut to the full online support material that MTB provides.
The MGI Group also maintains a community electronic bulletin board that serves as a discussion and announcement forum for issues relating to the genetics or biology of mouse and rat. The list is archived and can be searched using keywords (http://www.informatics.jax.org/mgihome/lists/lists.shtml). Anyone may search the archive, although only registered users may post messages to the list. Individuals may subscribe to this service on the Web at the Bulletin Board URL listed above.
CITATION OF MTB
Users of MTB are encouraged to cite this paper when referring to MTB in a publication. The following format is suggested when referring to specific data obtained from MTB:
Mouse Tumor Biology Database (MTB), Mouse Genome Informatics Group, The Jackson Laboratory, Bar Harbor, Maine, USA. World Wide Web (http://tumor.informatics.jax.org). [Include the date (month/year) when the data were retrieved.]
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank John Boddy for graphics support and Wesley Beamer, and Molly Bogue for critical reading of the manuscript. The MTB Database is supported by NCI grant CA89713. Funding to pay the Open Access publication charges for this article was provided by NCI grant CA89713.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bult C.J., Krupke D.M., Eppig J.T. Electronic access to mouse tumor data: the Mouse Tumor Biology Database (MTB) project. Nucleic Acids Res. 1999;27:99–105. doi: 10.1093/nar/27.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake J.A., Eppig J.A., Bult C.J., Kadin J.A., Richardson J.E., Mouse Genome Database Group The Mouse Genome Database (MGD): updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringwald M., Eppig J.T., Begley D.A., Corradi J.P., McCright I.J., Hayamizu T.F., Hill D.P., Kadin J.A., Richardson J.E. The Mouse Gene Expression Database (GXD) Nucleic Acids Res. 2001;29:98–101. doi: 10.1093/nar/29.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grubb S.C., Churchill G.A., Bogue M.A. A collaborative database of inbred mouse strain characteristics. Bioinformatics. 2004;20:2857–2859. doi: 10.1093/bioinformatics/bth299. [DOI] [PubMed] [Google Scholar]
- 5.Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F.W., Fisher M.C. Genealogies of mouse inbred strains. Nature Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 6.Cardiff R.D., Wagner U., Henninghausen L. Mammary Cancer in Humans and Mice: a tutorial for comparative pathology. Vet. Pathol. 2001;38:357–358. doi: 10.1354/vp.38-4-357. [DOI] [PubMed] [Google Scholar]
- 7.Xu F., Sahni H., Settnek S., Gupta A., Phillips J., Zhang D., Beasley J., De Coronado S., Wagner U., Rosso K., et al. Mouse models of human cancer web-based resources. AMIA Annu. Symp. Proc. 2003;2003:1056. [PMC free article] [PubMed] [Google Scholar]
- 8.Bult C.J., Krupke D.M., Sundberg J.P., Eppig J.T. Mouse Tumor Biology Database (MTB): enhancements and current status. Nucleic Acids Res. 2000;28:112–114. doi: 10.1093/nar/28.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bult C.J., Krupke D.M., Naf D., Sundberg J.P., Eppig J.T. Web-based access to mouse models of human cancers: the Mouse Tumor Biology (MTB) Database. Nucleic Acids Res. 2001;29:95–97. doi: 10.1093/nar/29.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krupke D.M., Naf D., Vincent M.J., Allio T., Mikaelian I., Sundberg J.P., Bult C.J., Eppig J.T. The Mouse Tumor Biology Database: integrated access to mouse cancer biology data. Exp. Lung Res. 2005;31:259–270. doi: 10.1080/01902140490495633. [DOI] [PubMed] [Google Scholar]
- 11.Mikaelian I., Nanney L.B., Parman K.S., Kusewitt D.F., Ward J.M., Näf D., Krupke D.M., Eppig J.T., Bult C.J., Seymour R., et al. Antibodies That Label Paraffin-Embedded Mouse Tissues: a collaborative endeavor. Toxicol. Pathol. 2004;32:181–191. doi: 10.1080/01926230490274335. [DOI] [PubMed] [Google Scholar]