Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? (original) (raw)
Abstract
Recent molecular analyses of leukemic blasts from pretreatment marrow or blood of patients with acute myeloid leukemia (AML) and a normal karyotype, the largest cytogenetic subset (ie, 40%-49%) of AML, have revealed a striking heterogeneity with regard to the presence of acquired gene mutations and changes in gene expression. Multiple submicroscopic genetic alterations with prognostic significance have been discovered, including internal tandem duplication of the FLT3 gene, mutations in the NPM1 gene, partial tandem duplication of the MLL gene, high expression of the BAALC gene, and mutations in the CEBPA gene. Application of gene-expression profiling has also identified a gene-expression signature that appears to separate cytogenetically normal AML patients into prognostic subgroups, although gene-expression signature-based classifiers predicting outcome for individual patients with greater accuracy are needed. These and similar future findings are likely to have a major impact on the clinical management of cytogenetically normal AML not only in prognostication but also in selection of appropriate treatment, since many of the identified genetic alterations already constitute or will potentially become targets for specific therapeutic intervention. In this report, we review prognostic genetic findings in karyotypically normal AML and discuss their clinical implications.
Introduction
The development of acute myeloid leukemia (AML) is associated with accumulation of acquired genetic alterations and epigenetic changes in hematopoietic progenitor cells that alter normal mechanisms of cell growth, proliferation, and differentiation. At diagnosis, most patients with AML harbor at least 1 chromosome aberration in their marrow blasts. Numerous recurrent structural and numeric cytogenetic aberrations have been identified and many of them not only are diagnostic markers for specific AML subtypes but also constitute independent prognostic factors for attainment of complete remission (CR), relapse risk, and overall survival (OS).1–5 However, in a sizable group of AML patients, 40% to 49% of adults and 25% of children, no microscopically detectable chromosome abnormality can be found on standard cytogenetic analysis.2–7
These cytogenetically normal (CN) patients have usually been classified in an intermediate-risk prognostic category because their CR rate, relapse risk, and survival are worse than those of adequately treated patients with such favorable aberrations as t(8;21)(q22;q22), inv(16)(p13q22)/t(16;16)(p13;q22), or t(15;17)(q22;q21) but better than those of patients with unfavorable cytogenetic findings [eg, −7, inv(3)(q21q26)/t(3;3)(q21;q26), balanced translocations involving 11q23 other than t(9;11)(p22;q23), or a complex karyotype].3–5 The outcome of CN patients has varied among studies, with 5-year survival rates between 24% and 42% reported.2,8 A recent Cancer and Leukemia Group B (CALGB) study found the relapse risk and disease-free survival (DFS) of these patients was improved by postremission treatment including 4 cycles of high-dose cytarabine (HDAC) or intermediate-dose cytarabine or 1 cycle of HDAC/etoposide followed by autologous stem-cell transplantation (ASCT) as opposed to regimens that include fewer cytarabine cycles or no ASCT.8 However, these therapeutic approaches do not improve outcome for all CN patients, likely because this cytogenetic subset is very heterogeneous at the molecular level. Indeed, during the last 12 years, several gene mutations9–12 and changes in gene expression13–16 have been discovered that strongly affect clinical outcome of CN AML patients.15–22 In this review, we will first discuss what constitutes a normal karyotype in AML and then those genetic alterations that are clinically relevant as both prognostic markers and potential targets for risk-adapted therapies in CN AML patients. We will begin with the internal tandem duplication (ITD) of the FLT3 gene, shown by many studies to be the most important prognostic factor, and follow with other genetic alterations discussed in order of the number of studies reporting their prognostic significance in CN AML.
The importance of ensuring that the patient's karyotype is truly normal
The proportion of adults with de novo CN AML has varied between 40% and 49% in the largest cytogenetic studies,3–7 although both lower and higher percentages have been reported.2 The differences among studies could relate to several factors including the number of older patients studied, since the proportion of CN cases increases with age,3,23 and differences in cytogenetic methodologies and the criteria used to consider a karyotype normal.
Leukemic blasts carrying AML-associated chromosome aberrations can constitute only a fraction of cells dividing in vitro. Occasionally, the cytogenetically abnormal clone is detectable in cells cultured in vitro for 24 to 48 hours but not in a direct preparation of the same sample (ie, when harvesting occurs immediately after the specimen is delivered to the cytogenetic laboratory). Moreover, a blood specimen can sometimes be cytogenetically normal when the marrow is abnormal. In the CALGB database, this was found in approximately 5% of AML patients whose marrow and blood specimens were studied simultaneously.24 Therefore, to reliably deem a patient as karyotypically normal it is recommended that full analysis of at least 20 metaphase cells originating from a marrow sample cultured in vitro for 24 to 48 hours is performed. Additionally, for multi-institutional research studies in which cytogenetic investigations are done in local laboratories, central karyotype review is important. In the CALGB experience, cases submitted as cytogenetically normal were found on central karyotype review to harbor such prognostically relevant chromosome aberrations as inv(3), t(9;11)(p22;q23), t(11;19)(q23;p13.1), and inv(16).25 Rarely, the opposite was true (ie, an institutionally reported abnormal karyotype was found to be normal on central review).
Although rare,26–28 there are patients who, despite having a normal karyotype on standard cytogenetic investigation, carry 1 of the fusion genes identical to those generated by recurrent translocations [eg, PML-RARA/t(15;17), RUNX1-RUNX1T1 (AML1-ETO)/t(8;21)] or inversions [_CBFB-MYH11/_inv(16)]. In most instances, these fusion genes are created by cryptic insertions of very small chromosome segments that do not alter the chromosome morphology.27,29,30 Patients with these cryptic rearrangements are usually categorized with and treated as patients with typical t(15;17), t(8;21), and inv(16). Both reverse transcriptase–polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) can be used to detect the presence of the aforementioned hidden rearrangements. Such testing is especially warranted in CN patients with FAB M3, M3v, and M4Eo marrow morphology but is otherwise not routinely recommended outside of a clinical trial.31 Studies employing spectral karyotyping,32,33 FISH with a comprehensive set of genomic DNA probes,34,35 and comparative genomic hybridization36 suggest that most patients with a normal karyotype on standard cytogenetic examination do not harbor other unrecognized chromosome aberrations. Instead, leukemic blasts of these patients carry acquired mutations and/or changes in gene expression that are unrelated to recurrent chromosome aberrations in AML but are important for predicting outcome and selecting treatment (Table 1)
Table 1.
Genes whose mutations or changes in expression occur recurrently in cytogenetically normal AML and have clinical significance
| Gene symbol(aliases and previous gene symbols) | Gene name | Chromosomal location |
|---|---|---|
| FLT3 (STK1, FLK2, CD135) | Fms-related tyrosine kinase 3 | 13q12 |
| NPM1 (B23)* | Nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 5q35 |
| MLL (ALL-1, HRX, HTRX1, CXXC7, TRX1, MLL1A) | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | 11q23 |
| BAALC | Brain and acute leukemia gene, cytoplasmic | 8q22.3 |
| CEBPA (CEBP) | CCAAT/enhancer binding protein (C/EBP), alpha | 19q13.1 |
| ERG | v-ets erythroblastosis virus E26 oncogene-like (avian) | 21q22.3 |
Mutations of the FMS-related tyrosine kinase 3 (FLT3) gene
The first report demonstrating that internal tandem duplications (ITDs) within the juxtamembrane domain (JMD) of the FLT3 gene are recurrent genetic alterations in AML was published in 1996.10 The FLT3 gene encodes a member of the class III receptor tyrosine kinase family that is normally expressed on the surface of bone marrow (BM) hematopoietic progenitor cells and plays an important role in the survival and/or differentiation of multipotent stem cells.41 Since 1996, _FLT3_-ITD has been shown to be among the most prevalent genetic abnormalities in AML, being especially frequent in t(15;17)–positive and CN patients.42 Among the latter, _FLT3_-ITD has been detected in 28% to 33% of patients.18,21,37,43–47 In addition, 2 types of point mutations in FLT3 have been reported: those localized within the activation loop of the tyrosine kinase domain (TKD), found in 5% to 14% of CN patients21,43,45,48; and those localized in the JMD, whose incidence is 2% among all AML patients but has not yet been established in CN AML.49,50
The _FLT3_-ITDs occur in exons 14 and 15 (previously exons 11 and 12) and vary in length of the duplicated JMD region from 3 to over 400 nucleotides.51 Despite such heterogeneity, and the fact that in some cases the _FLT3_-ITD is accompanied by insertions of additional nucleotides, the resultant transcripts are always in-frame. _FLT3_-ITD encodes an abnormal protein that undergoes ligand-independent receptor dimerization, autophosphorylation, and constitutive activation, which turns on downstream signaling pathways involved in cell proliferation, differentiation, and survival. These include the Janus kinase 2 (JAK2) signal transducer and activator of transcription 5 (STAT5) pathway and mitogen-activated protein kinase (MAPK) pathway.50,52,53 Similarly, both _FLT3_-TKD and JMD mutations bestow constitutive phosphorylation and activation of the FLT3 receptor, although the autophosphorylation of FLT3 and consequently transforming potential conferred by JMD mutations are weaker than those of _FLT3_-ITD and _FLT3_-TKD mutations.50 Furthermore, there is mounting evidence suggesting that the downstream cellular responses to FLT3-ITD and FLT3-TKD are not equivalent.52,53 In murine bone marrow transplantation (BMT) models, FLT3-ITD induces an oligoclonal myeloproliferative disease,54 and FLT3-TKD induces an oligoclonal lymphoid disorder with longer latency.52
Clinically, CN AML patients carrying _FLT3_-ITD differ from those without _FLT3_-ITD with regard to pretreatment characteristics and treatment outcome (Table 2) _FLT3_-ITD–positive patients present with increased white blood cell counts (WBCs)18,43,45,46 and percentages of blood and BM blasts43 and are more often diagnosed with de novo than secondary AML.43,45 Although the CR rates of _FLT3_-ITD–positive patients are often inferior, no study has observed a statistically significant difference between patients with and without _FLT3_-ITD (Table 2). However, an inferior prognosis for both remission duration and survival conferred by _FLT3_-ITD, initially reported by studies analyzing patients belonging to the intermediate-risk cytogenetic group that contained both CN patients and those with “intermediate-risk” chromosome abnormalities,42,56–58 has been firmly established in CN AML.18,37,43–46
Table 2.
Studies reporting clinical outcome of karyotypically normal AML patients according to the FLT3 status
| Prognostic significance | Independent prognostic factor on MVA | AML type | No. of pts (no. with/no. without alteration)* | Age range, y (median) | Median follow-up | Differences in pretreatment features | Source |
|---|---|---|---|---|---|---|---|
| _FLT3_-ITD+ versus _FLT3_-ITD− | |||||||
| CR rate: no significant difference (77% vs 86%) | — | De novo | 82 (23/59) | 20-59 (NR) | 1.7 y | Higher WBC | Whitman et al18 |
| OS: no significant difference (median, 11 vs 46 mo) | — | 82 (23/59) | |||||
| DFS: significantly shorter for _FLT3_-ITD+ pts (median, 8 vs 52 mo; P = .03) | ND | 68 (17/51) | |||||
| CR rate: no significant difference | — | De novo | 106 (33/73) | 17-65 (44) | NR | NR | Boissel et al37 |
| OS: no significant difference (6-year OS rates, 37% vs 43%) | — | 106 (33/73) | |||||
| EFS: significantly shorter for _FLT3_-ITD+ pts (5-year EFS rates, 19% vs 34%; P = .05) | No | 106 (33/73) | |||||
| CR rate: no significant difference (84% vs 85%) | — | De novo (84%), secondary (16%) | 67 (19/48) | 18-71 (52/48)‡ | 38 mo | Higher WBC, higher LDH | Bienz et al46 |
| OS: significantly shorter for _FLT3_-ITD+ pts (median, 10 vs 15.5 mo; P = .015) | Yes | 67 (19/48) | |||||
| DFS: significantly shorter for _FLT3_-ITD+ pts (median, 8 vs 12.5 mo; P = .033) | Yes | 57 (16/41) | |||||
| CR rate: no significant difference (50% vs 76%) | — | De novo | 53 (16/37) | Adults (58)§ | NR | Higher WBC, higher LDH, lower lactoferrin expression | Kainz et al44 |
| OS: significantly shorter for _FLT3_-ITD+ pts (5-year OS rates, 6% vs 28%; P < .003) | Yes | 53 (16/37) | |||||
| DFS: significantly shorter for _FLT3_-ITD+ pts (5-year DFS rates, 13% vs 41%; P < .02) | NR | 36 (8/28) | |||||
| _FLT3_-ITD+ versus _FLT3_-WT† | |||||||
| OS: significantly shorter for _FLT3_-ITD+ pts (P < .001) | Yes‖ | De novo and secondary | 192 (67/125) | 16-60 (47) | 34 mo¶ | Higher WBC, higher LDH, higher percentage of PB and BM blasts | Fröhling et al 43 |
| CRD: significantly shorter for _FLT3_-ITD+ pts (P = .007) | Yes‖ | 142 (47/95) | |||||
| CR rate: no significant difference | — | De novo and secondary | 84 (33/51) | 18-60 (NR) | NR | NR | Beran et al45 |
| OS: significantly shorter for _FLT3_-ITD+ pts (median, 32 vs 108 wk; P < .001) | ND | 84 (33/51) | |||||
| CRD: significantly shorter for _FLT3_-ITD+ pts (median, 42 vs 75 wk; P = .04) | ND | 58 (17/41) | |||||
| CR rate: no significant difference | — | De novo and secondary | 84 (20/64) | 61-82 (NR) | NR | NR | Beran et al45 |
| OS: no significant difference (median, 14 vs 34 wk) | — | 84 (20/64) | |||||
| CRD: no significant difference (median, 20 vs 45 wk) | — | 39 (11/28) | |||||
| _FLT3_ITD/− versus _FLT3_-ITD− | |||||||
| CR rate: no significant difference (75% vs 86%) | — | De novo | 67 (8/59) | 20-59 (37/45)# | 1.7 y | Higher WBC | Whitman et al18 |
| OS: significantly shorter for _FLT3_ITD/− pts (median, 7 vs 46 mo; P = .001) | Yes | 67 (8/59) | |||||
| DFS: significantly shorter for _FLT3_ITD/− pts (median, 4 vs 52 mo; P = .001) | Yes | 57 (6/51) | |||||
| _FLT3_-ITD+ versus _FLT3_-Asp835+ versus _FLT3_-WT | |||||||
| CR rate: no significant difference (65% vs 82% vs 76%) | — | De novo and secondary | 220 (67/28/125)** | 16-60 (47) | 34 mo | Secondary AML less Common in _FLT3_-ITD+ and _FLT3_-Asp835+ pts | Fröhling et al43 |
| _FLT3_-ITD+ versus _FLT3_-Asp835+ | |||||||
| OS: no significant difference | — | De novo and secondary | 95 (67/28) | 16-60 (47) | 34 mo¶ | Higher LDH, higher percentage of PB blasts | Fröhling et al43 |
| CRD: no significant difference | — | 70 (47/23) | |||||
| _FLT3_-Asp835+ versus _FLT3_-WT | |||||||
| OS: no significant difference | — | De novo and secondary | 153 (28/125) | 16-60 (47) | 34 mo¶ | Higher percentage of BM blasts | Fröhling et al43 |
| CRD: no significant difference | — | 118 (23/95) |
Several studies restricted to CN patients have demonstrated that patients harboring at least 1 allele with _FLT3_-ITD have a significantly worse complete remission duration (CRD), DFS, event-free survival (EFS), and OS than patients without _FLT3_-ITD (Table 2). Moreover, _FLT3_-ITD has been found to be an independent prognostic factor for OS,46,59 cumulative incidence of relapse (CIR),59 and CRD.19,46 Notably, in the first study limited to CN adult patients, _FLT3_-ITD–positive patients had a significantly shorter DFS than patients without _FLT3_-ITD.18 However, in one third of patients with _FLT3_-ITD, the FLT3 wild-type allele was not detectable. Their prognosis was particularly poor, with both DFS and OS significantly worse than those of patients without _FLT3_-ITD.18 In agreement with these results, Thiede et al48 reported that among AML patients with intermediate-risk cytogenetics, only cases with an _FLT3_-mutant/_FLT3_–wild-type allele ratio above the median value had a significantly shorter OS and DFS compared with patients without _FLT3_-ITD. Recent studies have shown that the absence of the FLT3 wild-type allele in this subset of patients with very poor prognosis likely results from acquired uniparental disomy, which could be either partial, due to somatic recombination between homologous chromosomes occurring after _FLT3_-ITD had been generated, or could involve a loss of the entire chromosome 13 with the FLT3 wild-type allele and subsequent duplication of the remaining homologous chromosome 13 with _FLT3_-ITD.60,61 The latter mechanism, however, seems less likely because in 2 patients studied by Whitman et al,18 the 2 homologous chromosomes 13 were heteromorphic for the chromosome 13 short arm (13p).
In contrast to _FLT3_-ITD, the _FLT3_-TKD has not been reported to have an adverse prognosis among either CN patients43 or those with intermediate-risk cytogenetics.48 Interestingly, a relatively small group of CN patients with _FLT3_-TKD who concurrently harbored a mutation in the NPM1 gene had significantly longer EFS than patients without these mutations.21 This finding requires corroboration, and further studies are necessary to determine the prognostic role of _FLT3_-TKD, especially in relation to other molecular prognostic factors in CN AML.
A recent study,62 not limited to CN AML, examined the prognostic impact of the length of the _FLT3_-ITD and found that patients with a duplicated segment consisting of at least 40 nucleotides had the worst relapse-free survival (RFS) compared with patients with smaller duplications and those without _FLT3_-ITD. Since length of the _FLT3_-ITD has not been correlated with level of aberrant autophosphorylation of the protein,63 it is unknown whether the different length of _FLT3_-ITDs can directly affect the protein's activity and response to treatment. It is also unknown whether the prognosis of CN patients is affected by _FLT3_-ITD length. Likewise, the adverse prognostic significance of overexpression of wild-type FLT3 alleles in _FLT3_-ITD– and _FLT3_-TKD–negative patients, suggested by a relatively small study of AML patients with diverse karyotypes,64 remains to be established in CN AML.65,66
In addition to being a prognostic marker, _FLT3_-ITD is a potential therapeutic target. Several small-molecule inhibitors of FLT3 tyrosine kinase activity are currently in clinical trials (Table 3)72 In patients with _FLT3_-ITD or _FLT3_-TKD mutations, clinical responses are usually manifested as a transient reduction in blood and, less frequently, BM blast counts, with few patients attaining a CR or a long-lasting partial response.67,68,70–72 Nevertheless, in vivo inhibition of FLT3 autophosphorylation by PKC41267 and CEP-70168 has been demonstrated in responding patients, supporting the further clinical development of these compounds. Notably, all these compounds are also capable of inhibiting multiple tyrosine kinases (Table 3), which may also explain clinical responses in AML patients without _FLT3_-ITD.72
Table 3.
FLT3 inhibitors currently in clinical trials for AML
| Compound | Chemical class | Manufacturer | Targeted kinases | FLT3 IC50,* nM | Selected clinical trials |
|---|---|---|---|---|---|
| PKC412 | Benzoyl-staurosporine | Novartis (Basel, Switzerland) | PKC, PDGFR, KDR, KIT, FLT3, ABL, FGFR1, FGFR3 | 528 | Stone et al67: Phase 2 trial in advanced MDS or relapsed or refractory AML with _FLT3_-ITD or _FLT3_-TKD |
| CEP-701 (lestaurtinib) | Indolocarbazole | Cephalon (Frazer, PA) | FLT3, TRKA, KDR, PKC, PDGFR, EGFR | 2-3 | Smith et al68; Phase 1/2 trial in refractory, relapsed, or poor-risk AML with _FLT3_-ITD or _FLT3_-TKD mutations |
| MLN518 (CT53518, tandutinib) | Piperazinyl quinazoline | Millennium (San Francisco, CA) | KIT, PDGFR, FLT3, FMS | 170-220 | De Angelo et al69: Phase 2 trial in relapsed, refractory, or untreated (not fit for induction therapy) AML with _FLT3_-ITD |
| SU11248 (sunitinib malate) | Indolinone | Pfizer (New York, NY; synthesized by Sugen, South San Francisco, CA) | FLT3, KDR, PDGFR, KIT, VEGFR | 10 | O'Farrell et al70: Phase 1 trial in refractory, relapsed, or untreated AML; Fiedler et al71: Phase 1 trial in relapsed or refractory AML with _FLT3_-ITD or _FLT3_-TKD |
Resistance to the tyrosine kinase inhibitors targeting FLT3-ITD has recently been observed in treated patients. Initial evidence suggests that such resistance is likely mediated by gain of additional point mutations in the _FLT3_-ITD gene that result in single–amino-acid substitutions within the ATP pocket of the protein, but confirmatory studies are required.76
The results of early clinical trials of FLT3 inhibitors used as single-agent therapy in heavily pretreated patients and the favorable toxicity profiles of these compounds67,68,70–72 have resulted in trials combining them with chemotherapy.77
Mutations of the nucleophosmin, member 1 (NPM1) gene
Mutations of NPM1, a partner in gene fusions generated by recurrent chromosome translocations [ie, t(2;5)(p23;q35) in anaplastic large-cell lymphoma, t(3;5)(q25;q35) in AML, and t(5;17)(q35;q21) in acute promyelocytic leukemia],38,39,78 are the most frequent gene mutations in CN AML patients, being reported in 46% to 62% of cases.12,21,37,47,79 The most common NPM1 mutation is a 4–base pair duplication, 956dupTCTG in exon 12 (called type A), that causes a shift in the reading frame leading to replacement of the last 7 amino acids by 11 different ones in the C-terminal portion of nucleophosmin. This and several other less-frequent mutations, which are always heterozygous, result in both loss of tryptophan residues 290 and 288 (or 290 only), critical for nucleolar protein localization, and acquisition of an additional nuclear export signal at the C-terminus of nucleophosmin, causing its aberrant cytoplasmic localization.40 Altered distribution in cell compartments likely interferes with the normal functions of nucleophosmin, which include preventing nucleolar protein aggregation, regulation of ribosomal protein assembly and the nucleocytoplasmic transport of these proteins, as well as the regulation of the ARF and p53 tumor-suppressor pathways.12
NPM1 mutations occur predominantly in CN patients12,47 and are associated with female sex21,79 and such pretreatment characteristics as higher WBCs21,37,47,79 and platelet counts,21,47,79 increased BM blast percentages,47,79 and low or absent CD34+ expression.12,21,47,79 Notably, patients with mutated NPM1 harbor _FLT3_-ITDs and _FLT3_-TKD mutations approximately twice as often as those with wild-type NPM1.12,21,47,79 Thiede et al47 provided evidence that NPM1 mutations usually precede the acquisition of _FLT3_-ITDs, suggesting that NPM1 mutations might constitute a primary event in leukemogenesis. CEBPA mutations occur with a similar incidence in patients with and without NPM1 mutations,21,79 whereas the partial tandem duplication (PTD) of MLL is extremely rare in patients with NPM1 mutations.21,47,79
The prognostic relevance of NPM1 mutations has been shown by most, but not all,37 recent studies that analyzed CN patients21,47,79 or those classified in the intermediate-risk cytogenetic category.80 Falini et al12 first reported that cytoplasmic localization of nucleophosmin, which is highly correlated with the presence of NPM1 mutation, was an independent favorable prognostic factor for achievement of CR. In subsequent studies, comparisons of the clinical outcome involving patients with and without NPM1 mutations regardless of the presence of other genetic rearrangements have yielded somewhat inconsistent results (Table 4) However, when _FLT3_-ITDs, which are detected in approximately 40% of patients with NPM1 mutations, have also been considered, all studies have found that NPM1 mutations are associated with a significantly improved outcome in the absence of _FLT3_-ITD. Patients with NPM1 mutations lacking _FLT3_-ITD had significantly better CR rates,47,79 EFS,21 RFS,79 DFS,47 and OS,21,47,79 whereas NPM1 mutations did not impact the poor outcome of patients harboring _FLT3_-ITD (Table 4). In multivariable analyses, the combined status of mutated NPM1 and the absence of _FLT3_-ITD constituted an independent favorable prognostic factor for the achievement of CR,79 DFS,47 RFS,79 EFS,21 and OS.47,79
Table 4.
Studies reporting clinical outcome of karyotypically normal AML patients according to the NPM1 mutational status
| Prognostic significance | Independent prognostic factor on MVA | AML type | No. of pts (no. with/no. without alteration)* | Age range, y (median) | Median follow-up | Differences in pretreatment features | Source |
|---|---|---|---|---|---|---|---|
| Cytoplasmic localization of NPM1 vs nucleolar localization of NPM1 | |||||||
| CR rate: no significant difference in univariable analysis (77% vs 62%): independent favorable prognostic factor for CR achievement (odds ratio 2.98, CI: 1.2-7.43; P = .019) | Yes | De novo | 126 (79/47) | 19-60 (52/42)† | NA | Older age | Falini et al12 |
| NPM1+ versus _NPM1_− | |||||||
| CR rate: significantly higher in NPM1+ pts (70.5% vs 54.7%; P = .003) | ND | De novo (93%), secondary (7%) | 401 (212/189) | 17-82 (56/58)† | 484 d | More often women; higher WBC; lower CD34, CD133, and MPO expression | Schnittger et al21 |
| OS: no significant difference (1012 vs 549 d) | — | 401 (212/189) | |||||
| EFS: significantly longer (median, 428 vs 336 d; P = .012). | Yes | 401 (212/189) | |||||
| RFS: no significant difference (median, 473 vs 386 d) | — | 226 (136/90) | |||||
| OS: no significant difference | — | De novo (86%), secondary (8%), unknown (6%) | 300 (145/155) | 16-60 (NR) | 46 mo | More often women, higher WBC, higher platelet counts, higher LDH, higher percentage of BM blasts, more FAB M4/M5, more extramedullary, involvement and lymphadenopathy, lower CD34 expression | Döhner et al79 |
| RFS: significantly longer for NPM1+ pts (P = .002) | NR | 217 (111/106) | |||||
| CR rate: no significant difference (86% vs 88%) | — | De novo | 106 (50/56) | 17-65 (43/46)† | NR | Higher WBC, more FAB M4/M5, less FAB M0, and less common CEBPA mutations | Boissel et al37 |
| OS: no significant difference (6-year OS rates, 43% vs 37%) | No | 106 (50/56) | |||||
| EFS: no significant difference (6-year EFS rates, 32% vs 26%) | No | 106 (50/56) | |||||
| RFS: no significant difference (6-year RFS rates, 47% vs 34%) | — | 92 (43/49) | |||||
| OS: no significant difference | — | De novo and secondary | 658 (298/360) | 15-87 (NR) | 20.2 mo‡ | More often women, lower CD34 expression‡ | Thiede et al47 |
| DFS: significantly longer for NPM1+ pts (P = .043) | NR | 301 (166/135) | |||||
| CR rate: significantly higher in NPM1+ pts (86.4% vs 64.3%; P = .037) | Yes | De novo | 79 (37/42) | 15-85 (58/47)†§ | NR | Older age, higher WBC, higher percentage of PB blasts, less FAB M2§ | Suzuki et al81 |
| OS: no significant difference | No | 79 (37/42) | |||||
| RR: significantly higher in NPM1+ pts (P = .032) | Yes | 59 (32/27) | |||||
| NPM1+/_FLT3_-ITD− versus NPM1+/_FLT3_-ITD+ versus _NPM1_−/_FLT3_-ITD+ versus _NPM1_−/_FLT3_-ITD− | |||||||
| CR rates: a significant difference according to the NPM1 and _FLT3_-ITD mutational status (86% vs 63% vs 76% vs 68.5%; P < .001) | Yes‖ | De novo (86%), secondary (8%), unknown (6%) | 300 (86/59/38/117)¶ | 16-60 (49/47/43/49)¶ | 46 mo | WBC, LDH, percentage of PB and BM blasts highest in NPM1+/_FLT3_-ITD+ pts, WBC, LDH, percentage of PB and BM blasts lowest in _NPM1_−-/_FLT3_-ITD− pts | Döhner et al79 |
| OS: significantly longer for NPM1+/_FLT3_-ITD− pts compared with the 3 other groups (P = .001) | Yes‖ | 300 (86/59/38/117)¶ | |||||
| RFS: significantly longer for NPM1+/_FLT3_-ITD− pts compared with the 3 other groups (P < .001) | Yes‖ | 217 (74/37/28/78)¶ | |||||
| CR rate: a significant difference according to the NPM1 and _FLT3_-ITD mutational status (61% vs 52.8% vs 50% vs 42.1%; P < .001) | NR | De novo and secondary | 709 (182/142/76/309)¶ | 15-87 (NR) | 20.2 mo‡ | NR | Thiede et al47 |
| OS: significantly longer for NPM1+/_FLT3_-ITD− pts compared with NPM1+/_FLT3_-ITD+ (P = .001) _NPM1_−/_FLT3_-ITD+ (P = .032) and _NPM1_−/_FLT3_-ITD− (P = .03) pts | Yes‖ | 658 (169/129/70/290)¶ | |||||
| DFS: significantly longer for NPM1+/_FLT3_-ITD− pts compared with NPM1+/_FLT3_-ITD+ (P = .04), _NPM1_−/_FLT3_-ITD+ (P < .001), and _NPM1_−/_FLT3_-ITD− (P = .04) pts | Yes‖ | 301 (103/63/31/104)¶ | |||||
| CR rate: no significant difference (68.8% vs 57.7% vs 68.9% vs 60.2%) | — | De novo | 352 | < 60 (NR) | 20.2 mo‡ | NR | Thiede et al47 |
| OS: significantly longer for NPM1+/_FLT3_-ITD− pts compared with NPM1+/_FLT3_-ITD+ (P = .003) and _NPM1_−/_FLT3_-ITD− (P = .02) pts; no difference between NPM1+/_FLT3_-ITD− and _NPM1_−/_FLT3_-ITD+ pts | Yes‖ | 359 (74/95/46/144)¶ | |||||
| DFS: significantly longer for NPM1+/_FLT3_-ITD− pts compared with NPM1+/_FLT3_-ITD+ (P = .02), _NPM1_−/_FLT3_-ITD+ (P < .001) and _NPM1_−/_FLT3_-ITD− (P = .006) pts | Yes‖ | 204 (43/67/26/68)¶ | |||||
| CIR: significantly reduced for NPM1+/_FLT3_-ITD− pts compared with the 3 other groups (40-mo CIR 25% vs 57.2% vs 51.3% vs 32.7%; P = .004) | NR | 204 (43/67/26/68)¶ | |||||
| NPM1+/_FLT3_-ITD− versus _NPM1_−/_FLT3_-ITD− | |||||||
| OS: significantly longer for NPM1+/_FLT3_-ITD− pts (median, 1183 vs 601 d; P = .022) | NR | De novo and secondary | 264 (126/138) | 17-82# (NR) | 484 d# | NR | Schnittger et al21 |
| EFS: significantly longer for NPM1+/_FLT3_-ITD− pts (median, 773 vs 365 d; P = .001) | NR | 264 (126/138) | |||||
| NPM1+/_FLT3_-ITD− versus NPM1+/_FLT3_-ITD+ | |||||||
| OS: significantly longer for NPM1+/_FLT3_-ITD− pts (median, 1183 vs 321 d; P = .014) | NR | De novo and secondary | 212 (126/86) | 17-82# (NR) | 484 d# | NR | Schnittger et al21 |
| EFS: significantly longer for NPM1+/_FLT3_-ITD− pts (median, 773 vs 234 d; P = .001) | NR | 212 (126/86) | |||||
| _NPM1_−/_FLT3_-ITD+ versus NPM1+/_FLT3_-ITD+ | |||||||
| OS: no significant difference (median, 405 vs 321 d) | — | De novo and secondary | 131 (45/86) | 17-82# (NR) | 484 d# | NR | Schnittger et al21 |
| EFS: no significant difference (median, 279 vs 234 d) | — | 131 (45/86) | |||||
| NPM1+/_FLT3_-TKD+ versus _NPM1_−/_FLT3_-TKD− | |||||||
| OS: no significant difference (median, not reached vs 676 d) | — | De novo and secondary | 173 (20/153) | 17-82# (NR) | 484 d# | NR | Schnittger et al21 |
| EFS: significantly longer for NPM1+/_FLT3_-ITD+ (median, not reached vs 336 d; P = .014) | NR | 173 (20/153) |
Partial tandem duplication (PTD) of the myeloid/lymphoid or mixed-lineage leukemia (MLL) gene
_MLL_-PTD was the first gene mutation shown to affect prognosis in CN AML patients. The discovery of _MLL_-PTD was made after Caligiuri et al,82 using Southern analysis, unexpectedly detected rearrangements of the MLL gene in patients who did not harbor structural chromosome aberrations involving band 11q23 and the MLL gene but had instead either an isolated trisomy of chromosome 11 or a normal karyotype.9,83 Among CN AML patients, _MLL_-PTD occurs in approximately 8% (range, 5%-11% among the largest studies17,84–86).
_MLL_-PTD most often involves a duplication of a genomic region spanning exons 5 through 11 and insertion of the duplicated region into intron 4 of the MLL gene; in a minority of cases the duplicated region encompasses exons 5 through 12. Both main types of _MLL_-PTD result in in-frame fusions of exons 11 or 12 upstream of exon 5 and a duplication of the N-terminal region of MLL, thereby producing an elongated protein. Unlike the MLL chimeric fusion proteins, the MLL-PTD retains all functional domains of the MLL wild-type protein, including the C-terminal SET domain that confers histone methyltransferase activity,87 along with duplication of the N-terminal AT hook DNA-binding motifs, a transcriptional repression domain, and a domain preferentially binding an unmethylated cytosine in cytidine phosphate guanosine (CpG) dinucleotides on DNA.88 The presence of _MLL_-PTD is concurrent with silencing of the MLL wild-type allele in AML blasts.88 The mechanism for the monoallelic gene silencing in _MLL_-PTD–positive AML appears to involve differential DNA methylation and histone modifications regulating either directly or indirectly the promoter of the MLL wild-type allele.
Pretreatment characteristics do not differ significantly between CN patients with and without _MLL_-PTD.17,85 Between 30% and 40% of _MLL_-PTD–positive patients also harbor _FLT3_-ITD,79,86 whereas coexistence of _MLL_-PTD with CEBPA19 or NPM121,79 mutations is rare. There is no significant difference in the probability of achieving a CR or OS (Table 5)17,85 However, in both the initial study on prognostic impact of the _MLL_-PTD17 and the largest series to date, comprising 221 CN patients,85 CRD was significantly shorter for _MLL_-PTD–positive patients. Likewise, RFS and EFS were also worse for CN patients with _MLL_-PTD in 2 smaller studies.84,89 In a multivariable analysis that did not include other molecular genetic markers, _MLL_-PTD status was the single prognostically significant factor for CRD.85
Table 5.
Studies reporting clinical outcome of karyotypically normal AML patients according to the _MLL_-PTD status: _MLL_-PTD+ versus _MLL_-WT
| Prognostic significance | Independent prognostic factor on MVA | AML type | No. of pts (no. with/no. without alteration)* | Age range, y (median) | Median follow-up, mo | Differences in pretreatment features | Source |
|---|---|---|---|---|---|---|---|
| CR rate: no significant difference (70% vs 71%) | — | De novo | 98 (11/87) | 18-84 (41/53)† | No significant differences | Caligiuri et al17 | |
| OS: no significant difference (median, 13.8 vs 20.1 mo) | — | 98 (11/87) | 27 | ||||
| CRD: significantly shorter for _MLL_-PTD+ pts (median, 7.1 vs 23.2 mo; P = .01) | ND | 68 (7/61) | 22 | ||||
| Median survival: significantly shorter for _MLL_-PTD+ pts (10 with normal and 5 with abnormal karyotype) compared with that of age-matched karyotypically normal control group (median, 5 vs 12 mo; P = .006) | ND | De novo and secondary | 45 (15/30) | 50-73‡ (61) | NR | NR | Schnittger et al84 |
| Relapse-free interval: significantly shorter for _MLL_-PTD+ pts (5 with normal and 4 with abnormal karyotype) compared with that of age-matched karyotypically normal control group (median, 4 mo vs median not reached; P < .001) | ND | 26 (9/17) | |||||
| CR rate: no significant difference (89% vs 78%) | — | De novo and secondary | 221 (18/203) | 16-60 (49/45)† | 19 | No significant differences | Döhner et al85 |
| OS: no significant difference (13.4 vs 20.8 mo) | — | 221 (18/203) | |||||
| CRD: significantly shorter for _MLL_-PTD+ pts (median, 7.75 vs 19 mo; P = .02) | Yes | 174 (16/158) | |||||
| DFS: no significant difference (median, 3.2 vs 19.6 mo) | — | De novo and secondary | 169 (8/161) | 16-60 (NR) | NR | NR | Steudel et al86 |
| EFS: significantly shorter for _MLL_-PTD+ pts (2-year EFS rates, 20% vs 66%; P = .03) | ND | De novo | 30 (5/25) | 16-60 (NR) | NR | NR | Muñoz et al89 |
Direct molecular targeting of MLL-PTD by pharmacologic agents has not been investigated, likely due to the acknowledged inherent difficulties in targeting a protein originally thought to be a transcription factor without enzymatic activity. However, Whitman et al88 have recently shown that the combination of decitabine (5′-aza-2′-deoxycytidine), a DNA methyltransferase inhibitor, and depsipeptide, a histone deacetylase inhibitor, can reactivate the transcription of the MLL wild-type allele in _MLL_-PTD–positive cells. Importantly, the induction of MLL wild-type expression was associated with enhanced cell death of _MLL_-PTD–positive leukemic blasts.88 These data suggest that therapy that includes DNA methyltransferase and/or histone deacetylase inhibitors should be explored in patients with _MLL_-PTD. However, until molecularly targeted therapies, such as the aforementioned drug combination or agents targeting the histone methyltransferase activity of MLL-PTD, are shown to be clinically effective, allogeneic SCT appears to be the best therapeutic option for CN AML patients with _MLL_-PTD.85
Overexpression of the BAALC gene
The BAALC gene is expressed primarily in neuroectoderm-derived tissues and hematopoietic precursors but not in mature marrow or blood mononuclear cells and encodes a protein with no homology to any known proteins or functional domains.13 In the initial report, high BAALC expression was detected in a subset of patients with AML, acute lymphoblastic leukemia, and chronic myelogenous leukemia (CML) in blast crisis but not in those with chronic-phase CML or chronic lymphocytic leukemia.13 In CN adults younger than 60 years with de novo AML, high BAALC expression (defined as expression above the median BAALC expression value) in blood at diagnosis negatively affected EFS, DFS, and OS in patients with wild-type FLT3 or those who harbored _FLT3_-ITD and concurrently expressed the wild-type FLT3 allele (Table 6)20
Table 6.
Studies reporting clinical outcome of karyotypically normal AML patients according to the changes in gene expression
| Prognostic significance | Independent prognostic factor on MVA | AML type | No. of pts (no. with/no. without alteration)* | Age range, y (median) | Median follow-up | Differences in pretreatment features | References |
|---|---|---|---|---|---|---|---|
| BAALC high expression versus BAALC low expression | |||||||
| CR rate: no significant difference (77% vs 81%) | — | De novo | 86 (43/43) | 18-59 (51/56)† | 3.3 y | Lower WBC, less often FAB M5 | Baldus et al20 |
| OS: significantly shorter for high BAALC pts (median, 1.7 vs 5.8 y; P = .02) | Yes | 86 (43/43) | |||||
| EFS: significantly shorter for high BAALC pts (median, 0.8 vs 4.9 y; P = .03) | Yes | 86 (43/43) | |||||
| DFS: significantly shorter for high BAALC pts (median, 1.4 vs 7.3 y; P = .03) | Yes | 68 (33/35) | |||||
| CR rate: no significant difference (82% vs 91%) | — | De novo (84%), secondary (16%) | 67 (44/23) | 18-71 (46/53)† | 38 mo | Higher CD34; lower CD11b, CD15, and MPO‡ | Bienz et al46 |
| OS: significantly shorter for high BAALC pts (median, 10 vs 21 mo; P = .021) | Yes | 67 (44/23) | |||||
| DFS: significantly shorter for high BAALC (median, 8.5 vs 21 mo; P = .015) | Yes | 57 (36/21) | |||||
| CR rate: significantly lower for high BAALC pts (62% vs 73%; P = .038) | No | De novo (91%), secondary (9%) | 307 (153/154) | 17-60 (46/50)† | 29 mo | Higher percentage of PB blasts, more FAB M0/M1, less FAB M5b, less gingival hyperplasia | Baldus et al59 |
| Primary resistant disease rate: significantly higher for high BAALC pts (16% vs 6%; P = .006) | Yes | 307 (153/154) | |||||
| OS: significantly shorter for high BAALC pts (3-year OS rates, 36% vs 54%; P = .001) | Yes | 307 (153/154) | |||||
| RR: significantly higher for high BAALC pts (43% vs 29%; P = .042) | ND | 208 (95/113) | |||||
| CIR: significantly higher for high BAALC pts (3-year CIR rates, 50% vs 32%; P = .018) | Yes | 208 (95/113) | |||||
| BAALC high expression/FLT3 high risk versus BAALC high expression/FLT3 low risk versus BAALC low expression/FLT3 high risk versus BAALC low expression/FLT3 low risk | |||||||
| CR rate: the lowest for BAALC high/FLT3 high-risk pts (57% vs 58% vs 83% vs 73%; P = .048) | ND | De novo and secondary | 268 (21/110/12/125)§ | 17-60 | 29 mo | NR | Baldus et al59 |
| OS: the worst for BAALC high/FLT3 high-risk pts (3-year OS rates, 0% vs 38% vs 22% vs 58%; P < .001) | ND | 268 (21/110/12/125)§ | |||||
| CIR: the highest for BAALC high/FLT3 high-risk pts (3-year CIR rates, 100% vs 44% vs 60% vs 28%; P < .001) | ND | 177 (12/64/10/91)§ | |||||
| ERG high expression versus ERG low expression | |||||||
| CR rate: no significant difference (76% vs 83%) | — | De novo | 48 (21/63) | 18-59 (48/47)‖ | 5.7 y | Higher percentage of PB and BM blasts, more often high BAALC expression | Marcucci et al16 |
| OS: significantly shorter for high ERG pts (median, 1.2 y vs not reached; P = .011) | Yes | 84 (21/63) | |||||
| CIR: significantly higher for high ERG pts (median, 0.7 y vs not reached, P < .001) | Yes | 68 (16/52) | |||||
| Wild-type FLT3 high expression versus wild-type FLT3 low expression | |||||||
| OS: a trend towards worse OS for high FLT3 pts (P = .059) | ND | De novo and secondary | 49 (21/28) | 17-84 (61)¶ | NR | NR | Kuchenbauer et al66 |
| EFS: a trend towards worse EFS for high FLT3 pts (P = .087) | ND | 50 (21/29) | |||||
| MN1 high expression versus MN1 low expression | |||||||
| CR rate: no significant difference (80 vs 89%) | — | De novo (87%), secondary (13%) | 142 (71/71) | 18-60 (46/45)# | 30 mo | Less often mutated NPM1, less often NPM1 mutated/_FLT3_-ITD absent, more often high CD34 expression | Heuser et al15 |
| OS: significantly shorter for high MN1 pts (3-year OS rates, 38.1% vs 58.8%; P = .03) | Yes | 142 (71/71) | |||||
| RFS: significantly shorter for high MN1 pts (3-year RFS rates, 23.4% vs. 51.4%; P = .002) | Yes | 120 (57/63) | |||||
| RR: significantly higher for high MN1 pts (56.1% vs 36.5%; P = .03) | — | 120 (57/63) |
In a second study of CN patients,46 who were also analyzed for the presence of _FLT3_-ITD and CEBPA mutations, high BAALC expression, measured in marrow in relation to the median value of BAALC expression in marrow from healthy volunteers, predicted for a shorter DFS and OS. A subgroup analysis, restricted to patients without _FLT3_-ITD or CEBPA mutations, also showed high BAALC expression to be associated with worse DFS and OS.46 Bienz et al46 concluded that BAALC expression appears to be particularly useful as a prognostic marker in CN AML patients lacking _FLT3_-ITD and CEBPA mutations.
In the largest study to date, comprising 307 CN adults aged 60 years or younger, high expression of BAALC in blood was an independent adverse prognostic factor for resistance to initial induction chemotherapy, CIR, and OS.59 On multivariable analysis, high BAALC expression and high _FLT3_-ITD/_FLT3_–wild-type ratio were the only factors predicting a high CIR and shorter OS. Notably, preliminary data from the same study also suggested that allogeneic SCT in first CR might overcome the higher relapse rate associated with high BAALC expression compared with ASCT or chemotherapy.59 Future studies confirming this are required.
Mutations of the CCAAT/enhancer-binding protein α (CEBPA) gene
Mutations of CEBPA, a gene encoding a protein member of the family of basic region leucine zipper (bZIP) transcription factors that plays an essential role in granulopoiesis, were discovered in AML in 2001 and associated with FAB M2 and M1 morphology and a normal karyotype at diagnosis.11 There are 2 main categories of CEBPA mutations: C-terminal mutations that occur in the bZIP domain, which are usually in-frame and predict mutant proteins lacking DNA binding and/or homodimerization activities; and N-terminal nonsense mutations that prevent expression of the full-length protein and result in truncated isoforms with dominant-negative activity. Some patients present with biallelic mutations at the C-terminus, whereas others are heterozygous for separate mutations or found to have a C-terminal mutation coexisting with a mutation in the N-terminus.19,90
The initial clinical studies revealed that CEBPA mutations were associated with a significantly better EFS,90,91 DFS,91 and OS90,91 among patients included in the cytogenetic intermediate-risk category. This favorable prognostic significance of CEBPA mutations was subsequently confirmed by investigations restricted to CN patients (Table 7), 15% to 19% of whom carry CEBPA mutations.19,37,46 At diagnosis, patients with CEBPA mutations had higher percentages of blood blasts, lower platelet counts, less lymphadenopathy and extramedullary involvement, less frequent _FLT3_-ITD and _FLT3_-TKD, and no _MLL_-PTD.19 CR rates did not differ between patients with and without CEBPA mutations,19,37,46 whereas CRD,19 DFS,46 EFS,37 and OS19,46 were significantly better for patients with CEBPA mutations. In multivariable analyses, CEBPA mutational status has added prognostic information to that provided by _MLL_-PTD and _FLT3_-ITD status, age, and resistant disease after the first course of induction therapy with regard to CRD and by _FLT3_-ITD status, age, and WBC for OS.19 Similar results were obtained in another study, where CEBPA mutations, BAALC, and _FLT3_-ITD status were prognostic for DFS, and CEBPA mutations, age, BAALC, and _FLT3_-ITD status were prognostic for OS.46
Table 7.
Studies reporting clinical outcome of karyotypically normal AML patients according to the CEBPA mutational status
| Prognostic significance | Independent prognostic factor on MVA | AML type | No. of pts (no. with/no. without alteration)* | Age range, y (median) | Median follow-up | Differences in pretreatment features | References |
|---|---|---|---|---|---|---|---|
| CEBPA+ versus _CEBPA_− | |||||||
| Rates of CR, resistant disease, and early or hypoplastic death: no significant differences | — | De novo and secondary | 236 (36/200) | 16-60 (47/47)† | 30 mo | Higher hemoglobin level, lower platelet counts, higher percentage of PB blasts, less lymphadenopathy, less extramedullary involvement | Fröhling et al19 |
| OS: significantly longer for CEBPA+ pts (median, not reached vs 19 mo; P = .05) | Yes | 236 (36/200) | |||||
| CRD: significantly longer for CEBPA+ pts (median, not reached vs 26 mo; P = .01) | Yes | 187 (31/156) | |||||
| CR rate: no significant difference (100% vs 82%) | — | De novo (84%), secondary (16%) | 67 (12/55) | 18-71 (46/49)† | 38 mo | Lower LDH, higher CD7 | Bienz et al46 |
| OS: significantly longer for CEBPA+ pts (median, 45.5 vs 12 mo; P < .001) | Yes | 67 (12/55) | |||||
| DFS: significantly longer for CEBPA+ pts (median, 33.5 vs 10 mo; P = .002) | Yes | 57 (12/45) | |||||
| CR rate: no significant difference | — | De novo | 106 (20/86) | 17-65 (44) | NR | NR | Boissel et al37 |
| OS: no significant difference (6-year OS rates, 53% vs 38%) | — | 106 (20/86) | |||||
| EFS: significantly longer for CEBPA+ pts (P = .01) | No | 106 (20/86) | |||||
| CEBPA N-terminal nonsense mutations versus other CEBPA mutations versus CEBPA wild-type | |||||||
| CRD: the longest for pts with CEBPA N-terminal nonsense mutations, the shortest for pts with _CEBPA_-WT (median, not reached vs 52 mo vs 26 mo; P = .04) | ND | De novo (88%), secondary (12%) | 187 | 16-60 | 30 mo | FAB M1/M2 more common in pts with N-terminal mutations than in those with other mutations | Fröhling et al19 |
Fröhling et al19 also analyzed clinical outcome separately for patients with N-terminal nonsense CEBPA mutations, those with other CEBPA mutation types, and those with wild-type CEBPA and found that the CRD of patients with N-terminal mutations was the longest, followed by CRD of patients with other mutations and of those without CEBPA mutations. However, there were no significant differences in CRD between patients with N-terminal mutations and those with other mutations, or between patients with other mutations and patients with wild-type CEBPA in pairwise comparisons.19 A larger study is needed to assess the prognostic role of different types of CEBPA mutations.
Interestingly, 3 patients with familial AML and a deletion of a cytosine residue at nucleotide 212 of the CEBPA gene, 2 of whom had a normal karyotype, also had an excellent prognosis.92
Overexpression of the ETS-related gene (ERG)
Overexpression of the ERG gene, a member of the ETS family of transcription factors that are involved in regulation of cell proliferation, differentiation, and apoptosis, was discovered in AML patients with prognostically unfavorable complex karyotypes that contained cryptic amplification of chromosome 21, first uncovered by spectral karyotyping.93,94 ERG overexpression was not always associated with genomic amplification and was also detected in CN AML,94 suggesting that it might bestow an adverse prognosis in CN AML patients.
Indeed, adults younger than 60 years with de novo AML, treated on CALGB 9621, whose high ERG expression in pretreatment blood placed them in the uppermost quartile of ERG expression, had a significantly worse CIR and OS than patients in the 3 quartiles with lower ERG expression.16 In multivariable analyses, high ERG expression and _FLT3_-ITD independently predicted worse CIR and OS. Analysis restricted to the more favorable subset of patients lacking _FLT3_-ITD or expressing both _FLT3_-ITD and the FLT3 wild-type allele revealed that high ERG expression and _MLL_-PTD both impacted on remission duration. With respect to OS, there was an interaction between ERG and BAALC expression, with ERG overexpression predicting shorter OS only in low-BAALC expressers.16 These results await independent corroboration.
Other prognostic factors in karyotypically normal AML
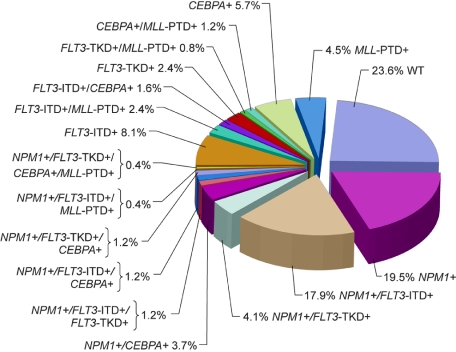
Although the majority of CN patients studied to date harbored at least 1 of the aforementioned genetic alterations, in a study by Döhner et al79 almost a quarter of patients did not carry _FLT3_-ITD, _FLT3_-TKD, _MLL_-PTD, or mutations in the CEBPA or NPM1 genes (Figure 1). Thus, it is likely that novel gene mutations and/or abnormal gene expression with prognostic significance will be discovered in the future. Expression of the meningioma 1 (MN1) gene might become such a novel prognostic factor. Heuser et al15 have reported high expression of the MN1 gene to bestow inferior RFS and OS, and a higher risk of relapse, in CN adults aged 60 years or younger with de novo or secondary AML. This observation requires confirmation.
Figure 1.
Pie chart based on 246 patients analyzed for the presence or absence of mutations in the NPM1 and CEBPA genes, _FLT3_-ITD, _FLT3_-TKD, and _MLL_-PTD indicating the coexistence of mutations in individual patients. WT indicates patients with only wild-type alleles of genes tested. Adapted by Kenneth X. Probst from Döhner et al79 with permission.
A recent study found a significantly shortened CRD of CN AML patients associated with overexpression of breast cancer resistance protein (BCRP) encoded by the ABCG2 gene.95 Interestingly, in another study that used gene-expression profiling to classify adult AML, overexpression of the ABCG2 gene was a consistent finding in a cluster of patients with the worst DFS and high induction failure rate, 42% of whom had a normal karyotype.96 Additionally, it will be of interest to determine if high expression of the EVI1 gene, hitherto reported to bestow adverse prognosis in patients classified in the intermediate-risk cytogenetic group,97 or overexpression of the WT1 gene, shown to impact unfavorably on clinical outcome of cytogenetically heterogeneous AML patients in some98 but not all99 studies, also confers adverse prognosis in CN AML. Likewise, although a recent study revealed no difference in outcome between CN patients with and without NRAS mutations,6 further studies of NRAS, KRAS, and KIT mutations seem warranted given the prospect of developing therapies with tyrosine kinase inhibitors targeting these mutations. Finally, future analyses should address potential prognostic relevance in CN AML of epigenetic changes that in some studies have already been correlated with clinical outcome in adult AML.100–103
Gene-expression profiling
Relatively little information on the clinical utility of gene-expression profiling in AML is currently available. However, this methodology, capable of determining expression levels of thousands of genes in 1 experiment, was recently used to ascertain patterns of gene activation and silencing (ie, gene-expression signatures) in CN AML with the goal of identifying patient subsets with diverse prognoses. Bullinger et al14 found that the majority of CN patients segregated into 2 gene-expression clusters with significantly different survival. Patients in cluster I, who had worse survival, more frequently harbored FLT3 aberrations and were more commonly diagnosed with AML FAB M1 and M2 and less frequently with AML FAB M4 and M5. Although these results suggested that gene-expression profiling might become useful as a tool in outcome prediction, they required confirmation, especially because in other studies CN patients segregated into several distinct clusters.96,104 A recent CALGB study22 has validated the prognostic significance of the gene-expression signature identified by Bullinger et al14 using a different microarray platform in a patient population treated on a different protocol and having longer follow-up (4.7 versus < 2 years). Moreover, this study increased the clinical applicability of the signature because Radmacher et al22 created a class-prediction algorithm that allows for the prediction of outcome for subsequent individual patients. This signature-based classifier identified groups with differences in DFS and OS.22 However, a strong association of the outcome classifier with the _FLT3_-ITD was observed that might explain the prognostic significance of the signature. Moreover, the prediction accuracy of the classifier for dichotomized outcome classes was modest, with OS and DFS of approximately 60% of the patients being correctly predicted. Of potential importance is the observation that the classifier showed some ability to identify a subset of patients with wild-type FLT3 who fare poorly.22 Future studies should refine this strategy and aim at identifying different classifiers that might predict outcome for individual CN AML patients with greater accuracy.
Conclusions and future directions
It is now clear that CN AML is very heterogeneous at the molecular level. We believe that upcoming modification to the World Health Organization (WHO) classification of AML should consider recurrent molecular genetic rearrangements discussed in this review. Moreover, since 2 or more genetic alterations are present simultaneously in many patients, it is important to devise a prioritized schema that stratifies patients to risk-adapted therapies using information on all known prognostic markers. While many questions still remain, we believe that the available data, discussed in this article, allow us to propose as a starting point for future clinical studies the molecular-risk classification, depicted in Figure 2. Based on the current evidence showing that CN patients with _FLT3_-ITD, and especially those with high _FLT3_-ITD/_FLT3_–wild-type ratios, have a particularly poor prognosis, which is only modestly influenced by the concomitant presence of other molecular markers tested so far, these patients should be considered as candidates for clinical trials such as those investigating compounds that directly inhibit FLT3 tyrosine kinase activity or that might facilitate ubiquitination and disposal of the mutant protein (ie, inhibitors of the chaperone activity of heat-shock protein such as 17 AAG) in combination with chemotherapy. Additionally, these patients might be considered as candidates for allogeneic SCT, although a definitive role for this treatment in first CR remains to be established.105 Likewise, aggressive treatment is recommended for patients with high BAALC expression and _MLL_-PTD; patients in the latter group could also potentially benefit from enrollment in clinical trials testing efficacy of histone deacetylase and/or DNA methyltransferase inhibitors.88 For patients without these molecular markers, the situation is less clear. _FLT3_-ITD–negative patients who harbor mutations in the NPM1 and/or CEBPA genes and have low BAALC and ERG expression appear to have a relatively favorable outcome and are likely to benefit from ASCT or HDAC-based therapy. On the other hand, no or little information is available on prognosis of patients harboring multiple mutations and/or gene overexpression with contradictory influence on clinical outcome (eg, concurrent presence of CEBPA and/or NPM1 mutation and high BAALC and/or ERG expression). Studies that would test all currently known molecular alterations concurrently to determine the relative impact of each of them are underway. In addition, while the recent results of gene-expression profiling are promising, it remains to be determined whether microarray technology will become more useful clinically and thus be able to replace testing for several individual genetic alterations with prognostic significance in CN AML patients.
Figure 2.
Proposed schema assigning AML patients with a normal karyotype to risk-adapted postremission therapies using information on currently known prognostic markers as a basis for designing future clinical studies. SCT denotes stem-cell transplantation; ASCT, autologous SCT; and HDAC, high-dose cytarabine. Illustration drawn by Kenneth X. Probst.
Acknowledgments
This work was supported in part by National Cancer Institute (Bethesda, MD) grants CA77658, CA101140, CA31946, CA102031, and CA16058 and The Coleman Leukemia Research Foundation.
Footnotes
Conflict-of-interest disclosure: the authors declare no competing financial interests.
References
- 1.Mitelman F, Johansson B, Mertens F, editors. Mitelman database of chromosome aberrations in cancer, 2006, 5/2006 update. [Accessed June 15, 2006]; http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- 2.Mrózek K, Heinonen K, Bloomfield CD. Clinical importance of cytogenetics in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:19–47. doi: 10.1053/beha.2000.0114. [DOI] [PubMed] [Google Scholar]
- 3.Grimwade D, Walker H, Harrison G, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–1320. doi: 10.1182/blood.v98.5.1312. [DOI] [PubMed] [Google Scholar]
- 4.Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 5.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 6.Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood. 2006;107:3847–3853. doi: 10.1182/blood-2005-08-3522. [DOI] [PubMed] [Google Scholar]
- 7.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 8.Farag SS, Ruppert AS, Mrózek K, et al. Outcome of induction and postremission therapy in younger adults with acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:482–493. doi: 10.1200/JCO.2005.06.090. [DOI] [PubMed] [Google Scholar]
- 9.Schichman SA, Caligiuri MA, Strout MP, et al. ALL-1 tandem duplication in acute myeloid leukemia with a normal karyotype involves homologous recombination between Alu elements. Cancer Res. 1994;54:4277–4280. [PubMed] [Google Scholar]
- 10.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 11.Pabst T, Mueller BU, Zhang P, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 12.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype [erratum appears in N Engl J Med. 2005;352:740]. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 13.Tanner SM, Austin JL, Leone G, et al. BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc Natl Acad Sci U S A. 2001;98:13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bullinger L, Döhner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 15.Heuser M, Beutel G, Krauter J, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–3905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 16.Marcucci G, Baldus CD, Ruppert AS, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 17.Caligiuri MA, Strout MP, Lawrence D, et al. Rearrangement of ALL1 (MLL) in acute myeloid leukemia with normal cytogenetics. Cancer Res. 1998;58:55–59. [PubMed] [Google Scholar]
- 18.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 19.Fröhling S, Schlenk RF, Stolze I, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 20.Baldus CD, Tanner SM, Ruppert AS, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood. 2003;102:1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 21.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 22.Radmacher MD, Marcucci G, Ruppert AS, et al. Independent confirmation of a prognostic gene-expression signature in adult acute myeloid leukemia with a normal karyotype: a Cancer and Leukemia Group B study. Blood. 2006;108:1677–1683. doi: 10.1182/blood-2006-02-005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacher U, Kern W, Schnittger S, Hiddemann W, Haferlach T, Schoch C. Population-based age-specific incidences of cytogenetic subgroups of acute myeloid leukemia. Haematologica. 2005;90:1502–1510. [PubMed] [Google Scholar]
- 24.Mrózek K, Marcucci G, Ruppert AS, et al. Molecular heterogeneity and its prognostic significance in acute myeloid leukemia (AML) with normal cytogenetics. Ann Hematol. 2006;85(suppl 1):114–117. [Google Scholar]
- 25.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is essential for cooperative group clinical and correlative studies of acute leukemia: the Cancer and Leukemia Group B (CALGB) 8461 experience [abstract]. Blood. 2004;104:307a. Abstract 1081. [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe D, Cotterill SJ, Ross FM, et al. Cytogenetically cryptic AML1-ETO and CBFbeta-MYH11 gene rearrangements: incidence in 412 cases of acute myeloid leukaemia. Br J Haematol. 2000;111:1051–1056. doi: 10.1046/j.1365-2141.2000.02474.x. [DOI] [PubMed] [Google Scholar]
- 27.Grimwade D, Biondi A, Mozziconacci M-J, et al. Characterization of acute promyelocytic leukemia cases lacking the classic t(15;17): results of the European Working Party. Blood. 2000;96:1297–1308. [PubMed] [Google Scholar]
- 28.Mrózek K, Prior TW, Edwards C, et al. Comparison of cytogenetic and molecular genetic detection of t(8;21) and inv(16) in a prospective series of adults with de novo acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2001;19:2482–2492. doi: 10.1200/JCO.2001.19.9.2482. [DOI] [PubMed] [Google Scholar]
- 29.Grimwade D, Gorman P, Duprez E, et al. Characterization of cryptic rearrangements and variant translocations in acute promyelocytic leukemia. Blood. 1997;90:4876–4885. [PubMed] [Google Scholar]
- 30.Gamerdinger U, Teigler-Schlegel A, Pils S, et al. Cryptic chromosomal aberrations leading to an AML1/ETO rearrangement are frequently caused by small insertions. Genes Chromosomes Cancer. 2003;36:261–272. doi: 10.1002/gcc.10168. [DOI] [PubMed] [Google Scholar]
- 31.National Comprehensive Cancer Network. Clinical Practice Guidelines in Oncology–v.1.2006: Acute Myeloid Leukemia. [Accessed July 29, 2006]; doi: 10.6004/jnccn.2003.0044. http://www.nccn.org/professionals/physician_gls/PDF/aml.pdf. [DOI] [PubMed] [Google Scholar]
- 32.Mohr B, Bornhauser M, Thiede C, et al. Comparison of spectral karyotyping and conventional cytogenetics in 39 patients with acute myeloid leukemia and myelodysplastic syndrome. Leukemia. 2000;14:1031–1038. doi: 10.1038/sj.leu.2401775. [DOI] [PubMed] [Google Scholar]
- 33.Preiss BS, Kerndrup GB, Pedersen RK, Hasle H, Pallisgaard N. Contribution of multiparameter genetic analysis to the detection of genetic alterations in hematologic neoplasia: an evaluation of combining G-band analysis, spectral karyotyping, and multiplex reverse-transcription polymerase chain reaction (multiplex RT-PCR). Cancer Genet Cytogenet. 2006;165:1–8. doi: 10.1016/j.cancergencyto.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Fröhling S, Skelin S, Liebisch C, et al. Comparison of cytogenetic and molecular cytogenetic detection of chromosome abnormalities in 240 consecutive adult patients with acute myeloid leukemia. J Clin Oncol. 2002;20:2480–2485. doi: 10.1200/JCO.2002.08.155. [DOI] [PubMed] [Google Scholar]
- 35.Cuneo A, Bigoni R, Cavazzini F, et al. Incidence and significance of cryptic chromosome aberrations detected by fluorescence in situ hybridization in acute myeloid leukemia with normal karyotype. Leukemia. 2002;16:1745–1751. doi: 10.1038/sj.leu.2402605. [DOI] [PubMed] [Google Scholar]
- 36.Casas S, Aventín A, Fuentes F, et al. Genetic diagnosis by comparative genomic hybridization in adult de novo acute myelocytic leukemia. Cancer Genet Cytogenet. 2004;153:16–25. doi: 10.1016/j.cancergencyto.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Boissel N, Renneville A, Biggio V, et al. Prevalence, clinical profile and prognosis of NPM mutations in AML with normal karyotype. Blood. 2005;106:3618–3620. doi: 10.1182/blood-2005-05-2174. [DOI] [PubMed] [Google Scholar]
- 38.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma [erratum appears in Science. 1995;267:316-317]. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 39.Yoneda-Kato N, Look AT, Kirstein MN, et al. The t(3;5)(q25.1;q34) of myelodysplastic syndrome and acute myeloid leukemia produces a novel fusion gene, NPM-MLF1. Oncogene. 1996;12:265–275. [PubMed] [Google Scholar]
- 40.Falini B, Bolli N, Shan J, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 41.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 42.Kottaridis PD, Gale RE, Frew ME, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 43.Fröhling S, Schlenk RF, Breitruck J, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002;100:4372–4380. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 44.Kainz B, Heintel D, Marculescu R, et al. Variable prognostic value of FLT3 internal tandem duplications in patients with de novo AML and a normal karyotype, t(15;17), t(8;21) or inv(16). Hematol J. 2002;3:283–289. doi: 10.1038/sj.thj.6200196. [DOI] [PubMed] [Google Scholar]
- 45.Beran M, Luthra R, Kantarjian H, Estey E. FLT3 mutation and response to intensive chemotherapy in young adult and elderly patients with normal karyotype. Leuk Res. 2004;28:547–550. doi: 10.1016/j.leukres.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Bienz M, Ludwig M, Oppliger Leibundgut E, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype [erratum appears in Clin Cancer Res. 2005;11:5659]. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 47.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 48.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 49.Stirewalt DL, Meshinchi S, Kussick SJ, et al. Novel FLT3 point mutations within exon 14 found in patients with acute myeloid leukaemia. Br J Haematol. 2004;124:481–484. doi: 10.1111/j.1365-2141.2004.04808.x. [DOI] [PubMed] [Google Scholar]
- 50.Reindl C, Bagrintseva K, Vempati S, et al. Point mutations in the juxtamembrane domain of FLT3 define a new class of activating mutations in AML. Blood. 2006;107:3700–3707. doi: 10.1182/blood-2005-06-2596. [DOI] [PubMed] [Google Scholar]
- 51.Schnittger S, Schoch C, Dugas M, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- 52.Grundler R, Miething C, Thiede C, Peschel C, Duyster J. FLT3-ITD and tyrosine kinase domain mutants induce 2 distinct phenotypes in a murine bone marrow transplantation model. Blood. 2005;105:4792–4799. doi: 10.1182/blood-2004-11-4430. [DOI] [PubMed] [Google Scholar]
- 53.Choudhary C, Schwäble J, Brandts C, et al. AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations. Blood. 2005;106:265–273. doi: 10.1182/blood-2004-07-2942. [DOI] [PubMed] [Google Scholar]
- 54.Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 55.Yoshimoto G, Nagafuji K, Miyamoto T, et al. FLT3 mutations in normal karyotype acute myeloid leukemia in first complete remission treated with autologous peripheral blood stem cell transplantation. Bone Marrow Transplant. 2005;36:977–983. doi: 10.1038/sj.bmt.1705169. [DOI] [PubMed] [Google Scholar]
- 56.Kiyoi H, Naoe T, Nakano Y, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–3080. [PubMed] [Google Scholar]
- 57.Rombouts WJC, Blokland I, Löwenberg B, Ploemacher RE. Biological characteristics and prognosis of adult acute myeloid leukemia with internal tandem duplications in the Flt3 gene. Leukemia. 2000;14:675–683. doi: 10.1038/sj.leu.2401731. [DOI] [PubMed] [Google Scholar]
- 58.Abu-Duhier FM, Goodeve AC, Wilson GA, et al. FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a high-risk group. Br J Haematol. 2000;111:190–195. doi: 10.1046/j.1365-2141.2000.02317.x. [DOI] [PubMed] [Google Scholar]
- 59.Baldus CD, Thiede C, Soucek S, Bloomfield CD, Thiel E, Ehninger G. BAALC expression and FLT3 internal tandem duplication mutations in acute myeloid leukemia patients with normal cytogenetics: prognostic implications. J Clin Oncol. 2006;24:790–797. doi: 10.1200/JCO.2005.01.6253. [DOI] [PubMed] [Google Scholar]
- 60.Fitzgibbon J, Smith L-L, Raghavan M, et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–9154. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 61.Griffiths M, Mason J, Rindl M, et al. Acquired isodisomy for chromosome 13 is common in AML, and associated with FLT3-itd mutations [letter]. Leukemia. 2005;19:2355–2358. doi: 10.1038/sj.leu.2403988. [DOI] [PubMed] [Google Scholar]
- 62.Stirewalt DL, Kopecky KJ, Meshinchi S, et al. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood. 2006;107:3724–3726. doi: 10.1182/blood-2005-08-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parcells BW, Ikeda AK, Simms-Waldrip T, Moore TB, Sakamoto KM. FMS-like tyrosine kinase 3 in normal hematopoiesis and acute myeloid leukemia. Stem Cells. 2006;24:1174–1184. doi: 10.1634/stemcells.2005-0519. [DOI] [PubMed] [Google Scholar]
- 64.Ozeki K, Kiyoi H, Hirose Y, et al. Biologic and clinical significance of the FLT3 transcript level in acute myeloid leukemia. Blood. 2004;103:1901–1908. doi: 10.1182/blood-2003-06-1845. [DOI] [PubMed] [Google Scholar]
- 65.Kainz B, Fonatsch C, Schwarzinger I, Sperr WR, Jäger U, Gaiger A. Limited value of FLT3 mRNA expression in the bone marrow for prognosis and monitoring of patients with acute myeloid leukemia [letter]. Haematologica. 2005;90:695–696. [PubMed] [Google Scholar]
- 66.Kuchenbauer F, Kern W, Schoch C, et al. Detailed analysis of FLT3 expression levels in acute myeloid leukemia. Haematologica. 2005;90:1617–1625. [PubMed] [Google Scholar]
- 67.Stone RM, DeAngelo DJ, Klimek V, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105:54–60. doi: 10.1182/blood-2004-03-0891. [DOI] [PubMed] [Google Scholar]
- 68.Smith BD, Levis M, Beran M, et al. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood. 2004;103:3669–3676. doi: 10.1182/blood-2003-11-3775. [DOI] [PubMed] [Google Scholar]
- 69.De Angelo DJ, Stone RM, Heaney ML, et al. Phase II evaluation of the tyrosine kinase inhibitor MLN518 in patients with acute myeloid leukemia (AML) bearing a FLT3 internal tandem duplication (ITD) mutation [abstract]. Blood. 2004;104:496a–497a. Abstract 1792. [Google Scholar]
- 70.O'Farrell A-M, Foran JM, Fiedler W, et al. An innovative phase I clinical study demonstrates inhibition of FLT3 phosphorylation by SU11248 in acute myeloid leukemia patients. Clin Cancer Res. 2003;9:5465–5476. [PubMed] [Google Scholar]
- 71.Fiedler W, Serve H, Döhner H, et al. A phase 1 study of SU11248 in the treatment of patients with refractory or resistant acute myeloid leukemia (AML) or not amenable to conventional therapy for the disease. Blood. 2005;105:986–993. doi: 10.1182/blood-2004-05-1846. [DOI] [PubMed] [Google Scholar]
- 72.Wadleigh M, DeAngelo DJ, Griffin JD, Stone RM. After chronic myelogenous leukemia: tyrosine kinase inhibitors in other hematologic malignancies. Blood. 2005;105:22–30. doi: 10.1182/blood-2003-11-3896. [DOI] [PubMed] [Google Scholar]
- 73.Chen J, Lee BH, Williams IR, et al. FGFR3 as a therapeutic target of the small molecule inhibitor PKC412 in hematopoietic malignancies. Oncogene. 2005;24:8259–8267. doi: 10.1038/sj.onc.1208989. [DOI] [PubMed] [Google Scholar]
- 74.Chen J, Deangelo DJ, Kutok JL, et al. PKC412 inhibits the zinc finger 198-fibroblast growth factor receptor 1 fusion tyrosine kinase and is active in treatment of stem cell myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:14479–14484. doi: 10.1073/pnas.0404438101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mendel DB, Laird AD, Xin X, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res. 2003;9:327–337. [PubMed] [Google Scholar]
- 76.Heidel F, Solem FK, Breitenbuecher F, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- 77.Giles F, Schiffer C, Kantarjian H, et al. Phase 1 study of PKC412, an oral FLT3 kinase inhibitor, in sequential and concomitant combinations with daunorubicin and cytarabine (DA) induction and high-dose cytarabine (HDAra-C) consolidation in newly diagnosed patients with AML [abstract]. Blood. 2004;104:78a. Abstract 262. [Google Scholar]
- 78.Redner RL, Rush EA, Faas S, Rudert WA, Corey SJ. The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood. 1996;87:882–886. [PubMed] [Google Scholar]
- 79.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 80.Verhaak RGW, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 81.Suzuki T, Kiyoi H, Ozeki K, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106:2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 82.Caligiuri MA, Schichman SA, Strout MP, et al. Molecular rearrangement of the ALL-1 gene in acute myeloid leukemia without cytogenetic evidence of 11q23 chromosomal translocations. Cancer Res. 1994;54:370–373. [PubMed] [Google Scholar]
- 83.Schichman SA, Caligiuri MA, Gu Y, et al. ALL-1 partial duplication in acute leukemia. Proc Natl Acad Sci U S A. 1994;91:6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schnittger S, Kinkelin U, Schoch C, et al. Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML. Leukemia. 2000;14:796–804. doi: 10.1038/sj.leu.2401773. [DOI] [PubMed] [Google Scholar]
- 85.Döhner K, Tobis K, Ulrich R, et al. Prognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: a study of the Acute Myeloid Leukemia Study Group Ulm. J Clin Oncol. 2002;20:3254–3261. doi: 10.1200/JCO.2002.09.088. [DOI] [PubMed] [Google Scholar]
- 86.Steudel C, Wermke M, Schaich M, et al. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer. 2003;37:237–251. doi: 10.1002/gcc.10219. [DOI] [PubMed] [Google Scholar]
- 87.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 88.Whitman SP, Liu S, Vukosavljevic T, et al. The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood. 2005;106:345–352. doi: 10.1182/blood-2005-01-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muñoz L, Nomdedéu JF, Villamor N, et al. Acute myeloid leukemia with MLL rearrangements: clinicobiological features, prognostic impact and value of flow cytometry in the detection of residual leukemic cells. Leukemia. 2003;17:76–82. doi: 10.1038/sj.leu.2402708. [DOI] [PubMed] [Google Scholar]
- 90.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Meijer J, et al. Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4:31–40. doi: 10.1038/sj.thj.6200216. [DOI] [PubMed] [Google Scholar]
- 91.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 92.Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med. 2004;351:2403–2407. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 93.Mrózek K, Heinonen K, Theil KS, Bloomfield CD. Spectral karyotyping in patients with acute myeloid leukemia and a complex karyotype shows hidden aberrations, including recurrent overrepresentation of 21q, 11q, and 22q. Genes Chromosomes Cancer. 2002;34:137–153. doi: 10.1002/gcc.10027. [DOI] [PubMed] [Google Scholar]
- 94.Baldus CD, Liyanarachchi S, Mrózek K, et al. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: amplification discloses overexpression of APP, ETS2, and ERG genes. Proc Natl Acad Sci U S A. 2004;101:3915–3920. doi: 10.1073/pnas.0400272101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Damiani D, Tiribelli M, Calistri E, et al. The prognostic value of P-glycoprotein (ABCB) and breast cancer resistance protein (ABCG2) in adults with de novo acute myeloid leukemia with normal karyotype. Haematologica. 2006;91:825–828. [PubMed] [Google Scholar]
- 96.Wilson CS, Davidson GS, Martin SB, et al. Gene expression profiling of adult acute myeloid leukemia identifies novel biologic clusters for risk classification and outcome prediction. Blood. 2006;108:685–696. doi: 10.1182/blood-2004-12-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WLJ, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–845. doi: 10.1182/blood-2002-05-1459. [DOI] [PubMed] [Google Scholar]
- 98.Barragán E, Cervera J, Bolufer P, et al. Prognostic implications of Wilms' tumor gene (WT1) expression in patients with de novo acute myeloid leukemia. Haematologica. 2004;89:926–933. [PubMed] [Google Scholar]
- 99.Schmid D, Heinze G, Linnerth B, et al. Prognostic significance of WT1 gene expression at diagnosis in adult de novo acute myeloid leukemia. Leukemia. 1997;11:639–643. doi: 10.1038/sj.leu.2400620. [DOI] [PubMed] [Google Scholar]
- 100.Li Q, Kopecky KJ, Mohan A, et al. Estrogen receptor methylation is associated with improved survival in adult acute myeloid leukemia. Clin Cancer Res. 1999;5:1077–1084. [PubMed] [Google Scholar]
- 101.Chim CS, Liang R, Tam CY, Kwong YL. Methylation of p15 and p16 genes in acute promyelocytic leukemia: potential diagnostic and prognostic significance. J Clin Oncol. 2001;19:2033–2040. doi: 10.1200/JCO.2001.19.7.2033. [DOI] [PubMed] [Google Scholar]
- 102.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2003;17:1813–1819. doi: 10.1038/sj.leu.2403054. [DOI] [PubMed] [Google Scholar]
- 103.Ropero S, Setien F, Espada J, et al. Epigenetic loss of the familial tumor-suppressor gene exostosin-1 (EXT1) disrupts heparan sulfate synthesis in cancer cells. Hum Mol Genet. 2004;13:2753–2765. doi: 10.1093/hmg/ddh298. [DOI] [PubMed] [Google Scholar]
- 104.Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 105.Gale RE, Hills R, Kottaridis PD, et al. No evidence that FLT3 status should be considered as an indicator for transplantation in acute myeloid leukemia (AML): an analysis of 1135 patients, excluding acute promyelocytic leukemia, from the UK MRC AML10 and 12 trials. Blood. 2005;106:3658–3665. doi: 10.1182/blood-2005-03-1323. [DOI] [PubMed] [Google Scholar]