Evolutionary Strata on the X Chromosomes of the Dioecious Plant Silene latifolia: Evidence From New Sex-Linked Genes (original) (raw)
Abstract
Despite its recent evolutionary origin, the sex chromosome system of the plant Silene latifolia shows signs of progressive suppression of recombination having created evolutionary strata of different X–Y divergence on sex chromosomes. However, even after 8 years of effort, this result is based on analyses of five sex-linked gene sequences, and the maximum divergence (and thus the age of this plant's sex chromosome system) has remained uncertain. More genes are therefore needed. Here, by segregation analysis of intron size variants (ISVS) and single nucleotide polymorphisms (SNPs), we identify three new Y-linked genes, one being duplicated on the Y chromosome, and test for evolutionary strata. All the new genes have homologs on the X and Y chromosomes. Synonymous divergence estimated between the X and Y homolog pairs is within the range of those already reported. Genetic mapping of the new X-linked loci shows that the map is the same in all three families that have been studied so far and that X–Y divergence increases with genetic distance from the pseudoautosomal region. We can now conclude that the divergence value is saturated, confirming the cessation of X–Y recombination in the evolution of the sex chromosomes at ∼10–20 MYA.
SILENE latifolia is a model system for the study of the evolution of plant sex chromosomes. The sex chromosomes of dioecious Silene species have several striking similarities with those of animals, including mammals (Guttman and Charlesworth 1998; Filatov 2005c; Nicolas et al. 2005), but they evolved independently and much more recently. The recent origin in a largely hermaphroditic plant genus, and the evidence of synteny of sex-linked genes and their orthologs in the hermaphroditic species S. vulgaris (Filatov 2005a), show clearly that, like mammalian sex chromosomes, those in S. latifolia evolved from a pair of ordinary chromosomes.
Due to its recent origin, the S. latifolia Y chromosome is probably in an early stage of degeneration. It is large in size, and it has been suggested that this reflects a large gene content (Negrutiu et al. 2001). However, this must be tested; an alternative is that the large size of the Y might reflect a highly repetitive DNA content, suggesting a stage in the degeneration process when repetitive sequences, including transposable elements, have probably accumulated, but before the stage in which most individual genes have lost function and can be deleted. The Y chromosome indeed appears to have accumulated chloroplast sequences (Kejnovsky et al. 2006), and there is also evidence of repetitive sequences and transposons in the S. latifolia genome (Pritham et al. 2003), but the extent of male-specific (Y-linked) sequence accumulation is not yet clear, although Y-specific sequences certainly exist (Donnison and Grant 1999). Similarly, the small Y chromosome-like region surrounding the sex-determining region in papaya (which may possibly have evolved more recently than the S. latifolia sex chromosomes) has a higher repetitive sequence content (and thus a lower gene density) than the genome as a whole (Liu et al. 2004).
To make progress in understanding sex chromosome evolution and organization in plants, and to test for genetic degeneration of Y chromosomes, sex-linked genetic markers are required. Several kinds of Y-linked genetic markers have been developed in S. latifolia, including anonymous markers such as AFLPs and RAPDs (Di Stilio et al. 1998; Nakao et al. 2002; Obara et al. 2002). Although it is straightforward to develop such anonymous markers, which are useful for obtaining genetic maps of the sex chromosomes (Lebel-Hardenack et al. 2002; Moore et al. 2003; Zluvova et al. 2005; Scotti and Delph 2006), these markers provide no information about the age of the sex chromosome system or the times since recombination between the X and Y stopped in different regions of these chromosomes, nor about whether the Y chromosome is genetically degenerated or degenerating. Genic markers are thus potentially much more valuable than anonymous ones. Such markers provide access to the gene coding sequences, containing synonymous and nonsynonymous sites that are subject to different selective constraints in functional copies (Gillespie 1991; Ohta 1995), so that it becomes possible to estimate the divergence time between the X and Y copies and to test for genetic degeneration of Y-linked copies (Guttman and Charlesworth 1998; Nicolas et al. 2005). Such studies are progressing rapidly for the neo-Y chromosome of Drosophila miranda (Bachtrog 2003; Bachtrog 2004).
Only seven genes on the Y chromosome of S. latifolia have been described after almost a decade of work (Guttman and Charlesworth 1998; Delichere et al. 1999; Atanassov et al. 2001; Matsunaga et al. 2003; Moore et al. 2003; Filatov 2005c; Nicolas et al. 2005). One of these has no X counterpart, being duplicated from an autosomal gene (Matsunaga et al. 2003), and only one is degenerated (Guttman and Charlesworth 1998). The five X-linked genes so far mapped are arranged along a gradient of X–Y synonymous divergence (Filatov 2005a), increasing with distance from the pseudoautosomal region (Filatov 2005a; Nicolas et al. 2005), although neither family allowed mapping all these genes. These findings suggest progressive steps in the cessation of recombination between the X and Y chromosomes, thus creating “evolutionary strata” on the sex chromosomes, similar to those described in mammalian X and Y chromosomes (Lahn and Page 1999). In the S. latifolia sex chromosomes, three divergence levels have been suggested. The two genes, SlX3/Y3 and SlX4/Y4, with the highest divergence have synonymous site divergence (_K_s) >15%, while, for the least diverged pair, SlX1/Y1, _K_s is only 3.6% (and intron divergence ∼2%), and two gene pairs, DD44X/DD44Y and SlSSX/SlSSY, have intermediate divergence (_K_s ∼7–8%). With only five loci, discrete groupings of _K_s values cannot be statistically significant, and the number is too low to formally test the ordering along the X chromosome of genes with different X–Y divergence in “evolutionary strata.”
To answer these questions, and to help understand the evolution of sex chromosomes, we use straightforward genetic approaches to identify sex-linked genes in S. latifolia, based on cDNA sequences obtained from this species. By using segregation analysis of ISVS and single nucleotide polymorphisms (SNPs), we identify four new Y-linked loci with homologs on the X chromosome. Comparison of silent site divergence between pairs of X/Y homologs, together with genetic mapping of the X-linked copies, confirm the existence of a gradient in divergence (evolutionary strata) of genes in this sex chromosome system, which is much younger than the sex chromosomes of mammals or birds.
MATERIALS AND METHODS
S. latifolia families and DNA samples:
Male and female S. latifolia plants were grown from seeds collected from European natural populations (Table 1). Progeny from a total of four F1 families generated by between-population cross-pollination of females were used for segregation analyses. Genetic mapping (described in detail below) was done using an F2 family of 92 plants generated by crossing two members of the F1 family H2005-1, a full sibship from crossing a female plant from France with a male from The Netherlands (Table 1). Genomic DNA from S. latifolia individuals was obtained from fresh leaves using the FastDNA kit (Q-biogene) following the manufacturer's instructions.
TABLE 1.
S. latifolia families used in this study
| No. of progeny plants | Maternal plant | Population of origin | Pollen donor | Population of origin | ||
|---|---|---|---|---|---|---|
| Families | Males | Females | ||||
| F2005-4 | 7 | 4 | E2004 17-3 | The Netherlands | E2004 15-1 | Serre de Nogere (Portugal) |
| F2005-5 | 4 | 5 | E2004 11-1 | Canche (France) | E2004 12-1 | Vandy (United States) |
| H2005-1 | 7 | 7 | E2004 11-1 | Canche (France) | E2004 17-1 | The Netherlands |
| H2005-2 | 5 | 5 | E2004 11-1 | Canche (France) | E2004 15-1 | Serre de Nogere (Portugal) |
The ISVS method:
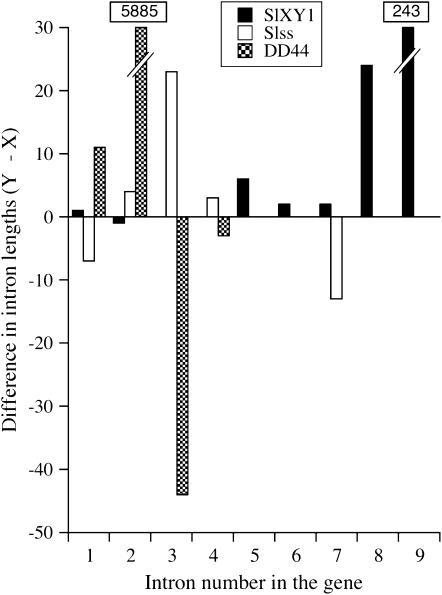
A size comparison of homologous introns in previously described X–Y gene pairs in S. latifolia revealed that length variants are common (Figure 1). Among homologous introns in the SlX1/SlY1, SlssX/SlssY, and DD44X/DD44Y gene pairs (Atanassov et al. 2001; Moore et al. 2003; Filatov 2005c), most have fixed size differences between the X and Y copies (in seven of the nine SlX1/SlY1 introns, five of the seven introns from the sequenced region of the SlssX/SlssY gene pair and four of the five DD44X/DD44Y introns). These presumably represent indel substitutions accumulated after recombination had ceased between the sex chromosomes. Even in the gene pair with the lowest X–Y divergence so far found in S. latifolia, SlX1/SlY1 (Atanassov et al. 2001; Nicolas et al. 2005), most of the homologous introns differ in size. We therefore expected that a large proportion of introns might exhibit size polymorphism. To develop molecular markers to test for sex linkage, we used a method for analyzing their segregation in S. latifolia and for detecting intron size variants in loci whose sequences we obtained from a cDNA library. The general approach of using length variants has been employed previously (Feltus et al. 2006), but we have refined the method so that it is highly sensitive but inexpensive. We refer to this method as “ISVS.”
Figure 1.—
Fixed intron-size differences (in base pairs) among three S. latifolia sex-linked homologous genes: SlX1/SlY1, SlSSX/SlSSY, and DD44X/DD44Y. Intron-size differences exceeding the _y_-axis scale are given in boxes.
To identify potential intron positions in our cDNA sequences, we used genome sequence data from Arabidopsis thaliana (At) and Oryza sativa (Os). Intron positions conserved in the genomes of these two distantly related plants are also likely to be maintained in S. latifolia (Inada et al. 2003). Primers were designed in exonic regions flanking likely intron sites (supplemental Table 1 at http://www.genetics.org/supplemental/) according to Palumbi and Baker (1994). To increase our ability to detect small length differences, we used capillary electrophoresis, which can discriminate PCR products differing by as little as a single base pair (Swerdlow and Gesteland 1990; Strege and Lagu 1991).
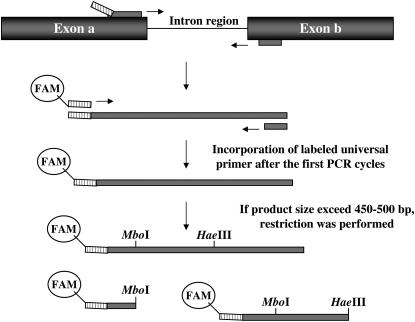
To reduce the cost of primer synthesis, we used the approach originally designed for microsatellite genotyping (Schuelke 2000), with a universal forward primer labeled with a fluorescent dye, a specific forward primer with a 5′ tail complementary to the forward universal primer, and a specific reverse primer (Figure 2). The universal primer used in this study was 5′-GGTTGGAGCTAGTGTTGTG-3′, which has an annealing temperature of 4°–5° below those of the specific forward and reverse primers (Schuelke 2000). To maximize the efficiency of the TAQ polymerase transferase activity, reverse primers were designed to start with a G at the 5′-end (Vos et al. 1995), and the final 72° extension step was extended to 20 min. This guarantees that single, instead of double, peaks were observed in the capillary electrophoregrams.
Figure 2.—
The ISVS method. Primers (shown as narrow bars) were designed in exonic regions close to intron boundaries. The forward (specific) primer contains a 5′ 20-bp tail complementary to a universally labeled primer and is added to the PCR reaction at limiting concentrations. This guarantees the incorporation of the universally labeled primer after a few PCR cycles. If the PCR product was >500 bp, restriction with a panel of restriction enzymes was performed before capillary electrophoresis analysis.
The fluorescently labeled PCR products were separated by capillary electrophoresis and length differences were detected in an ABI 3730 capillary sequencer (Applied Biosystems, Foster City, CA) using the GeneScan 500 LIZ (Applied Biosystems) size standard. Genotypes were determined using the Genemapper software package 3.7 (Applied Biosystem). When a PCR amplicon size exceeded the limit of resolution of the size standard (500 bp), restriction enzyme digestion was performed with a panel of endonucleases (_Hae_III, _Hin_fI, _Mbo_I, _Mse_I, _Msp_I, _Rsa_I, _Scr_FI) in a final volume of 10 μl, including buffer 4 1× (New England Biolabs, Beverly, MA), 1 unit restriction enzyme, and 4 μl of PCR product. Reactions were kept at 37° for 20 min and 65° for 5 min and analyzed in the capillary sequencer to find an enzyme giving fragments in a suitable size range. Restriction digests are normally desalted before capillary electrophoresis because of the interference of ions (especially chloride ions) with the uptake of DNA samples into the capillary (Moeseneder et al. 1999). We found that using a buffer for the restriction digestions containing acetate instead of chloride ions [buffer 4 from New England Biolabs and MULTI-CORE buffer from Promega (Madison, WI)] and extending the injection time to 20 sec eliminated the need for the column desalinization step.
Segregation of SNPs for inferring sex linkage:
cDNA sequences that did not have significant amino acid identity with either of the At and Os genomes or lacked the predicted introns could not be analyzed by ISVS. For these, sex linkage was preliminarily tested by amplifying PCR products with primers designed from S. latifolia cDNA sequences and using PCR–RFLP with a panel of restriction enzymes (_Hae_III, _Hin_fI, _Mbo_I, _Mse_I, _Msp_I, _Rsa_I, and _Scr_FI). If no suitable polymorphism was detected, direct sequencing of PCR products was done according to Filatov (2005c) to detect SNPs for use in segregation analyses. Primers for all loci studied are listed in supplemental Table 1 at http://www.genetics.org/supplemental/.
Amplification of 5′ and 3′ cDNA ends:
To obtain complete coding sequences of the sex-linked genes, we performed 5′- and 3′-RACE reactions. Total RNA was extracted from young leaves of the male plant E2004-17-1 from sibship E2004-17, using the RNeasy plant mini kit (QIAGEN, Chatsworth, CA). mRNA was purified using poly(T)-tailed Dyna-beads (Invitrogen, San Diego). First-strand cDNA synthesis was accomplished starting from 0.5 to 1 μg poly(A) RNA by using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and the oligo(dT) primer (T)15VN. Two microliters of this reaction was used to obtain 3′ cDNA ends by PCR using forward specific primers and the poly(T) primer. For the 5′-RACE reactions, a modified protocol of the CapSelect technique (Schmidt and Mueller 1999) was used to enrich the cDNA library for complete mRNA 5′-ends. Details of the modifications can be provided on request to R. Bergero.
Sequence analysis:
PCR products of interest (sex linked or belonging to sets of paralogous loci) were cloned into a T-tailed pBSKS+ vector (Marchuk et al. 1991) and sequenced in an ABI 3730 capillary sequencer (Applied Biosystems). Specific primers were used to obtain separate X- and Y-linked partial sequences (see below). Genomic sequences of the sex-linked gene pairs were obtained from male E2004-17-1 (see Table 1), and exon/intron boundaries were inferred on the basis of comparison with the cDNA sequences and gene structures of orthologous At genes.
Sequences were first aligned with Sequencer v. 4.5 (GeneCodes, Ann Arbor, MI) and then manually adjusted using the sequence alignment editor Se-Al v. 2.0a11 (Se-Al: Sequence Alignment Editor, http://evolve.zoo.ox.ac.uk/). Sequence divergence values were computed using the package DnaSP v. 3.99 (Rozas and Rozas 1999). Sequence data from cDNA clones and sex-linked loci were deposited in GenBank (accession nos. DV768193–DV768364 and EF408657–EF408664).
Genetic mapping:
Segregation of molecular markers (indels and SNPs) located within X-linked loci and polymorphic in the maternal genotype were scored in the male and female parents and in 92 progeny plants of an F2 family (see above) and used to build a linkage map of the newly identified loci together with the previously mapped ones. The pseudoautosomal marker OPA (Filatov 2005a; Nicolas et al. 2005) was mapped in our F2 family, as well as the SlX4 locus, the most distantly located gene at the other end of the X chromosome in previous maps of two families (Filatov 2005a; Nicolas et al. 2005); we also included SlX3. The Kosambi mapping function was used for computing genetic distances in centimorgans.
RESULTS
Screening of cDNA sequences for sex linkage:
A total of 39 cDNA sequences were tested for sex linkage as described above, using intron length variants (the clones are denoted in what follows by our numbers, “SlS1-,” followed by an individual clone number; here we give only the clone numbers) or SNP segregation. In total, 22 cDNAs had introns suitable for analysis by intron length analysis, and we analyzed most of these using ISVS.
Intron size variants were first scored in the parental individuals of several families (Table 1). Sixty-eight percent of the introns had a size polymorphism in at least one parental male and one parental female (supplemental Table 2 at http://www.genetics.org/supplemental/). Segregation patterns of these variants were scored in the F1 families to distinguish sex-linked and autosomal loci. In a few cases, paralogous loci amplified, and the progeny did not show a simple biallelic pattern. For two of them, no clear interpretation of segregation results was evident (clones 09B11 and 11D10, where a band common to both parents was not found in some of the offspring). The others were analyzed as small gene families, which is straightforward using ISVS (see supplemental Table 2 at http://www.genetics.org/supplemental/).
For those EST sequences that could not be analyzed by ISVS, sex linkage was tested by SNP segregation using PCR–RFLP analysis. Inference of sex linkage or autosomal inheritance (supplemental Table 3 at http://www.genetics.org/supplemental/) was conclusive for only 47% of the genes tested. For the other clones, there was either no polymorphism or the analysis was inconclusive, often because of amplification of related loci, a common difficulty when attempting to analyze a genome as large and complex as this; the 1C DNA content is estimated to be 2646 Mb (Costich et al. 1991). Overall, ISVS was thus superior to SNP segregation analysis in efficacy and simplicity for testing sex linkage, particularly because it allows segregation analysis in the presence of paralogous loci.
Two Y-linked genes identified by ISVS:
SlCypX/Y:
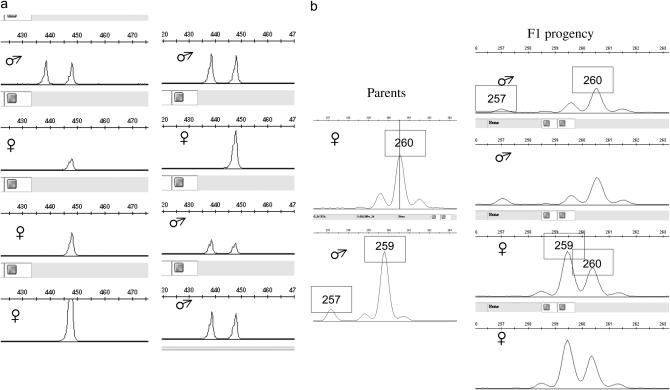
Clone 09C11 yielded a sequence containing an ORF of 623 amino acids, showing 80% amino acid identity to the A. thaliana peptidyl-prolyl _cis_-trans isomerase locus, Cyp2 (At3g44600). ISVS using primers flanking introns 2 and 3 showed clear segregation patterns consistent with X and Y linkage (Figure 3; supplemental Table 2 at http://www.genetics.org/supplemental/). Coding sequences of the Y-linked genomic fragments unambiguously matched the cDNA sequence 09C11 (_K_s = 0.2%, _K_a = 0), showing that the Y-homolog is actively transcribed. This gene was named SlCypY and its X-linked counterpart was named SlCypX.
Figure 3.—
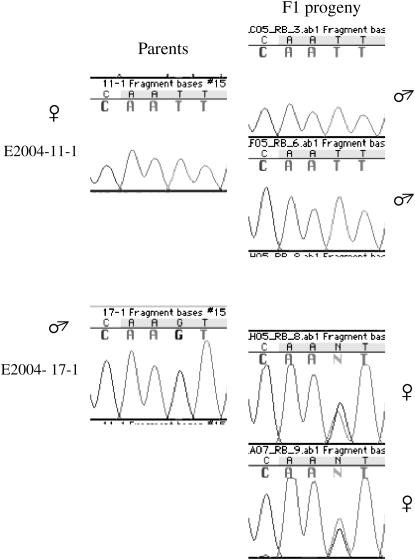
Inference of X and Y linkage for locus SlCypX/SlCypY. (a) Intron 3 variants. All S. latifolia individuals from several natural populations yielded a 447-bp band, and male plants have an extra 438-bp variant. (b) Segregation of intron 2 variants in the H2005-1 family after restriction with _Hae_III. The male parent has 257- and 259-bp variants, whereas the female parent is homozygous for a 260-bp variant. The segregation of these variants follows a clear X linkage pattern in the F1. All males inherit the 260-bp variant from the mother, while all females are heterozygous, inheriting both maternal and paternal variants. Moreover, because only the F1 males inherit the 257-bp variant, this must correspond to the Y-linked intron variant.
SlX6/SlY6:
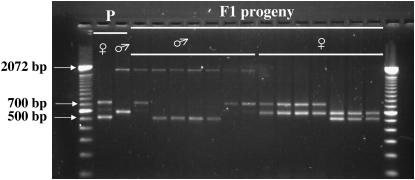
ISVS based on the cDNA sequence 10C04, containing an ORF of 281 amino acids with 39% amino acid identity with an At unknown protein (AT2G34570), produced differently sized bands, readily distinguishable by gel analysis. The consistent presence of two bands found only in male plants suggested duplicated Y-linked loci, although the putative orthologs in the two hermaphroditic species A. thaliana and O. sativa are single copy. Segregation of four amplicons (510, 590, 730, and 1830 bp) obtained with primer pairs designed upstream of intron 1 and within intron 2 clearly showed X and Y linkage of the PCR products (Figure 4; supplemental Table 2 at http://www.genetics.org/supplemental/). Large insertions were found in both introns 1 and 2 (Figure 6). This X- and Y-linked gene pair was named SlX6a/SlY6a.
Figure 4.—
Inference of X and Y linkage for locus SlX6a/SlY6a. Forward and reverse primers were designed within exon 1 and intron 2, respectively. Segregation of four amplicons (510 and 730 bp from the maternal plant and 590 and 1830 bp from the paternal plant) in the H2005-1 family is clearly consistent with X and Y linkage of the PCR size variants. All F1 males, and no F1 female, inherited the Y-linked 1830 bp from the parental male. All F1 female plants inherited the 590-bp band from the father and either the 510- or the 730-bp band from the mother. Conversely, all F1 males did inherit either the 510- or the 730-bp band from the mother, and not the 590-bp band from the father. An extra faint band observed in the maternal plant (size 610 bp) cosegregated with the 730-bp band in F1 males. This probably represents an X-linked paralogous locus.
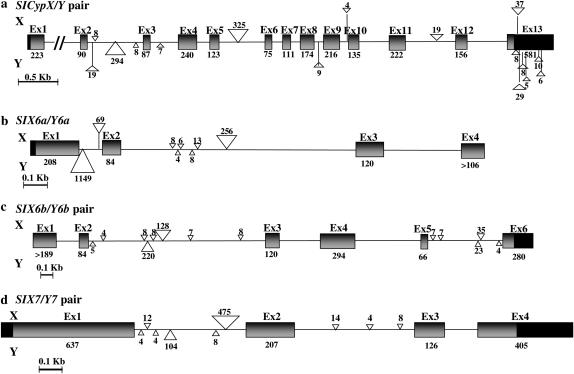
Figure 6.—
Gene structures of the new X- and Y-linked gene pairs. (a) SlCypX/SlCypY. (b) SlX6a/SlY6a. (c) SlX6b/SlY6b. (d) SlX7/SlY7. For the SlX6a/SlY6a pair, only partial genomic sequences are known (almost to the end of exon 3 of SlX6a and of exon 4 of SlY6a). The X and Y homologs exhibit strict conservation in gene structure, but the intron sizes often differ, with some large indels. Intronic insertions >3 bp are represented by triangles. Sizes in base pairs of exons and intronic insertions are given. Solid boxes represent the 5′ and 3′ untranslated regions.
ISVS from amplification of intron 2 (data not shown) clearly demonstrated the existence of paralogous loci and their X and Y linkage (SlX6b/SlY6b).
A third Y-linked gene, SlX7/SlY7, identified by SNP analysis:
A SNP scorable by the restriction enzyme _Hsp92_II was detected in partial genomic sequences amplified with primers designed on the sequence of the cDNA clone 11F03. This corresponds to an ORF of 348 amino acids showing 75% amino acid identity with an A. thaliana unknown protein (locus AT5G48020) for 95% of its ORF. Segregation analysis of a _Hsp92_II restriction site showed Y linkage of sequences containing the _Hsp92_II restriction site. Segregation of a SNP causing a nonsynonymous substitution (I → S) in sequences lacking the _Hsp92_II restriction site indicated X linkage (Figure 5). Coding sequences of the X-linked genomic fragments matched the cDNA sequence 11F03 (1% divergence). This X- and Y-linked gene pair was named SlX7/SlY7.
Figure 5.—
Inference of X linkage for locus SlX7. X linkage was inferred on the basis of an SNP causing a nonsynonymous substitution (I → S) at position +434 in H2005-1 family. The maternal plant was homozygous for T whereas the paternal plant was hemizygous for G. All male plants in the F1 progeny were hemizygous for T, inheriting the maternal SNP, whereas all females belonging to the F1 progeny were heterozygous (G/T), inheriting both the paternal and the maternal SNPs.
Gene structure and gene sequence comparisons:
The gene structures of SlCypY and SlY7 and their respective X counterparts were inferred by assembling PCR-amplified genomic partial sequences and sequences obtained by 5′- and 3′-RACE reactions. The gene structures of the X and Y homologs of the two sex-linked genes are very similar, with the same number of introns in conserved positions. The introns differ in size, and large indels are seen in either the X or Y copies (Figure 6).
For the Y homologs of SlCyp and Sl7, the sequences do not suggest any evidence of sequence degeneration, such as premature stop codons or indels causing frameshifts. Sequence divergence estimates between X/Y pairs are shown in Table 2. The synonymous site divergence (_K_s) between SlX7 and SlY7 is estimated to be 24.0%, similar to that of the most diverged S. latifolia sex-linked gene pair so far reported (SlX4/SlY4); the value for silent sites (both synonymous coding sequence sites and noncoding sequences) was lower (_K_silent = 14.9%). For SlCypX vs. SlCypY, _K_s is 6.1% (Table 2). Nonsynonymous site divergence for both genes was much lower than synonymous site divergence (Table 2), suggesting that the Y-linked copies are functional.
TABLE 2.
Sequence divergence of the four new S. latifolia X/Y gene pairs
| Best BLAST hit to A. thaliana genome | Divergence in coding sequence | Silent site (synonymous and noncoding) divergence | ||||||
|---|---|---|---|---|---|---|---|---|
| Locus | Locus name | Locus tag | % amino acid identity | No. of codons analyzed | _K_s (%) | _K_a (%) | No. of sites analyzed (bp) | Nucleotide divergence (%) |
| SlCypX/SlCypY | Cyp2 | At3G44600 | 80 | 553 | 6.1 | 0.8 | 3785 | 6.4 |
| SlX6a/SlY6a | Unknown protein | At2G34570 | 39 | 86 | 20.3 | 0.5 | 1165 | 18.1 |
| SlX6b/SlY6b | Unknown protein | At2G34570 | 41 | 200 | 4.5 | 0.4 | 2903 | 5.7 |
| SlX7/SlY7 | Unknown protein | At5G48020 | 75 | 316 | 24.0 | 2.6 | 1395 | 14.9 |
Synonymous site divergence between the two other Y-linked sequences, SlY6a and SlY6b, is 18.2%. Together with the pairwise _K_s values in Table 2, we can infer the respective X–Y orthologous pairs. One of the X sequences is quite similar to SlY6b (_K_s = 4.5%) and much more diverged from SlY6a (_K_s = 12.6%); this identifies this X sequence as the SlY6b ortholog. _K_s between the two X-linked sequences (SlX6a and SlX6b) is also equal to 12.6%, consistent with paralogy.
Between either SlX6a and SlY6a or SlX6b and SlY6b, _K_a is low (Table 2), indicating selective constraint on both sequences, so that even the highly diverged Y-linked gene must encode a functional protein; again, no premature stop codons or frameshifts were found in the almost complete coding sequences obtained for the locus SlY6b and the partial coding sequence obtained for SlY6a. Neither SlX6a nor SlY6a matched the cDNA sequence (_K_s's are 12.6 and 21.8%, respectively), whereas there is only one synonymous substitution between the cDNA sequence and the SlX6b coding sequence. This suggests that the SlX6b locus is the source of the cDNA sequence.
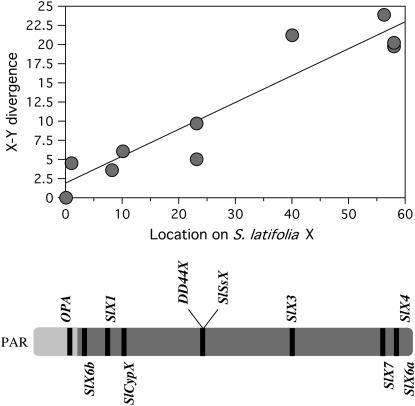
All eight loci now mapped on the X chromosome are shown in Figure 7, including both of the two most extreme markers so far mapped (the pseudoautosomal marker and SlX4, at the other end of the X; see Nicolas et al. 2005), and the map includes a large genetic distance (58 cM). The two loci with the highest X–Y divergence (S_lX7_ and SlX6a) are closely linked to SlX4, which has a similar X–Y K_s. This establishes a reliable upper limit for X–Y divergence (i.e., for the region of oldest X–Y divergence, or evolutionary stratum I of the X). The SlCypX locus, with an intermediate X–Y divergence, maps distantly from these genes, and only 10 cM from the pseudoautosomal marker (Nicolas et al. 2005), close to the S_lX1 gene. The X–Y divergence values differ significantly between these two groups of loci (the 95% confidence intervals are 3.89–8.15 and 19.6–23.2, respectively), and divergence from the Y loci (_K_s) suggests two distinctly separated clusters (Figure 7), possibly corresponding to distinct strata. With the current number of loci, we did not test for discontinuity between the strata.
Figure 7.—
Genetic map of the S. latifolia X chromosome (bottom), and the relationship with synonymous site divergence (_K_s) of X-linked loci from their Y counterparts (top). The regression is highly significant (P = 0.001 for a linear regression or 0.012 with a Wilcoxon signed rank test). Map positions, in centimorgans, from the pseudoautosomal marker (OPA) are indicated. The markers shown below the diagram of the X chromosome are the new ones, and the previously mapped markers are shown above the diagram. Two of the three previously described markers, OPA, SlX3, and SlX4, were also mapped in our family, while the SlSSX, DD44X, and SlX1 map positions are based on previously published maps.
DISCUSSION
Sex-linked genes:
With the development of straightforward genetic approaches such as ISVS, there is now no major obstacle preventing isolation of a large set of sex-linked genes from S. latifolia or from other plant and animal young sex chromosome systems, such as those of papaya (Liu et al. 2004; Ma et al. 2004) and fish (Kondo et al. 2004; Peichel et al. 2004). All our new Y-linked genes have homologs on the X; this includes the duplicates of the gene denoted above by SlY6, which also appears to be duplicated on the X. Indeed, so far, all except two sex-linked genes identified from S. latifolia have X and Y counterparts, and the Y-linked copies of most are probably functional (reviewed in Filatov 2005b; Nicolas et al. 2005). The only degenerated Y-linked gene so far discovered in S. latifolia, _MROS3_-Y (Guttman and Charlesworth 1998), might not have degenerated as a direct consequence of Y linkage, because there are multiple copies on the X (Kejnovsky et al. 2001) and duplicate genes are often likely to degenerate (Walsh 1995; Wolfe and Shields 1997).
The lack of known degenerate Y-linked genes does not, however, necessarily imply that the S. latifolia X and Y chromosomes have similar gene content or that the Y has not lost genes that are present on the X. To estimate the proportion of genes carried on the X chromosome that are also present on the Y, the proportion of these that have degenerated copies, and the forms of degeneration that have occurred, genes need to be ascertained from X linkage or from unbiased mapping of S. latifolia. X linkage is, however, more difficult to establish than Y linkage, because informative markers distinguishing different X-linked alleles in mapping families are scarce. In contrast, Y-specific molecular markers (indels and SNPs) are much more common, because the evolutionary time since recombination has ceased between the X and Y in most species is long enough that divergence is considerable (several percent, for most S. latifolia X–Y pairs; see Figure 7 and Filatov 2005b; Nicolas et al. 2005). X-linked genes with deleted or silenced Y counterparts could therefore easily escape detection in segregation analyses of short genomic sequences of only a few hundred base pairs.
Our results nevertheless support previous evidence that there is considerable homology between the S. latifolia X and Y chromosomes. So far, only one Y-linked gene, SlAp3, has been reported to have only an autosomal homolog (Matsunaga et al. 2003). The situation is thus different from that in the fruit fly, Drosophila melanogaster, in which all Y-linked genes appear to have autosomal origins (Carvalho 2002), and in the primate Y chromosomes, which carry both X-homologous and transposed genes (Vallender and Lahn 2004). The ascertainment of X-linked genes and searches for homologous Y-linked sequences are necessary to clarify the relative frequencies of the two components in the S. latifolia Y chromosome.
Evolutionary strata on the sex chromosomes:
The eight X-linked genes now mapped are arranged along a gradient of X–Y silent site divergence, increasing with distance from the pseudoautosomal region, as was previously suggested (Filatov 2005a; Nicolas et al. 2005); our results show that this pattern is statistically significant and that it is consistent in three S. latifolia mapping families. The slight differences in map intervals in the different families are within the range expected from sampling error, given the family sizes, and do not suggest any differences in the gene orders in different plants of this species, whereas the X of S. dioica has a region inverted relative to the S. latifolia arrangement (Filatov 2005a; Nicolas et al. 2005).
These findings suggest progressive steps in the cessation of recombination between the X and Y chromosomes, thus creating “evolutionary strata” on the sex chromosomes, like those in mammalian X and Y chromosomes (Lahn and Page 1999) and in the Z and W chromosomes of the chicken (Lawson-Handley et al. 2004). Our divergence results support previous evidence that the S. latifolia sex chromosomes are of much more recent origin than the youngest of the human X chromosome evolutionary strata (>30 MY), dating the cessation of recombination between the Y and the X to ∼10–20 MYA (Filatov 2005a; Nicolas et al. 2005). If an inversion was involved in stopping X–Y recombination, a sharp break in divergence should characterize the “strata.” More genes will probably be needed before it becomes clear whether these “strata” are truly discontinuous. Even in the human sex chromosomes, some boundaries are not distinct (Skaletsky et al. 2003).
Our divergence estimates between the paralogous loci SlX6a/Y6a and SlX6b/Y6b suggest that the two gene pairs stopped recombining at different times, and, for both, the time is consistent with their location in the X chromosome genetic map. The divergence between the two SlY6 copies (SlY6a and SlY6b) is similar to the raw divergence between the highly diverged SlX6a/Y6a pair, suggesting similar divergence times. Thus the duplication that created SlY6a and SlY6b probably occurred soon after the X and Y chromosomes ceased recombining in the region where the SlY6a gene is located (although the standard errors of these estimates imply some uncertainty, and data on diversity among the SlX6a and among the SlY6a and SlY6b sequences will be necessary to estimate net divergence and to better assess the X–Y divergence times). If so, this event may have been associated with the formation of the Y chromosome, perhaps increasing its size, but at present it is unknown whether other loci were also duplicated.
It will be of interest to verify whether recombination suppression was caused by chromosomal rearrangements such as inversions on the Y or by other mechanisms. Deletion mapping of Y-linked genes will help to further test the possibility of an inversion on the S. latifolia Y with respect to the X chromosome arrangement (Zluvova et al. 2005). Any duplications, such as that found for the SlX6/Y6 genes (which, as just suggested, may have involved other loci), will cause difficulties when attempting to develop a Y deletion map, and it will be important to use sensitive methods, like those developed here, to distinguish between paralogous copies of loci to be mapped. This suggests that genic markers will be necessary to map the Y.
Markers for autosomal loci and genes in gene families:
ISVS and other similar approaches recently reported (Feltus et al. 2006) will be valuable for obtaining markers in nonmodel systems. In addition to efficiently allowing us to infer sex linkage, ISVS yielded polymorphic autosomal loci. Sixty-four percent of these followed a typical biallelic pattern, representing single-copy codominant markers ideal for genetic mapping. Multiple bands, however, were sometimes obtained, and segregation analysis then clearly indicated independently segregating markers, probably representing PCR amplicons from paralogous genes. The ability to work with gene families is a great advantage of the ISVS approach. Discrimination between paralogues and allelic sequence variants is important when developing markers for genetic mapping, but also in other situations. Incorrect phylogenies can be generated by using gene sequences from unrecognized paralogues (reviewed in Shakhnovich and Koonin 2006). A method for checking for the presence of paralogous genes in PCR amplification products is thus often needed. Multiple copies differing in length by a few base pairs can often be missed entirely by just examining PCR bands in agarose gel. Moreover, recombinant sequences can be obtained during PCR amplification (Bradley and Hillis 1997) and cloning of these recombinant artifacts can readily cause confusion between alleles and paralogous sequences unless segregation analysis is performed.
When analyzing plants with large genomes and with highly repetitive sequence content, such as S. latifolia (Lengerova et al. 2004), it is important to check genetically that a sequence is single copy to obtain markers that can be scored reliably for genetic mapping. Because paralogous genes often accumulate indel differences in introns during the course of evolution (since they may recombine with one another rarely or not at all), typing by length differences can provide a simple way to get markers for such genetic tests. Gene conversion between paralogues may hinder analysis of some gene families, especially when their sequences are similar and when they are tandemly duplicated (Gao and Innan 2004; Thomas 2006), but ISVS analysis can be very helpful provided that concerted evolution has not homogenized the sequences of paralogues. In S. latifolia, ISVS could quickly distinguish genes with good marker potential (single-copy genes or small gene families) from cDNAs whose primers yielded too many bands for analysis. Intron-spanning primers designed from less conserved amino acid regions sometimes increased specificity and amplified fewer loci, and again ISVS analysis provided a quick way to test such alternative primers.
The ISVS approach is, of course, limited by the availability of indels. Indels are frequent in noncoding sequences of genes sampled from outcrossing plant populations (Liu et al. 1998, 1999) and in many other organisms (reviewed in Mills et al. 2006), but length differences are not guaranteed in any given intron between alleles or paralogues, especially recent duplicates. This approach should nevertheless be useful in other nonmodel organisms where it can help with the important but laborious task of distinguishing single-copy loci from gene families.
Acknowledgments
We are grateful to Sarah Grant for providing the cDNA library, Dario Beraldi and Angus Davison for technical advice, and the Natural Environment Research Council of the United Kingdom for funding (grant no. NE/B504230/1).
References
- Atanassov, I., C. Delichere, D. A. Filatov, D. Charlesworth, I. Negrutiu et al., 2001. Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol. Biol. Evol. 18**:** 2162–2168. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., 2003. Protein evolution and codon usage bias on the neo-sex chromosomes of Drosophila miranda. Genetics 165**:** 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog, D., 2004. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat. Genet. 36**:** 518–522. [DOI] [PubMed] [Google Scholar]
- Bradley, R. D., and D. M. Hillis, 1997. Recombinant DNA sequences generated by PCR amplification. Mol. Biol. Evol. 14**:** 592–593. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B., 2002. Origin and evolution of the Drosophila Y chromosome. Curr. Opin. Genet. Dev. 12**:** 664–668. [DOI] [PubMed] [Google Scholar]
- Costich, D. E., T. R. Meagher and E. J. Yurkow, 1991. A rapid means of sex identification in Silene latifolia by use of flow cytometry. Plant Mol. Biol. Rep. 9**:** 359–370. [Google Scholar]
- Delichere, C., J. Veuskens, M. Hernould, N. Barbacar, A. Mouras et al., 1999. SlY1, the first active gene cloned from a plant Y chromosome, encodes a WD-repeat protein. EMBO J. 18**:** 4169–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stilio, V. S., R. V. Kesseli and D. L. Mulcahy, 1998. A pseudoautosomal random amplified polymorphic DNA marker for the sex chromosomes of Silene dioica. Genetics 149**:** 2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnison, I. S., and S. R. Grant, 1999. Male sex-specific DNA in Silene latifolia and other dioecious plant species, pp. 73–87 in Sex Determination in Plants, edited by C. C. Ainsworth. BIOS, Oxford.
- Feltus, F. A., H. P. Singh, H. C. Lohithaswa, S. R. Schulze, T. D. Silva et al., 2006. A comparative genomics strategy for targeted discovery of single-nucleotide polymorphisms and conserved-noncoding sequences in orphan crops. Plant Physiol. 140**:** 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov, D. A., 2005. a Evolutionary history of Silene latifolia sex chromosomes revealed by genetic mapping of four genes. Genetics 170**:** 975–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov, D. A., 2005. b Isolation of genes from plant Y chromosomes. Methods Enzymol. 395**:** 418–442. [DOI] [PubMed] [Google Scholar]
- Filatov, D. A., 2005. c Substitution rates in a new Silene latifolia sex-linked gene, SlssX/Y. Mol. Biol. Evol. 22**:** 402–408. [DOI] [PubMed] [Google Scholar]
- Gao, L. Z., and H. Innan, 2004. Very low gene duplication rate in the yeast genome. Science 306**:** 1367–1370. [DOI] [PubMed] [Google Scholar]
- Gillespie, J., 1991. The Causes of Molecular Evolution. Oxford University Press, Oxford.
- Guttman, D. S., and D. Charlesworth, 1998. An X-linked gene with a degenerate Y-linked homologue in a dioecious plant. Nature 393**:** 263–266. [DOI] [PubMed] [Google Scholar]
- Inada, D. C., A. Bashir, C. Lee, B. C. Thomas, C. Ko et al., 2003. Conserved noncoding sequences in the grasses. Genome Res. 13**:** 2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky, E., J. Vrana, S. Matsunaga, P. Soucek, J. Siroky et al., 2001. Localization of male-specifically expressed MROS genes of Silene latifolia by PCR on flow-sorted sex chromosomes and autosomes. Genetics 158**:** 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky, E., Z. Kubat, R. Hobza, M. Lengerova, H. Sato et al., 2006. Accumulation of chloroplast DNA sequences on the Y chromosome of Silene latifolia. Genetica 128**:** 167–175. [DOI] [PubMed] [Google Scholar]
- Kondo, M., I. Nanda, U. Hornung, M. Schmid and M. Schartl, 2004. Evolutionary origin of the medaka Y chromosome. Curr. Biol. 14**:** 1664–1669. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999. Four evolutionary strata on the human X chromosome. Science 286**:** 964–967. [DOI] [PubMed] [Google Scholar]
- Lawson-Handley, L. J., H. Ceplitis and and H. Ellegren, 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167**:** 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel-Hardenack, S., E. Hauser, T. F. Law, J. Schmid and S. R. Grant, 2002. Mapping of sex determination loci on the white campion (Silene latifolia) Y chromosome using amplified fragment length polymorphism. Genetics 160**:** 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerova, M., E. Kejnovsky, R. Hobza, J. Macas, S. R. Grant et al., 2004. Multicolor FISH mapping of the dioecious model plant, Silene latifolia. Theor. Appl. Genet. 108**:** 1193–1199. [DOI] [PubMed] [Google Scholar]
- Liu, F., L. Zhang and D. Charlesworth, 1998. Genetic diversity in Leavenworthia populations with different inbreeding levels. Proc. Biol. Sci. 265**:** 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., D. Charlesworth and M. Kreitman, 1999. The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in the plant genus Leavenworthia. Genetics 151**:** 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., P. H. Moore, H. Ma, C. M. Ackerman, M. Ragiba et al., 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427**:** 348–352. [DOI] [PubMed] [Google Scholar]
- Ma, H., P. H. Moore, Z. Liu, M. S. Kim, Q. Yu et al., 2004. High-density linkage mapping revealed suppression of recombination at the sex determination locus in papaya. Genetics 166**:** 419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk, D., M. Drumm, A. Saulino and F. S. Collins, 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19**:** 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, S., E. Isono, E. Kejnovsky, B. Vyskot, J. Dolezel et al., 2003. Duplicative transfer of a MADS box gene to a plant Y chromosome. Mol. Biol. Evol. 20**:** 1062–1069. [DOI] [PubMed] [Google Scholar]
- Mills, R. E., C. T. Luttig, C. E. Larkins, A. Beauchamp, C. Tsui et al., 2006. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 16**:** 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter and G. J. Herndl, 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65**:** 3518–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. C., O. Kozyreva, S. Lebel-Hardenack, J. Siroky, R. Hobza et al., 2003. Genetic and functional analysis of DD44, a sex-linked gene from the dioecious plant Silene latifolia, provides clues to early events in sex chromosome evolution. Genetics 163**:** 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao, S., S. Matsunaga, A. Sakai, T. Kuroiwa and S. Kawano, 2002. RAPD isolation of a Y chromosome specific ORF in a dioecious plant, Silene latifolia. Genome 45**:** 413–420. [DOI] [PubMed] [Google Scholar]
- Negrutiu, I., B. Vyskot, N. Barbacar, S. Georgiev and F. Moneger, 2001. Dioecious plants. A key to the early events of sex chromosome evolution. Plant Physiol. 127**:** 1418–1424. [PMC free article] [PubMed] [Google Scholar]
- Nicolas, M., G. Marais, V. Hykelova, B. Janousek, V. Laporte et al., 2005. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3**:** e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara, M., S. Matsunaga, S. Nakao and S. Kawano, 2002. A plant Y chromosome-STS marker encoding a degenerate retrotransposon. Genes Genet. Syst. 77**:** 393–398. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 1995. Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. J. Mol. Evol. 40**:** 56–63. [DOI] [PubMed] [Google Scholar]
- Palumbi, S. R., and C. S. Baker, 1994. Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol. Biol. Evol. 11**:** 426–435. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L., J. A. Ross, C. K. Matson, M. Dickson, J. Grimwood et al., 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14**:** 1416–1424. [DOI] [PubMed] [Google Scholar]
- Pritham, E. J., Y. H. Zhang, C. Feschotte and R. V. Kesseli, 2003. An Ac-like transposable element family with transcriptionally active Y-linked copies in the white campion, Silene latifolia. Genetics 165**:** 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15**:** 174–175. [DOI] [PubMed] [Google Scholar]
- Schmidt, W. M., and M. W. Mueller, 1999. CapSelect: a highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 27: e31. [DOI] [PMC free article] [PubMed]
- Schuelke, M., 2000. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18**:** 233–234. [DOI] [PubMed] [Google Scholar]
- Scotti, I., and L. F. Delph, 2006. Selective trade-offs and sex-chromosome evolution in Silene latifolia. Evolution Int. J. Org. Evolution 60**:** 1793–1800. [PubMed] [Google Scholar]
- Shakhnovich, B. E., and E. V. Koonin, 2006. Origins and impact of constraints in evolution of gene families. Genome Res. 16**:** 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky, H., T. Kuroda-Kawaguchi, P. J. Minx, H. S. Cordum, L. Hillier et al., 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423**:** 825–837. [DOI] [PubMed] [Google Scholar]
- Strege, M., and A. Lagu, 1991. Separation of DNA restriction fragments by capillary electrophoresis using coated fused silica capillaries. Anal. Chem. 63**:** 1233–1236. [DOI] [PubMed] [Google Scholar]
- Swerdlow, H., and R. Gesteland, 1990. Capillary gel electrophoresis for rapid, high resolution DNA sequencing. Nucleic Acids Res. 18**:** 1415–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. H., 2006. Concerted evolution of two novel protein families in Caenorhabditis species. Genetics 172**:** 2269–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender, E. J., and B. T. Lahn, 2004. How mammalian sex chromosomes acquired their peculiar gene content. BioEssays 26**:** 159–169. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 21**:** 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J. B., 1995. How often do duplicated genes evolve new functions? Genetics 139**:** 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387**:** 708–713. [DOI] [PubMed] [Google Scholar]
- Zluvova, J., B. Janousek, I. Negrutiu and B. Vyskot, 2005. Comparison of the X and Y chromosome organization in Silene latifolia. Genetics 170**:** 1431–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]