Transcriptional Regulation of the Stem Cell Leukemia Gene (SCL) — Comparative Analysis of Five Vertebrate SCL Loci (original) (raw)
Abstract
The stem cell leukemia (SCL) gene encodes a bHLH transcription factor with a pivotal role in hematopoiesis and vasculogenesis and a pattern of expression that is highly conserved between mammals and zebrafish. Here we report the isolation and characterization of the zebrafish SCL locus together with the identification of three neighboring genes, IER5, MAP17, and MUPP1. This region spans 68 kb and comprises the longest zebrafish genomic sequence currently available for comparison with mammalian, chicken, and pufferfish sequences. Our data show conserved synteny between zebrafish and mammalian SCL and MAP17 loci, thus suggesting the likely genomic domain necessary for the conserved pattern of SCL expression. Long-range comparative sequence analysis/phylogenetic footprinting was used to identify noncoding conserved sequences representing candidate transcriptional regulatory elements. The SCL promoter/enhancer, exon 1, and the poly(A) region were highly conserved, but no homology to other known mouse SCL enhancers was detected in the zebrafish sequence. A combined homology/structure analysis of the poly(A) region predicted consistent structural features, suggesting a conserved functional role in mRNA regulation. Analysis of the SCL promoter/enhancer revealed five motifs, which were conserved from zebrafish to mammals, and each of which is essential for the appropriate pattern or level of SCL transcription.
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: N. Tanese.]
The SCL gene (also known as Tal1) encodes a basic helix-loop-helix (bHLH) transcription factor with a critical role in hematopoiesis and vasculogenesis. It was identified by virtue of its disruption in T-cell acute leukemia and rearrangements of the SCL locus are perhaps the most frequent molecular pathology associated with this tumor (Begley and Green 1999; Orkin et al. 1999). Targeted mutation of the SCL gene has shown that it is essential for the development of all hemopoietic lineages (Porcher et al. 1996; Robb et al. 1996), and also for normal yolk sac angiogenesis (Visvader et al. 1998). Ectopic SCL expression in zebrafish embryos specifies hemangioblast development from early mesoderm, with a consequent excessive production of blood and endothelial progenitors, and can also partially rescue endothelial and hemopoietic phenotypes of the cloche mutant (Gering et al. 1998; Liao et al. 1998). A crucial role for SCL in haemopoietic and endothelial development has also been revealed in an analysis of the in vitro differentiation potential of SCL−/− embryonic stem cells (Robertson et al. 2000).
SCL is normally expressed in hemopoietic cells, endothelium, and within specific regions of the central nervous system (CNS). This pattern of expression is highly conserved throughout vertebrates from mammals to teleost fish (Green et al. 1992; Kallianpur et al. 1994; Gering et al. 1998; Liao et al. 1998; Mead et al. 1998; Sinclair et al. 1999; Drake and Fleming 2000). Murine and human SCL expression is tightly regulated and involves two lineage-specific promoters (Lecointe et al. 1994; Bockamp et al. 1995, 1997; Bockamp et al. 1998). In addition, a detailed analysis of the chromatin structure of the mouse SCL locus identified a number of DNaseI hypersensitive sites associated with enhancer or silencer activity (Göttgens et al. 1997). More recently, studies using transgenic mice have identified five separate enhancers, which direct reporter gene expression in vivo to endothelium, midbrain, hindbrain/spinal cord, or hemopoietic progenitor cells, all subdomains of the normal SCL expression pattern (Sanchez et al. 1999; Sinclair et al. 1999; Göttgens et al. 2000).
We have recently cloned and sequenced the SCL locus from human, mouse, chicken, and pufferfish (Göttgens et al. 2000, 2001; Barton et al. 2001). Comparative sequence analysis of the human and mouse loci showed that all known regulatory regions are highly conserved, and revealed a number of additional conserved noncoding regions that represent candidate gene regulatory elements (Göttgens et al. 2001). Inclusion of chicken sequences into multiple sequence alignments allowed us to prioritize some of these regions for functional studies, but failed to detect chicken homologs for other known enhancers (Göttgens et al. 2000). The pufferfish SCL locus exhibits considerable genomic compression, and a 10.5-kb region containing the SCL gene and extending to the immediate flanking genes was sufficient to produce appropriate expression in zebrafish embryos (Barton et al. 2001). These results suggest that all of the regulatory elements necessary for conserved embryonic expression are present in this construct. However, this approach would not detect regulatory elements necessary for expression in adult tissues and is also limited by the paucity of information on the pattern of SCL expression in pufferfish.
Zebrafish represent a powerful model organism for studies of vertebrate development (Driever et al. 1994), and zebrafish studies have provided considerable insight into hematopoiesis (Amatruda and Zon 1999). Plans for a zebrafish genome sequencing project are well advanced (Duyk and Schmitt 2001), but little information is available on the utility of zebrafish/mammalian genomic sequence comparisons (for examples of conserved enhancer sequences, see Beckers et al. 1996; Zerucha et al. 2000). The pattern of SCL expression is highly conserved between mammals and zebrafish (Gering et al. 1998; Liao et al. 1998; Elefanty et al. 1999; Sinclair et al. 1999) and we therefore reasoned that comparison of the zebrafish SCL locus with the other four available vertebrate SCL loci would be likely to illuminate the transcriptional regulation of the SCL gene. In this work, we describe the characterization of the zebrafish SCL locus and its comparison with the SCL loci from man, mouse, chicken, and pufferfish.
RESULTS
Structure of the Zebrafish SCL Gene
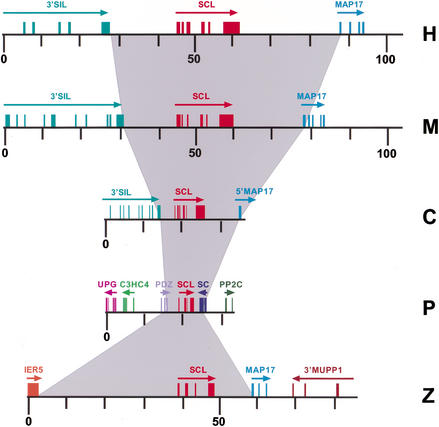
As large-scale gene duplications have been shown in zebrafish (Postlethwait et al. 2000), it was important to determine that only one SCL gene was present within the zebrafish genome. To address this, genomic DNA from six individual and pooled fish was subjected to Southern analysis. Digestion with four restriction enzymes each resulted in a single band hybridizing to a zebrafish SCL cDNA probe, and a fifth enzyme identified a restriction fragment length polymorphism (data not shown). These results strongly suggest the presence of a single SCL locus within the zebrafish genome, and are consistent with previously reported mapping studies (Liao et al. 1998). We next screened a Zebrafish genomic PAC library and identified three positive clones. Southern analysis showed that the SCL gene was centrally situated in one clone (data not shown). This clone was completely sequenced and annotated as described previously (Göttgens et al. 1998). The insert size was 84,489 bp with the start and stop codons of the SCL gene at 41,051 and 47,373 bp, respectively.
The boundaries of _SCL_-coding exons were obtained by aligning the zebrafish cDNA to the genomic sequence. The position of noncoding exons was determined by RT–PCR using total RNA derived from whole adult zebrafish. A forward primer was designed from a putative first exon immediately downstream of a sequence highly homologous to mouse promoter 1a, and a reverse primer was designed from a sequence within the coding region. A single product of 350 bp was generated and sequenced. Sequence analysis revealed a single 5′ untranslated exon immediately downstream of the promoter 1a homology region (data not shown). Therefore, as in pufferfish, no exon homologous to mammalian exon 1b was identified. The size of the zebrafish SCL gene from transcriptional start site to poly(A) site was 9.2 kb, compared with 16.5 kb for human, 15.5 kb for mouse, 8.4 kb for chicken, and 4.8 kb for pufferfish (Fig. 1). By comparison, the sizes for the vertebrate SCL RNA sequences are 4.7 kb for human, 4.3 kb for mouse, 3.3 kb for chicken, 2.7 kb for pufferfish, and 2.8 kb for zebrafish, with the different lengths being due to variations in the length of the 3′ UTR. A total of 20.7% of our zebrafish sequence was masked by RepeatMasker compared with only 5.6% for the pufferfish sequence. Less abundant fish repetitive elements may yet be identified. Nevertheless, our data suggest that a mere lack of repetitive DNA may not account for the full compression of the pufferfish genome.
Figure 1.
Structure of the human (H), mouse (M), chicken (C), pufferfish (P), and zebrafish (Z) SCL loci. All loci are drawn to the same scale. Boxes represent exons and arrows indicate gene orientation. The gray shading illustrates the variation in size of the five vertebrate SCL loci. Numbering refers to distances in kilobases. Only the 3′ ends of the human, mouse, and chicken SIL genes are shown.
Gene Content of the Zebrafish SCL PAC Clone
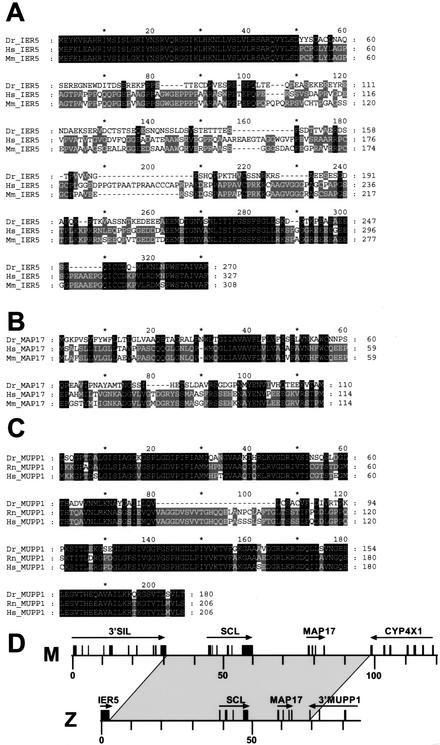
Three new genes flanking zebrafish SCL were identified using exon predictions, BLAST database searches, and where required, RT–PCR to confirm exon/intron structure. At the 5′ extent of the SCL PAC, a single exon was predicted (195–1004 bp) with high homology to human and mouse immediate early response gene IER5, which are also intronless (Williams et al. 1999; Figs. 1 and 2). The second gene identified lay 3′ to SCL (58,174–61,906 bp; Figs. 1 and 2) and showed homology to human and mouse MAP17 (Kocher et al. 1996). MAP17 is known to lie downstream of SCL in the human, mouse, and chicken genomes (Göttgens et al. 2000, 2001). A putative zebrafish MAP17 cDNA sequence was assembled from the predicted exons and confirmed by RT–PCR, followed by direct sequencing of RT–PCR products. At the 3′ extent of the zebrafish contig (68,676–82,972 bp), we identified the last four exons of a gene highly homologous to the human multi-PDZ protein MUPP1 (Ullmer et al. 1998; Figs. 1,2). Conserved gene order between zebrafish and human/mouse therefore does not extend beyond the MAP17 genes (Fig. 2D).
Figure 2.
Newly identified genes flanking zebrafish SCL. (A) Alignment of predicted zebrafish IER5 protein (Dr_IER5) to the corresponding human (Hs_IER5) and mouse (Mm_IER5) proteins. (B) Alignment of zebrafish MAP17 protein (Dr_MAP17) to the respective human (Hs_MAP17) and mouse (Mm_MAP17) proteins. (C) Alignment of predicted zebrafish MUPP1 protein (Dr_MUPP1) to the respective rat (Rn_MUPP1) and human (Hs_MUPP1) proteins. (D) Diagram indicating the region of conserved gene order in the mouse (M) and zebrafish (Z) SCL loci.
Within the zebrafish SCL PAC clone, we failed to identify any homology to either the SIL gene, which is upstream of human, mouse, and chicken SCL (Göttgens et al. 2000), or the PDZ and RING finger genes upstream of pufferfish SCL (Barton et al. 2001). This suggests that the region of conserved gene order ends between SCL and the immediate upstream gene. However, the interval between IER5 and SCL is relatively large (∼39 kb), and we therefore carried out a detailed analysis of this region (see Methods), yet found no evidence of additional genes. The expression pattern of SCL is highly conserved from zebrafish to mammals (Gering et al. 1998; Liao et al. 1998; Elefanty et al. 1999; Sinclair et al. 1999). Our synteny data therefore suggest that the murine genomic domain, which contains all regulatory elements necessary for the conserved pattern of SCL expression, lies within a region extending from the end of SIL to the beginning of CYP4x1 (Fig. 2D).
Comparative Sequence Analysis Identified Three Regions of Noncoding Homology
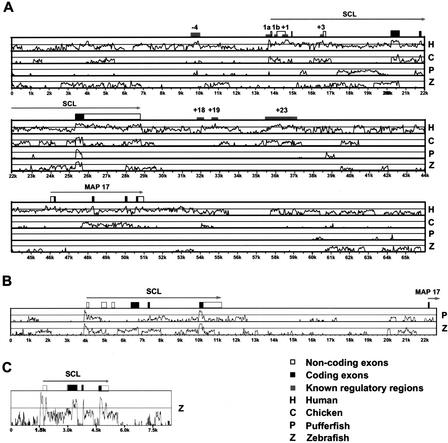
The availability of five orthologous SCL genomic sequences covering a wide evolutionary range allowed us to explore the potential usefulness of different vertebrate genome sequences for comparative analysis/phylogenetic footprinting. By use of currently available programs (VISTA, BLAST, PipMaker, DiAlign, and Synplot) (Altschul et al. 1997; Morgenstern et al. 1998; Mayor et al. 2000; Schwartz et al. 2000; Göttgens et al. 2001), we performed pairwise alignments using the mouse sequence as a reference sequence (Fig. 3A). This analysis demonstrated that mouse/human alignments showed high sequence similarity for all coding exons and all known regulatory regions (Fig. 3A). Mouse/chicken alignments identified similarity with all coding exons and also five known regulatory regions (promoters 1a and 1b, +1, +3, +23), but failed to identify significant similarity to three other known murine regulatory regions (−4, +18, +19; see Fig. 3A). Pairwise mouse/pufferfish and mouse/zebrafish alignments failed to identify some coding exons (exon 4 in pufferfish and exon 5 in zebrafish) and found similarity for only two of the eight known regulatory regions in pufferfish (−4 region and promoter 1a), and no significant similarity for any known regulatory region in the zebrafish sequence. Because mouse/zebrafish synteny comprises the SCL and MAP17 genes, our analysis could potentially have revealed _MAP17_-conserved noncoding sequences. However, as for the SCL locus, no such sequences were identified from mouse/zebrafish comparisons. Moreover, these comparisons failed to identify any of the MAP17 coding exons, even though these were clearly identifiable using the BLASTX program. Our results therefore suggest that the zebrafish genome sequence may be of limited use for the identification of mammalian gene regulatory regions using simple pairwise comparisons.
Figure 3.
Comparative sequence analysis of vertebrate SCL loci. (A) VISTA global alignment plots displaying similarity between the mouse locus (used as the reference sequence) and the human (H), chicken (C), pufferfish (P), and zebrafish (Z) loci. Gray boxes indicate the positions of known mouse regulatory regions. Black and white boxes depict exons with the coding part shaded red. (B) VISTA global alignment plots displaying sequence similarity of chicken SCL locus (used as the reference sequence) and pufferfish (P) and zebrafish (Z) SCL loci. (C) VISTA global alignment plot displaying sequence similarity between the pufferfish locus (used as the reference sequence) and the zebrafish (Z) SCL locus. VISTA plots (Mayor et al. 2000) depicted in A to C were calculated using a 50-bp window and displayed using a 25% lower cut-off limit.
Pairwise comparisons of pufferfish and zebrafish with chicken sequences revealed higher similarity than the comparisons with the mouse sequences described above (Fig. 3B). Only one coding exon was not identified (exon 4 in pufferfish), and high sequence similarity was detected for the promoter 1a region in all three species. This sequence similarity extended to a region near the 3′ end of the first noncoding exon. Moreover, pairwise comparisons of the pufferfish with the zebrafish genomic sequence clearly identified all coding sequences. Furthermore, high similarity was seen for the promoter 1a and exon 1 regions as well as a region just upstream of the poly(A) site (Fig. 3C). This analysis showed that pairwise comparisons of two teleost genomic sequences can identify noncoding conserved sequences missed by mammalian/teleost comparisons. Moreover, multiple sequence alignments of the promoter 1a, exon1, and poly(A) sequences from all five species showed that all three were, in fact, highly conserved between all species. This suggests that whereas current pairwise global alignment programs may struggle to identify conserved noncoding sequences between fish and mammalian sequences, inclusion of additional genomic sequences can be instrumental in identifying meaningful regions of homology.
The Polyadenylation Sequence Lies Within a Highly Conserved Region
A 200-bp stretch of sequence surrounding the AAUAAA polyadenylation sequence showed conservation across all five species (Fig. 4A). The GU- or U-rich sequence usually found downstream of the AAUAAA poly(A) signal (Levitt et al. 1989) could not be identified. In genes lacking this sequence, efficient polyadenylation has been shown to be dependent on an AU-rich region upstream of the poly(A) signal (Huarte et al. 1992). As can be seen from the alignment in Figure 4A, all five SCL sequences share an AU-rich motif upstream of the poly(A) signal.
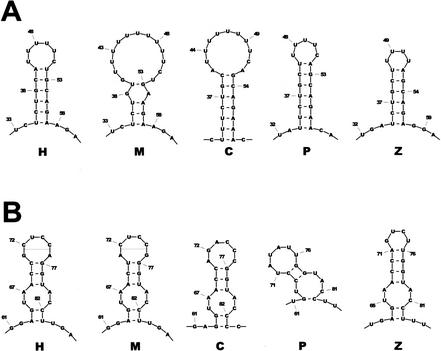
Figure 4.
: Conserved noncoding sequences from vertebrate SCL loci. (A) Alignment of the conserved sequences upstream of the poly(A) signal. The two predicted stem-loop structures, the AU-rich region and the poly(A) signal are indicated. (B) Alignment of the conserved sequences in exon 1. The conserved YY1 site is indicated. (C) Alignment of the conserved SCL promoter/enhancer sequences. The three motifs studied previously (SKN1 and two GATA sites) and the two newly identified conserved sequences (CS1 and CS2) are indicated. The vertical arrow indicates the transcriptional start site.
The conserved 3′ UTR sequences are likely to reflect conservation of a secondary structure necessary for interaction with RNA-binding proteins. However, secondary structure predictions for an individual RNA sequence are often unreliable, as potential structures may have similar minimum energy (ΔG) values. However, prediction accuracy can be greatly improved by taking comparative sequence data into consideration (Juan and Wilson 1999). Therefore, we used a set of algorithms that incorporates evolutionary history into the process of predicting conserved structures (Knudsen and Hein 1999), and thereby allows identification of conserved regions with highly significant secondary structure predictions. By use of this approach, a 50-bp stretch (position 32 to 86, Fig. 4A) was predicted to form two stem-loop structures in all five sequences (see Figs. 4 and 5), suggesting that this structure is important for some conserved aspect of SCL RNA regulation. Interestingly, the first loop is made up entirely of U residues and the second stem loop contains a bulge structure composed of a perfect palindrome (GGUACC). Both of these motifs have been shown to interact with RNA-binding proteins (Peng et al. 1998; Afouda et al. 1999; Anant and Davidson 2000).
Figure 5.
Predicted conserved stem-loop structures for the human (H), mouse (M), chicken (C), pufferfish (P), and zebrafish (Z) poly(A) regions. (A) Stem loop 1 from Fig. 4. (B) Stem loop 2 from Fig. 4.
Sequence Conservation in Exon1
Within the first noncoding exon, 58/74 nucleotides (78%) were conserved between pufferfish and zebrafish, and within this sequence, two blocks of 19 and 15 nucleotides were identical (Fig. 4B). Furthermore, an 18-bp stretch of sequence was conserved across all five species and contained a consensus-binding motif for the transcription fac-tor YY1 (5′-AANATGGC-3′) (Hyde-DeRuyscher et al. 1995). YY1 is a ubiquitously expressed zinc finger transcription factor belonging to the GL1-Kruppel family members (Thomas and Seto 1999), suggesting that this region may act as a transcriptional enhancer on the nearby promoter 1a. However, because the sequence conservation is within a transcribed region (i.e., the 5′ UTR), our data do not exclude the possibility that the observed sequence conservation reflects the recognition sequence for an RNA rather than a DNA-binding protein, and thus indicates a possible conserved post-transcriptional function for this region.
Sequence Conservation Immediately Upstream of Exon 1a
A 170-bp region upstream of SCL exon 1 in zebrafish (38,869 bp–39,024 bp) was highly homologous to regions upstream of the first exons of human, mouse, chicken, and pufferfish SCL genes (Fig. 4C). This region has promoter activity in hemopoietic cell lines and also contains a midbrain enhancer (Bockamp et al. 1995, 1997, 1998; Sinclair et al. 1999). A high level of human/mouse homology is seen throughout this region, but with inclusion of pufferfish, zebrafish, and chicken sequences, five blocks were defined that were conserved across all five species. Three of the conserved blocks, two GATA sites, and a putative SKN1 site have been shown to be important for promoter activity in hematopoietic cell lines or for activity of the midbrain enhancer in transgenic mice (Lecointe et al. 1994; Bockamp et al. 1995, 1997, 1998; Sinclair Sinclair et al. 1999). Functionally significant regulatory elements have therefore been conserved within this SCL promoter/enhancer from fish to man and can be identified by phylogenetic footprinting. The remaining two conserved sequences (CS1 and CS2) were unrecognized previously and represent candidate binding sites for additional transcription factors.
Functional Analysis of the CS1 and CS2 Motifs in Transgenic Mice
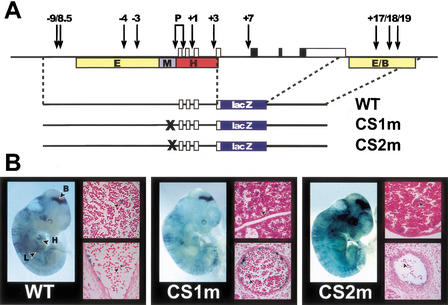
The functional significance of CS1 and CS2 was investigated in transgenic mice. Each site was mutated in the context of the –10E3/lacZ/3′enh reporter construct, which contains mouse genomic sequences corresponding to both SCL promoters (1a and 1b) together with four SCL enhancers responsible for directing expression to endothelium, midbrain, hindbrain/spinal cord, and hematopoietic progenitors (Göttgens et al. 1997; Sanchez et al. 1999; Sinclair et al. 1999). Following oocyte microinjection, embryos were examined at day 11.5 post coitum (E11.5), at a time when endogenous SCL is expressed in yolk sac hemopoietic progenitors, fetal liver, brain, and endothelium (Green et al. 1992; Kallianpur et al. 1994).
The expression pattern of the –10E3/lacZ/3'enh construct was consistent with the known expression patterns of the –10E3/lacZ transgene and transgenes containing the 3′ enhancer (Sanchez et al. 1999; Sinclair et al. 1999). Strong expression of β-galactosidase was evident in the midbrain and endothelium in 2/2 embryos and sections showed lacZ expression in endothelial cells (endocardium, dorsal aorta, intersomitic vessels, capillary networks, and vitelline vessels) as well as in a subset of fetal liver and circulating blood cells (see arrowhead, Fig. 6B). Mutation of either the CS1 or CS2 site did not alter the pattern of expression. Following mutation of the CS1 site, 2/2 transgenic embryos showed a wild-type pattern of expression in brain, yolk sac, embryonic endothelium, and blood cells (Fig. 6B). Following mutation of the CS2 site, 3/5 transgenic embryos displayed the same pattern of expression. In the two remaining embryos, staining was seen in brain and fetal liver, but was not evident in endothelium. The lack of detectable endothelial staining in the latter two embryos may reflect position effects that differentially influence the endothelial elements more than the brain regulatory elements. These data therefore show that neither CS1 nor CS2 are necessary for the staining pattern observed with the –10E3/lacZ/3′enh cassette at E11.5. However, our results do not exclude a critical role for these motifs either at other stages of development or in regulating the level of SCL transcription.
Figure 6.
Transgenic analysis of the CS1 and CS2 motifs. (A) Diagram of the mouse SCL locus with the constructs used for transgenic analysis shown underneath. Vertical arrows indicate previously mapped DNaseI hypersensitive sites. Colored boxes specify enhancer regions mapped previously in transgenic mice [E = endothelial, M = midbrain, (H) hindbrain/spinal cord, (E/B) endothelial/blood]. (B) β-galactosidase activity of constructs shown in A in E11.5 transgenic mouse embryos. A whole-mount view of an E11.5 embryo together with sections of fetal liver (top) and dorsal aorta (bottom) are shown for each construct. Arrowheads in sections indicate _lacZ_-positive hematopoietic cells. (B) Brain; (H) heart; (L) fetal liver.
CS1 and CS2 Are Both Necessary for Full SCL Promoter Activity in Erythroid Cells
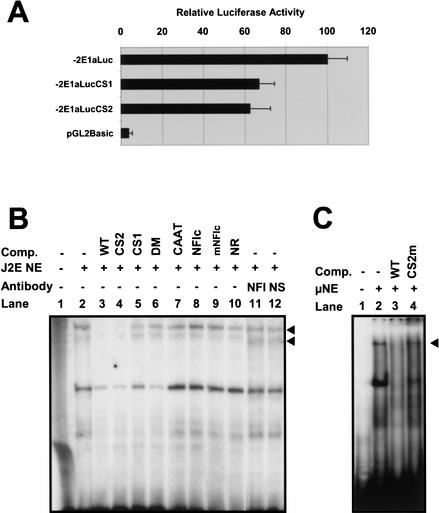
To investigate whether CS1 or CS2 are important for the level of SCL transcription, we analyzed the effect of corresponding mutations on the activity of SCL promoter 1a in a transient transfection assay. Mutation at CS1 or CS2 reduced promoter activity to 67% and 63%, respectively in the erythroid cell line, J2E (Fig. 7A). This degree of reduction is comparable with that seen with a mutation in the –69GATA site (Bockamp et al. 1998). Our results indicate that both sites contribute to the level of SCL transcription in erythroid cells.
Figure 7.
Functional analysis of the CS1 and CS2 motifs in the J2E erythroid cell line. (A) Relative luciferase activity of a wild-type mouse reporter construct containing 2 kb upstream of exon 1a (−2E1aLuc) compared with a CS1 mutant (−2E1aLucCS1), CS2 mutant (−2E1aLucCS2), and promoterless control construct (pGL2Basic). Results represent the mean of four independent experiments (a minimum of 10 luciferase values for each construct) using two different DNA preparations. (B) Gel-shift analysis identifies two specific complexes (arrowheads) binding to the CS1 motif — see text for details. (Comp) Competitor oligonucleotides; (NE) nuclear extract; (NFI) antibody to NFI; (NS) nonspecific control antibody . (C) Gel-shift analysis using a rapid protocol for nuclear extract preparation identifies a high molecular weight complex (arrowhead) binding to the CS2 site. (Comp) Competitor oligonucleotides; (NE) nuclear extract. Details of oligos used in B and C can be found in the Methods section.
Band shift analysis was performed to identify protein complexes binding at CS1 and CS2 in J2E cells (Fig. 7B,C). An oligonucleotide was used that spanned the region of promoter 1a containing both CS1 and CS2. Two specific complexes (arrowheads, Fig. 7B) bound the wild-type oligonucleotide (see Fig. 7B, lane 2). Binding of both complexes was competed by an excess of unlabeled wild-type (see Fig. 7B, lane 3) or CS2 mutant (see Fig. 7B, lane 4) competitor oligonucleotides but not by an oligonucleotide with a mutation at CS1 (see Fig. 7B, lane 5), an oligonucleotide with mutations at both sites (see Fig. 7B, lane 6), or an oligonucleotide of unrelated sequence (see Fig. 7B, lane 10). These results show that both complexes bound specifically to the CS1 site and that binding of both was prevented by the mutation of this motif.
CS1 contains an NFI (Nuclear Factor I) consensus half-site and a variant C/EBP site. However, the complex could neither be competed by an oligonucleotide containing a NFI consensus site known to bind all NFI family members (Baumeister et al. 1999) (see Fig. 7B, lane 8, mutant NFI oligo control, see Fig. 7B, lane 9), nor supershifted using an NFI antiserum (see Fig. 7B, lane 11). Similarly, no competition was seen using an oligonucleotide carrying the CAAT consensus sequence (see Fig. 7B, lane 7) known to bind C/EBP α, β, and δ (Zhang et al. 1996). These results therefore suggest that the complexes binding to CS1 do not contain NFI or C/EBP family members.
CS2 is a 7-nucleotide sequence precisely conserved across all five species (Fig. 4C). The TAAT sequence represents the core-binding motif of many homeobox transcription factors (Mann 1995), and is used preferentially by members of the Ultrabithorax, Antennapedia, Sex combs reduced, and Deformed families of proteins (Ekker et al. 1994), together with their mammalian homologs (Krumlauf 1994). However, the extended CS2 sequence (5′-ATAATGC-3′) has not been reported to bind any specific vertebrate homeobox protein. No complexes could be seen binding to CS2 using standard nuclear extracts and an oligonucleotide containing both CS1 and CS2 sites. An oligonucleotide was therefore designed with CS2 centrally positioned, and nuclear extracts were prepared using a modified rapid protocol (see Methods) to minimize protein degradation. Using this approach, a specific low mobility complex was detected that was competed by wild-type but not CS2 mutant oligos (see Fig. 7C, lanes 2–4). Given the large number of possible candidate proteins that might bind to the TAAT core of CS2, further studies will be needed to identify the nature of the protein(s) present in this complex.
DISCUSSION
A Genomic Domain Necessary for the Conserved Pattern of SCL Transcription
Chromosomal rearrangements occuring during evolution may shed light on the location of transcriptional elements necessary to produce conserved patterns of gene expression. We have previously characterized and sequenced the SCL genomic loci from the pufferfish, fugu rubripes (Barton et al. 2001) and have found that the genes immediately flanking pufferfish SCL were unrelated to those known to flank both avian and mammalian SCL genes. These results imply that SCL regulatory elements might be confined to the region between the upstream and downstream flanking genes, a region of 65 kb in human and 8.5 kb in pufferfish. Consistent with this concept, a 10.4-kb fragment of pufferfish genomic DNA, containing the SCL gene and extending to the 5′ and 3′ flanking genes, directed appropriate expression to hematopoietic and neural tissue in transgenic zebrafish embryos (Barton et al. 2001). However, three caveats need to be borne in mind when considering the pufferfish synteny data. Firstly, expression of the pufferfish transgene was assessed at a single time point in zebrafish embryos. The transgene may therefore lack elements necessary for expression in adult fish or at other developmental timepoints. Secondly, compression of the pufferfish genome may have been accompanied by a simplification of gene regulatory mechanisms (Rothenberg 2001), which, if true, would limit the ability of pufferfish/mammalian sequence comparisons to shed light on mammalian gene regulation. Thirdly, the pattern of SCL expression in pufferfish has been assessed by RT–PCR and not by in situ hybridization. It is therefore possible that the expression pattern of pufferfish SCL may differ from other vertebrates, possibly as a consequence of the compact pufferfish genome.
In contrast, the zebrafish genome is much less compressed (1800 Mbp compared with 400 Mbp), and the pattern of SCL expression as assessed by in situ techniques is highly conserved between zebrafish and mammals, both during embryogenesis and, where it has been assessed, also in adults. In both zebrafish and mouse, SCL is expressed in specific cells within the anterior horns of the spinal cord, in the ventral midbrain, in the hindbrain, in progenitors of blood and endothelium, and in erythroid cells (Gering et al. 1998; Liao et al. 1998; Elefanty et al. 1999; Sinclair et al. 1999). The results presented here show the existence of a region of synteny conserved between zebrafish and mouse extending from the 3′ end of the gene immediately upstream of SCL to the beginning of the gene immediately downstream of MAP17. The comparative synteny data presented here, therefore, raise the possibility that additional SCL regulatory elements may be found between the beginning of the MAP17 gene and the start of the gene immediately downstream, a region known to contain several peaks of human/mouse sequence homology (Göttgens et al. 2001), but which has not yet been assessed for the presence of SCL regulatory elements. However, it is important to bear in mind that comparative mapping studies of zebrafish and human genes have shown that gene order within syntenic regions may be inverted or transposed (Postlethwait et al. 2000). It therefore remains possible that as yet unsequenced regions neighboring the SCL locus may exhibit conserved synteny with the SCL gene, and could conceivably harbor long-range regulatory elements.
Comparative Sequence Analysis of Zebrafish and Mammalian Loci
The zebrafish SCL gene (from upstream to downstream flanking genes) is 59 kb, mainly as a result of the large interval between Dr-IER5 and zebrafish SCL. The zebrafish SCL gene is therefore similar in size to its mouse or human homologs (∼60 and 65 kb, repectively) and seven times larger than the pufferfish gene. Our data show that the mammalian and zebrafish SCL loci all have MAP17 downstream of SCL and that this is therefore likely to represent the ancestral pattern of gene order. Conservation of gene order between mammals and zebrafish does not extend downstream of the MAP17 gene. In contrast, the pufferfish SCL locus does not share immediate 5′ or 3′ flanking genes with those present at zebrafish or mammalian SCL loci. Moreover, none of the genes present either side of zebrafish SCL flank the pufferfish SCL gene. These results are consistent with the suggestion that gene shuffling may have been especially prevalent during the speciation of bony fish (Ohta et al. 2000).
Most studies comparing mammalian and fish sequences have focused on the pufferfish because of its compressed genome. Conservation of regulatory elements between pufferfish and mammalian homologs has been found in a number of genes. In some, but not all cases, the regulatory elements are functionally equivalent. For example, comparative analysis of pufferfish and mouse hox gene loci identified conserved enhancers and the pufferfish enhancers functioned appropriately in transgenic mice (Aparicio et al. 1995; Popperl et al. 1995). In contrast, a comparison of the pattern of activity in transgenic mice exhibited by conserved pufferfish and mouse Wnt1 regulatory elements revealed consistent differences (Rowitch et al. 1998). There are also several examples of genes in which sequence comparisons between pufferfish and mammalian homologs have not revealed any noncoding sequence homology, despite the genes having highly conserved expression patterns and functionally homologous regulatory elements (Sathasivam et al. 1997; Gellner and Brenner 1999). In this situation, it seems likely that current computational methods are not sensitive enough to detect individual conserved transcription factor binding sites in a background of extensive sequence divergence (Flint et al. 2001).
There has been relatively little comparative analysis of zebrafish regulatory elements. Here we show that comparisons of the zebrafish SCL locus with mammalian SCL loci did not identify zebrafish elements homologous with the majority of known murine enhancers (e.g., enhancers corresponding to −4/3, +3, +17, +18, +19 DNase1 hypersensitive sites). In contrast, all known mouse regulatory regions were highly conserved in human, whereas significant similarity for half of them could be identified in chicken. We suspect that once the transcription factors binding to these enhancers are identified, it will prove possible to detect corresponding conserved motifs within the zebrafish sequence. In any case, our results did reveal three regions of noncoding sequence conservation, two of which are of particular interest.
First, the poly(A) region contained a stretch of conserved sequence homology. Conserved stretches of 3′ UTR sequence can be identified in 36% or 17% of orthologous genes in pair-wise comparisons between mammals and birds or mammals and fish, respectively (Duret et al. 1993). These regions have been implicated in the regulation of mRNA stability and translation (Spicher et al. 1998). The conserved sequences in the SCL 3′ UTR predict the formation of two stem-loop structures, and contain motifs implicated in interactions with RNA-binding proteins (Peng et al. 1998; Afouda et al. 1999; Anant and Davidson 2000). Further studies will be needed to ascertain whether these sequences play a role in the post-transcriptional regulation of SCL expression that has been reported previously (Murrell et al. 1995; Prasad et al. 1995).
The second conserved region of particular interest was located immediately upstream of the first SCL exon. This region functions as a promoter in hematopoietic cell lines (Bockamp et al. 1995) and as a powerful midbrain enhancer (Sinclair et al. 1999). Five short blocks of homology were identified which were conserved between all five vertebrates studied, and each of which corresponded to a transcription factor binding motif. Two of these motifs are important for promoter activity in hematopoietic cells (Bockamp et al. 1995) and for the pattern of SCL expression in the brain (Sinclair et al. 1999). The remaining three motifs are necessary for the level of SCL promoter activity in hematopoietic cells (this study; Bockamp et al. 1998). These results are consistent with the concept that within modular promoters and enhancers, only a minority of transcription factor motifs influence the pattern of gene expression, whereas the majority will only be detected by quantitative assays (Yuh et al. 1998).
Zebrafish has emerged as one of the most powerful experimental models to study vertebrate development and organogenesis. This has culminated recently in the announcement of plans to sequence its entire genome (Duyk and Schmitt 2001). We have described previously the genomic loci of human, mouse, chicken, and pufferfish SCL (Göttgens et al. 2000, 2001; Barton et al. 2001), and are therefore in a unique position to place zebrafish genomic sequence data into a wider comparative context. The results presented here represent the largest segment of zebrafish genomic DNA so far subjected to comparative sequence analysis. Our analysis suggests that comparative analysis of zebrafish and mammalian genomic sequences may be of limited value for the identification of functionally significant noncoding sequences. However, comparison of zebrafish with pufferfish sequences revealed conserved noncoding sequences missed by the zebrafish/mammalian comparisons, emphasizing the potential benefits of having multiple fish genome sequences. Moreover, the zebrafish genome sequence will greatly accelerate the characterization of genes controlling vertebrate organogenesis identified in large-scale mutation screens.
METHODS
Isolation and Sequencing of Zebrafish SCL Locus
A gridded zebrafish genomic PAC library (library no. 706) from the RZPD (German Human Genome Project) was screened using a zebrafish SCL cDNA probe. The three positive clones (BUMSP706C0952Q3, BUMSP706B2050Q3, and BUMSP706I22129Q3) were further analyzed by Southern analysis following standard protocols. The insert of clone BUMSP706I22129Q3 was fully sequenced as described (Göttgens et al. 1998).
Sequence Analysis
Interactive annotation of genomic sequences was performed within ACeDB (Eeckman and Durbin 1995) as described (Göttgens et al. 1998). Long-range sequence comparisons were performed using DiAlign version 1 (Morgenstern et al. 1998), dotter (Sonnhammer and Durbin 1995), and PipMaker (Schwartz et al. 2000). Small-scale DNA alignments were performed and displayed as described (Göttgens et al. 1998). RNA secondary structure predictions were performed using the programs described in Knudsen and Hein (1999). Repeat content was analyzed using RepeatMasker. In an attempt to determine whether any exons predicted 5′ of zebrafish SCL were, in fact, part of a gene, RT–PCR was performed using total RNA from whole adult zebrafish with multiple primers. Where possible, primers were designed from exons that were predicted by more than one program. Whenever RT–PCR products were obtained, these were sequenced and shown to result from mispriming of one or both of the primers used, and thus contained no spliced exons (data not shown).
Transgenic Analysis
Mutations into the CS1 and CS2 motifs were introduced as described (Bockamp et al. 1995). The CS1 mutation converted the wild-type GCCAAAT sequence to GCTAGCT, whereas the CS2 mutation converted ATAATG to ATGGTG. The transgenic reporter constructs contained a 139-kb _Bgl_II/_Xcm_I fragment of the mouse SCL locus ranging from 10 kb upstream of the promoter to exon 3 upstream of a lacZ reporter gene, which was followed by a 5.5-kb _Bgl_II fragment containing the 3′ enhancer (Sanchez et al. 1999). F0 transgenic mouse embryos were prepared and analyzed as described (Sanchez et al. 1999).
Reporter Assays and EMSA Analysis
J2E cells were grown as described (Bockamp et al. 1998). Transient transfections and luciferase assays were performed as described previously (Göttgens et al. 1997). For the rapid nuclear extract preparation, 2 × 106 cells were collected by centrifugation, washed in PBS, and resuspended in 400 μL of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 0.1 mM EGTA, 1 mM DTT, 0.5 mM PMSF), and incubated at 4°C for 15 min. After adding 25 μL of 10% NP40, the sample was mixed and briefly centrifuged. The pellet was resuspended in 25 μL of buffer C (10 mM HEPES at pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF), and incubated for 30 min followed by a 5-min centrifugation. A total of 1 μL of the resultant supernatant was used per track for EMSA analysis. Standard nuclear extracts were prepared and EMSAs performed as described (Bockamp et al. 1998). Oligonucleotides used in Figure 7B are as follows: WT, (GAAATTGCCAAATTAAAAT GAATCATTTGGCCCATAATGGCCGA); CS1, (GAAATTGC TAGCTTAAAATGAATCATTTGGCCCATAATGGCCGA); CS2, (GAAATTGCCAAATTAAAATGAATCATTTGGCCC AT GGTGGCCGA); DM, (GAAATTGCTAGCTTAAAATGAAT CATTTGGCCCATGGTGGCCGA); CAAT, (TGCAGATTGCG CAATCTGCA); NFIc (AGGTCTGGCTTTGGGCCAAGAGC CGC); mNFIc, (AGGTCTCGCTTTGGGCCAAGAGCCGC); NR, (AGGCGGCTCCTTATCTCGGC). Oligonucleotides used in Figure 7C were as follows: WT, (ATCATTTGGCCCATAATG GCCGAGGCGCTTATCGGGGGCG); CS2m, (ATCATTTGGC CCATGGTGGCCGAGGCGCTTATCGGGGGCG).
WEB SITE REFERENCES
www.http://ftp.genome.washington.edu/RM/RepeatMasker.html; A resource for masking repetitive DNA in genomic sequences.
Acknowledgments
Work in the authors' laboratories is supported by the Wellcome Trust and the Kay Kendall Leukemia Fund. The NFI antibody was kindly provided by Dr. N. Tanese (NYU Medical Center). We would also like to acknowledge the support of sequencing team 47 at the Sanger Centre
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bg200@cam.ac.uk; FAX 44-1223-762670.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.45502.
REFERENCES
- Afouda AB, Reynaud-Deonauth S, Mohun T, Spohr G. Localized XId3 mRNA activation in Xenopus embryos by cytoplasmic polyadenylation. Mech Dev. 1999;88:15–31. doi: 10.1016/s0925-4773(99)00166-5. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Zon LI. Dissecting hematopoiesis and disease using the zebrafish. Dev Biol. 1999;216:1–15. doi: 10.1006/dbio.1999.9462. [DOI] [PubMed] [Google Scholar]
- Anant S, Davidson NO. An AU-rich sequence element (UUUN[A/U]U) downstream of the edited C in apolipoprotein B mRNA is a high-affinity binding site for Apobec-1: Binding of Apobec-1 to this motif in the 3′ untranslated region of c- myc increases mRNA stability. Mol Cell Biol. 2000;20:1982–1992. doi: 10.1128/mcb.20.6.1982-1992.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc Natl Acad Sci. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton LM, Göttgens B, Gering M, Gilbert JG, Grafham D, Rogers J, Bentley D, Patient R, Green AR. Regulation of the stem cell leukemia (SCL) gene: A tale of two fishes. Proc Natl Acad Sci. 2001;98:6747–6752. doi: 10.1073/pnas.101532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister H, Gronostajski RM, Lyons GE, Margolis FL. Identification of NFI-binding sites and cloning of NFI-cDNAs suggest a regulatory role for NFI transcription factors in olfactory neuron gene expression. Brain Res Mol Brain Res. 1999;72:65–79. doi: 10.1016/s0169-328x(99)00210-7. [DOI] [PubMed] [Google Scholar]
- Beckers J, Gerard M, Duboule D. Transgenic analysis of a potential Hoxd-11 limb regulatory element present in tetrapods and fish. Dev Biol. 1996;180:543–553. doi: 10.1006/dbio.1996.0327. [DOI] [PubMed] [Google Scholar]
- Begley CG, Green AR. The SCL gene: From case report to critical hematopoietic regulator. Blood. 1999;93:2760–2770. [Google Scholar]
- Bockamp E-O, McLaughlin F, Murrell AM, Göttgens B, Robb L, Begley CG, Green AR. Lineage-restricted regulation of the murine SCL/TAL-1 promoter. Blood. 1995;86:1502–1514. [PubMed] [Google Scholar]
- Bockamp EO, McLaughlin F, Göttgens B, Murrell AM, Elefanty AG, Green AR. Distinct mechanisms direct SCL/tal-1 expression in erythroid cells and CD34 positive primitive myeloid cells. J Biol Chem. 1997;272:8781–8790. doi: 10.1074/jbc.272.13.8781. [DOI] [PubMed] [Google Scholar]
- Bockamp EO, Fordham JL, Göttgens B, Murrell AM, Sanchez M-J, Green AR. Transcriptional regulation of the stem cell leukemia gene by PU.1 and Elf-1. J Biol Chem. 1998;273:29032–29042. doi: 10.1074/jbc.273.44.29032. [DOI] [PubMed] [Google Scholar]
- Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- Driever W, Stemple D, Schier A, Solnica-Krezel L. Zebrafish: Genetic tools for studying vertebrate development. Trends Genet. 1994;10:152–159. doi: 10.1016/0168-9525(94)90091-4. [DOI] [PubMed] [Google Scholar]
- Duret L, Dorkeld F, Gautier C. Strong conservation of non-coding sequences during vertebrates evolution: Potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 1993;21:2315–2322. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G, Schmitt K. Fish x 3. Nat Genet. 2001;27:8–9. doi: 10.1038/83807. [DOI] [PubMed] [Google Scholar]
- Eeckman FH, Durbin R. ACeDB and macace. Methods Cell Biol. 1995;48:583–605. [PubMed] [Google Scholar]
- Ekker SC, Jackson DG, von Kessler DP, Sun BI, Young KE, Beachy PA. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994;13:3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefanty AG, Begley CG, Hartley L, Papaevangeliou B, Robb L. SCL expression in the mouse embryo detected with a targeted lacZ reporter gene demonstrates its localization to hematopoietic, vascular, and neural tissues. Blood. 1999;94:3754–3763. [PubMed] [Google Scholar]
- Flint J, Tufarelli C, Peden J, Clark K, Daniels RJ, Hardison R, Miller W, Philipsen S, Tan-Un KC, McMorrow T, et al. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the α globin cluster. Hum Mol Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- Gellner K, Brenner S. Analysis of 148 kb of genomic DNA around the wnt1 locus of Fugu rubripes. Genome Res. 1999;9:251–258. [PMC free article] [PubMed] [Google Scholar]
- Gering M, Rodaway ARF, Göttgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttgens B, McLaughlin F, Bockamp EO, Fordham JL, Begley CG, Kosmopoulos K, Elefanty AG, Green AR. Transcription of the SCL gene in erythroid and CD34 positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene. 1997;15:2419–2428. doi: 10.1038/sj.onc.1201426. [DOI] [PubMed] [Google Scholar]
- Göttgens B, Gilbert JGR, Barton LM, Aparicio S, Hawker K, Mistry S, Vaudin M, King A, Bentley D, Elgar G, et al. The pufferfish SLP-1 gene, a new member of the SCL/TAL-1 family of transcription factors. Genomics. 1998;48:52–62. doi: 10.1006/geno.1997.5162. [DOI] [PubMed] [Google Scholar]
- Göttgens B, Barton LM, Gilbert JG, Bench AJ, Sanchez MJ, Bahn S, Mistry S, Grafham D, McMurray A, Vaudin M, et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- Göttgens B, Gilbert JG, Barton LM, Grafham D, Rogers J, Bentley DR, Green AR. Long-range comparison of human and mouse SCL loci: Localized regions of sensitivity to restriction endonucleases correspond precisely with peaks of conserved noncoding sequences. Genome Res. 2001;11:87–97. doi: 10.1101/gr.153001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AR, Visvader J, Lints T, Harvey R, Begley CG. SCL is co-expressed with GATA-1 in haemopoietic cells but is also expressed in developing brain. Oncogene. 1992;7:653–660. [PubMed] [Google Scholar]
- Huarte J, Stutz A, O'Connell ML, Gubler P, Belin D, Darrow AL, Strickland S, Vassalli JD. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan V, Wilson C. RNA secondary structure prediction based on free energy and phylogenetic analysis. J Mol Biol. 1999;289:935–947. doi: 10.1006/jmbi.1999.2801. [DOI] [PubMed] [Google Scholar]
- Kallianpur AR, Jordan JE, Brandt SJ. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood. 1994;83:1200–1208. [PubMed] [Google Scholar]
- Knudsen B, Hein J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. BioInformatics. 1999;15:446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- Kocher O, Cheresh P, Lee SW. Identification and partial characterization of a novel membrane-associated protein (MAP17) up-regulated in human carcinomas and modulating cell replication and tumor growth. Am J Pathol. 1996;149:493–500. [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lecointe N, Bernard O, Naert K, Joulin V, Larsen CJ, Romeo PH, Mathieu-Mahul D. GATA- and SP1-binding sites are required for the full activity of the tissue-specific promoter of the tal-1 gene. Oncogene. 1994;9:2623–2632. [PubMed] [Google Scholar]
- Levitt N, Briggs D, Gil A, Proudfoot NJ. Definition of an efficient synthetic poly(A) site. Genes & Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstrem of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes & Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS. The specificity of homeotic gene function. BioEssays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. BioInformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Mead PE, Kelley CM, Hahn PS, Piedad O, Zon LI. SCL specifies hematopoietic mesoderm in Xenopus embryos. Development. 1998;125:2611–2620. doi: 10.1242/dev.125.14.2611. [DOI] [PubMed] [Google Scholar]
- Morgenstern B, Frech K, Dress A, Werner T. DIALIGN: Finding local similarities by multiple sequence alignment. BioInformatics. 1998;14:290–294. doi: 10.1093/bioinformatics/14.3.290. [DOI] [PubMed] [Google Scholar]
- Murrell AM, Bockamp E-O, Göttgens B, Chan YS, Goss MA, Heyworth CM, Green AR. Discordant regulation of SCL/TAL-1 mRNA and protein during erythroid differentiation. Oncogene. 1995;11:131–139. [PubMed] [Google Scholar]
- Ohta Y, Okamura K, McKinney EC, Bartl S, Hashimoto K, Flajnik MF. Primitive synteny of vertebrate major histocompatibility complex class I and class II genes. Proc Natl Acad Sci. 2000;97:4712–4717. doi: 10.1073/pnas.97.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Porcher C, Fujiwara Y, Visvader J, Wang LC. Intersections between blood cell development and leukemia genes. Cancer Res. 1999;59:1784s–1787s. [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Prasad KS, Jordan JE, Koury MJ, Bondurant MC, Brandt SJ. Erythropoietin stimulates transcription of the TAL1/SCL gene and phosphorylation of its protein products. J Biol Chem. 1995;270:11603–11611. doi: 10.1074/jbc.270.19.11603. [DOI] [PubMed] [Google Scholar]
- Robb L, Elwood NJ, Elefanty AG, Köntgen F, Li R, Barnett LD, Begley CG. The SCL gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- Robertson SM, Kennedy M, Shannon JM, Keller G. A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-1. Development. 2000;127:2447–2459. doi: 10.1242/dev.127.11.2447. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Mapping of complex regulatory elements by pufferfish/zebrafish transgenesis. Proc Natl Acad Sci. 2001;98:6540–6542. doi: 10.1073/pnas.131199098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, Echelard Y, Danielian PS, Gellner K, Brenner S, McMahon AP. Identification of an evolutionary conserved 110 base-pair cis-acting regulatory sequence that governs wnt-1 expressino in the murine neural plate. Development. 1998;125:2735–2746. doi: 10.1242/dev.125.14.2735. [DOI] [PubMed] [Google Scholar]
- Sanchez MJ, Göttgens B, Sinclair AM, Stanley M, Begley CG, Hunter S, Green AR. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult hematopoietic progenitors. Development. 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Baxendale S, Mangiarini L, Bertaux F, Hetherington C, Kanazawa I, Lehrach H, Bates GP. Aberrant processing of the Fugu HD (FrHD) mRNA in mouse cells and in transgenic mice. Hum Mol Genet. 1997;6:2141–2149. doi: 10.1093/hmg/6.12.2141. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker-A web server for aligning two genomic DNA sequences . Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AM, Göttgens B, Barton LM, Aparicio S, Bahn S, Sanchez M-J, Fordham J, Stanley ML, Green AR. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal chord and endothelium; conserved neural expression is GATA factor dependant. Dev Biol. 1999;209:128–142. doi: 10.1006/dbio.1999.9236. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Spicher A, Guicherit OM, Duret L, Aslanian A, Sanjines EM, Denko NC, Giaccia AJ, Blau HM. Highly conserved RNA sequences that are sensors of environmental stress. Mol Cell Biol. 1998;18:7371–7382. doi: 10.1128/mcb.18.12.7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Ullmer C, Schmuck K, Figge A, Lubbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Fujiwara Y, Orkin SH. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes & Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M, Lyu MS, Yang YL, Lin EP, Dunbrack R, Birren B, Cunningham J, Hunter K. Ier5, a novel member of the slow-kinetics immediate-early genes. Genomics. 1999;55:327–334. doi: 10.1006/geno.1998.5679. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: Experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DE, Hetherington CJ, Meyers S, Rhoades KL, Larson CJ, Chen HM, Hiebert SW, Tenen DG. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]