Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo (original) (raw)
Abstract
Critical to our understanding of the developmental potential of stem cells and subsequent control of their differentiation in vitro and in vivo is a thorough understanding of the genes that control stem cell fate. Here, we report that Foxd3, a member of the forkhead family of transcriptional regulators, is required for maintenance of embryonic cells of the early mouse embryo. _Foxd3_−/− embryos die after implantation at approximately 6.5 days postcoitum with a loss of epiblast cells, expansion of proximal extraembryonic tissues, and a distal, mislocalized anterior organizing center. Moreover, it has not been possible to establish _Foxd3_−/− ES cell lines or to generate _Foxd3_−/− teratocarcinomas. Chimera analysis reveals that Foxd3 function is required in the epiblast and that _Foxd3_−/− embryos can be rescued by a small number of wild-type cells. _Foxd3_−/− mutant blastocysts appear morphologically normal and express Oct4, Sox2, and Fgf4, but when placed in vitro the inner cell mass initially proliferates and then fails to expand even when Fgf4 is added. These results establish Foxd3 as a factor required for the maintenance of progenitor cells in the mammalian embryo.
Keywords: mouse embryogenesis, stem cells, Foxd3, winged helix gene
Multipotent progenitor cells exist transiently in the mammalian embryo in several tissues including the preimplantation stage blastocyst, the gastrulating epiblast, and the neural crest. Each of these cell types can be cultured in vitro to generate multipotent stem cell lines, and when grafted ectopically, blastocysts and epiblast tissue will produce teratocarcinomas containing multipotent stem cells. Understanding the common molecular regulatory mechanisms of different stem cell types is critical to an understanding of how multipotency is maintained in vivo and how differentiation is controlled.
Mammalian embryonic development initiates with a series of cleavage divisions, and at 2.5 days postcoitum (dpc) the free-floating mouse embryo undergoes compaction: cell boundaries become tightly apposed to one another and cells are no longer equivalent. Inner cells contribute to the inner cell mass (ICM) and embryo proper, whereas outer cells contribute to the trophectoderm, a tissue essential for implantation. After implantation, the ICM proliferates and extends distally to form the cup-like egg cylinder (at ∼5.5 dpc) consisting of an inner layer of epiblast cells and an outer layer of extraembryonic visceral endoderm (for review, see Hogan et al. 1994).
Few genes have been identified that are required for the maintenance of the epiblast cell population and the establishment of ICM-derived embryonic stem (ES) cells in vitro. _Oct4_−/− embryos, mutant for this POU family transcription factor, die around 5.0 dpc, shortly after implantation but before the egg cylinder is formed. _Oct4_−/− ES cells cannot be established, as the ICM cells fail to survive (Nichols et al. 1998). Survival of multipotent cells of the ICM is highly sensitive to Oct4 expression levels; low Oct4 levels result in the differentiation of ES cells into trophoblast, and high levels result in differentiation into primitive endoderm and mesoderm (Niwa et al. 1998). Fibroblast growth factor 4 (Fgf4) is also required for formation of the egg cylinder; _Fgf4_−/− embryos fail to develop after implantation, and cells of the ICM do not proliferate in vitro (Feldman et al. 1995). Fgf4 transcription is directly regulated by Oct4 and Sox2 binding to an Fgf4 enhancer element (Yuan et al. 1995). Establishment of _Fgf4_−/− ES cells is possible with addition of recombinant Fgf4 to the culture medium (Wilder et al. 1997). The complexities of maintaining pluripotent cells in the embryo are likely to involve additional key regulatory factors.
Foxd3 (previously published as Hfh2) was isolated in a low-stringency screen for forkhead family genes (Labosky and Kaestner 1998). Originally termed “_Genesis,_” Foxd3 was reported to be expressed only in mouse ES cells and their malignant equivalents (Sutton et al. 1996). In addition, Foxd3 expression is one of several diagnostic markers characteristic of human ES cell lines (M.F. Pera, pers. comm.). However, Foxd3 expression is not as restricted as first suggested and is also detected during early embryogenesis in the epiblast and later in neural crest cells (Labosky and Kaestner 1998; Hromas et al. 1999; Dottori et al. 2001). Foxd3 has been implicated in the control of differentiation in multiple systems: Overexpression of Foxd3 in a myeloid cell line prevents appropriate maturation of these cells into granulocytes (Xu et al. 1998), ectopic expression of Foxd3 in the chick neural tube changes the fate of those cells into neural crest and can interfere with the subsequent differentiation of crest derivatives (Dottori et al. 2001; Kos et al. 2001), and Foxd3 mRNA can induce the formation of mesoderm in Xenopus (D.S. Kessler, unpubl.). We demonstrate here that Foxd3 is required for the maintenance of pluripotent cells in the preimplantation and peri-implantation stages of mouse embryogenesis.
Results
Expression of Foxd3 in early mouse embryos
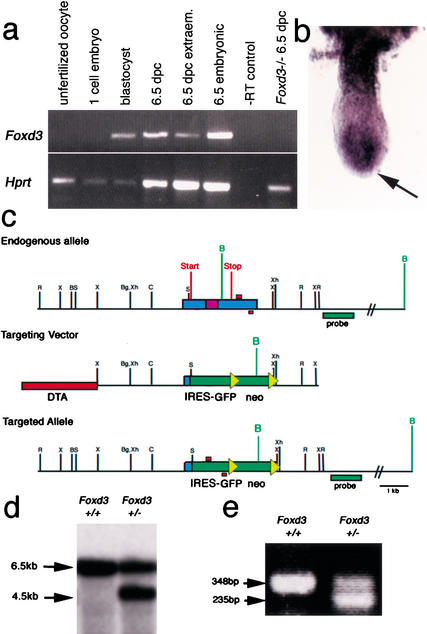
Foxd3 was previously shown to be expressed in premigratory neural crest cells, and expression is downregulated in all differentiated derivatives with the exception of Schwann cells (Labosky and Kaestner 1998; Dottori et al. 2001), a cell type that can give rise to multipotent stem cells in vitro (Stemple and Anderson 1992). To assess Foxd3 expression at earlier stages of development, we used RT-PCR to analyze expression in unfertilized oocytes through early implantation-stage embryos. Foxd3 is not expressed in the unfertilized oocyte or fertilized one-cell embryos, but transcripts are detected in blastocyst-stage embryos, after the time when zygotic transcription is initiated in the mouse embryo (Fig. 1a). Whole mount and section in situ hybridization detected expression of Foxd3 throughout the epiblast of the 6.5-dpc embryo with faint expression in the extraembryonic region (Fig. 1b, data not shown). The extraembryonic expression was confirmed by RT-PCR analysis of dissected extraembryonic and embryonic portions of embryos (Fig. 1a).
Figure 1.
Embryonic expression of Foxd3 and generation of Foxd3 mutant allele. (a) RT-PCR analysis of staged mouse embryos shows that Foxd3 is not expressed in unfertilized oocytes or one-cell embryos but can be detected in the blastocyst at 3.5 dpc. Foxd3 mRNA is also detected at 6.5 dpc in both the extraembryonic and the embryonic portions of the embryo. Expression is undetectable in a 6.5-dpc _Foxd3_−/− embryo. Negative controls with no reverse transcriptase added show no amplification of the Foxd3 message. RT-PCR for the hypoxanthine phosphoribosyl transferase gene (Hprt) was used to monitor the integrity of the cDNA produced by reverse transcription. (b) Whole mount in situ hybridization of a 6.5-dpc embryo shows that Foxd3 transcripts are present in the layer of epiblast cells, but not in the outer layer of endoderm (arrow). (c) The Foxd3 null mutation was generated using a targeting vector shown here. PCR primers for genotyping are shown as red boxes, and the external 3′ probe used to screen ES cells is shown in green. The loxP sites flanking the neo cassette are yellow triangles. (d) Southern analysis of ES cells was performed by digesting genomic DNAs with _BamH_1 as diagrammed in c. (e) PCR was used to genotype Foxd3 mice and embryos.
Targeted deletion of the Foxd3 locus
To determine the role of Foxd3 during embryogenesis, we replaced the entire coding region with a neo resistance cassette and a Gfp gene, generating a null mutation (Fig. 1c–e). No Foxd3 mRNA was detected in _Foxd3_−/− embryos, confirming that this is a null mutation (Fig. 1a). Southern analysis with 5′ (data not shown) and 3′ (Fig. 1d) flanking probes confirmed the structure of the targeted allele as shown. The neo cassette was flanked by loxP sites for subsequent CRE deletion. We generated three alleles of Foxd3: the first with an insertion of a Gfp gene with an internal ribosomal entry site (Ires-Gfp), the second with a Gfp gene insertion, and the third with the Ires-Gfp with the neo cassette removed. All three alleles of Foxd3 (Foxd3Ires-Gfp, Foxd3Gfp, and Foxd3Ires-GfpΔneo) gave the same phenotype, and most of the studies reported here were carried out with the Foxd3Ires-Gfp allele, hereafter called _Foxd3_−.
Embryonic lethality of Foxd3−/− embryos
Foxd3+/− mice derived from targeted ES cell lines were normal and healthy, but no _Foxd3_−/− mice were born from Foxd3+/− intercrosses with over 300 mice weaned and analyzed, indicating that _Foxd3_−/− embryos did not survive to birth. Timed matings were carried out and no _Foxd3_−/− embryos were detected at 13.5 dpc; abnormal embryos consisting primarily of an empty yolk sac were present at 9.5 dpc, and PCR genotyping revealed these to be _Foxd3_−/− (data not shown). Morphological and histological analysis at 5.5 dpc revealed no overtly abnormal embryos in multiple litters. Further analysis revealed that abnormalities in _Foxd3_−/− embryos were apparent as early as 6.5 dpc (Figs. 2, 3, see below). At this time, _Foxd3_−/− embryos do not have a smooth rounded distal end characteristic of wild-type embryos. Instead, cells resembling extraembryonic endoderm extend in multiple finger-like protrusions at the distal tip (Fig. 2, cf. a,i and b,j). Normally, the proximal extraembryonic endoderm is columnar with large vacuoles, whereas distal extraembryonic endoderm is squamous in character (Fig. 2c,e). In _Foxd3_−/− embryos, this distinction is not seen; columnar and vacuolated extraembryonic endoderm extends nearly to the distal tip of the mutant embryo (yellow arrows in Fig. 2d,f). Significantly, the epiblast of _Foxd3_−/− embryos is reduced in size, and histological analysis revealed the absence of a primitive streak. These observations suggest that either the epiblast is converted to extraembryonic tissue or that upon failure of the epiblast the extraembryonic tissue expands distally.
Figure 2.
Gastrulation of _Foxd3_−/− embryos is disrupted. (a,b) Morphology of embryos dissected at 6.5 dpc shows that _Foxd3_−/− embryos have an abnormal distal epiblast region and no evidence of a primitive streak. (c) A sagittal section of a normal 6.5-dpc embryo from a Foxd3+/− intercross. Note the clear distinction between the extraembryonic (ex) and embryonic (em) regions. The proamnionic cavity (pac) extends the length of the embryonic region, while the exocoelomic cavity (xc) is restricted to the extraembryonic region. (d) A sagittal section of an abnormal 6.5-dpc embryo from the same cross. Remnants of a proamniotic cavity are visible, and visceral endoderm (ve), extraembryonic ectoderm (ee), and embryonic ectoderm (e) can be identified. (e,f) The normal and abnormal visceral endoderm in c and d are shown at higher magnification. The region of the section is indicated in c and d with yellow arrows. (g,h) Phosphohistone H3 is detected at 6.5 dpc, and although several epiblast cells of the normal embryo are strongly stained, no internal distal cells of a _Foxd3_−/− embryo react with the antibody for phosphohistone H3. Positive cells are indicated with red arrows. (i,j) TUNEL staining to detect cells undergoing apoptosis shows no appreciable cell death at 6.5 dpc in a normal embryo, with some cell death in a _Foxd3_−/− embryo. By 7.5 dpc (k,l), there is very little embryonic tissue remaining in the _Foxd3_−/− embryo. e, embryonic ectoderm; ec, ectoplacental cone; ee, extraembryonic ectoderm; em, embryonic region; ex, extraembryonic region; pac, proamnionic cavity; ps, primitive streak; ve, visceral endoderm; xc, exocoelomic cavity.
Figure 3.
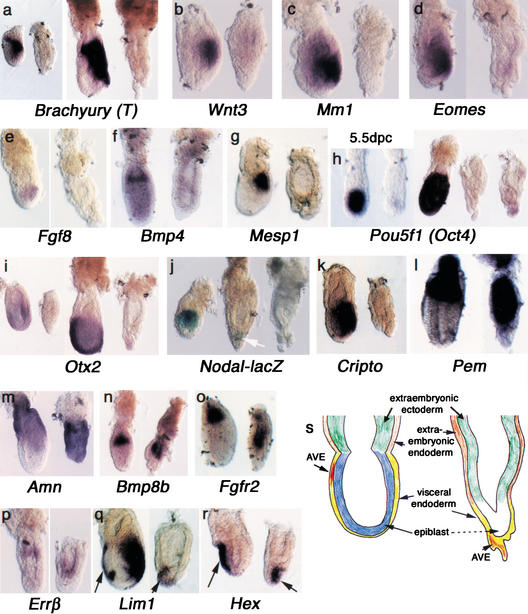
Molecular marker analysis of Foxd3 mutant embryos. Whole mount in situ analysis of normal littermates is shown on the left side of each set of embryos. All embryos are 6.5 dpc except where noted. Anterior (when discernible) is to the left. (a) Brachyury (or T) is expressed in the proximal epiblast of early 6.5-dpc embryos and in all cells of the primitive streak at later stages. T expression is never detected in _Foxd3_−/− embryos. (b) Wnt3, also normally expressed in the proximal epiblast and later in the primitive streak, is not expressed in mutants. (c_–_e) Mm1, Eomes, and Fgf8, markers of mesodermal cells of the primitive streak, are not expressed in _Foxd3_−/− embryos. (f,g) Extraembryonic mesodermal markers Bmp4 and Mesp1 are not detected in mutant embryos. (h) Pou5f1/Oct4 expression is detected throughout the epiblast in a normal embryo at 5.5 dpc, but in _Foxd3_−/− embryos, expression is missing at 5.5 dpc. At 6.5 dpc, Pou5f1/Oct4 is expressed robustly in normal embryos (middle embryo) but is either missing or only weakly expressed in scattered cells (the two embryos on the right). (i) Otx2 expression is normally detected throughout the epiblast of the early streak-stage embryo, but is missing in mutant littermates (first pair of embryos). Otx2 expression in an early headfold-stage embryo (∼7 dpc) is concentrated in the anterior half of the embryo (third embryo). A small amount of Otx2 expression is detected in the distal tip of the mutant littermate (far right embryo). (j) Nodal-lacZ expression is detected in the proximal epiblast of the early streak-stage embryo (left embryo). However, Nodal-lacZ expression was not detected in most _Foxd3_−/− embryos (far right embryo). An exception is shown here with a low level of β-galactosidase activity restricted to the distal tip (arrow, middle embryo). (k) The Nodal cofactor Cripto is not expressed in mutant embryos. (l,m) Extraembryonic visceral endoderm, as indicated by Pem and Amn, is distally expanded in the mutants. (n_–_p) Extraembryonic ectoderm is also distally expanded as shown by expression of Bmp8b, Fgfr2, and _Err_β. (q,r) The AVE signaling center, indicated by the arrows, is located distally in _Foxd3_−/− embryos as shown by the expression of Lim1 and Hex. The posterior Lim1 expression in q corresponds to the mesodermal expression domain of this gene. (s) Schematic illustrating the morphological changes in _Foxd3_−/− embryos.
To determine whether loss of epiblast was a result of decreased cell proliferation, we used histone H3 phosphorylation to assess cells in M phase. A marked decrease in M-phase cells was observed in the inner and distal cells of mutant embryos (Fig. 2g,h). To quantify these results, we counted positive cells located in the internal portion of 6.5-dpc embryos but excluded from the external layer of extraembryonic endoderm. The number of phospho-H3-positive cells in normal 6.5-dpc embryos averaged 6.53 (n = 17 embryos) versus 2.64 (n = 22 embryos) in abnormal (presumably _Foxd3_−/−) embryos. A two-tailed T-test demonstrated that this difference is statistically significant with P < 0.05. There was a similar decrease in the number of S-phase cells as shown by BrdU incorporation (data not shown). TUNEL labeling showed a slight increase in cell death in the 6.5-dpc distal embryo (Fig. 2i,j), but by 7.5 dpc (head fold stage), the entire distal region of the mutant embryo was severely truncated and thus cell death measurements were not informative (Fig. 2k,l). Together, these data indicate that cell proliferation is reduced and cell death is increased in the distal embryonic portion of 6.5-dpc _Foxd3_−/− embryos compared to the wild type.
Molecular analysis of Foxd3−/− embryos
To define the molecular basis of the developmental defects, we analyzed lineage-specific gene expression (Fig. 3). In all of the panels of Figure 3, wild-type expression of the marker analyzed is shown on the left with _Foxd3_−/− embryos to the right. Because ectopic expression of Foxd3 in Xenopus embryos (D. Kessler, unpubl.) can cause the formation of mesoderm and our histological analyses of _Foxd3_−/− embryos indicated a disruption in the primitive streak, we first wished to determine whether any mesoderm was present in _Foxd3_−/− embryos. Expression of the T gene (Brachyury), normally expressed in mesodermal cells (Herrmann 1991), was not detected in early or late 6.5-dpc mutant embryos (Fig. 3a), suggesting that mesodermal cells are not being generated and that gastrulation is not occurring. The absence of expression of other embryonic and extraembryonic mesoderm markers, including Wnt3, Mm1, Eomesodermin, Fgf8, Bmp4, and Mesp1 (Fig. 3b–g; Crossley and Martin 1995; Saga et al. 1996; Lawson et al. 1999; Liu et al. 1999; Pearce and Evans 1999) supports this conclusion.
Pou5f1, encoding the transcription factor Oct4, is expressed throughout the epiblast in wild-type embryos and is essential for the survival of epiblast cells (Rosner et al. 1990; Nichols et al. 1998). Despite the absence of morphological differences at 5.5 dpc, Pou5f1 expression was not detected in 22% of embryos dissected from Foxd3+/− intercrosses (Fig. 3h; total number of embryos = 27). At 6.5 dpc, most _Foxd3_−/− embryos showed no expression of Pou5f1, although weak expression in cells scattered throughout the embryo was sometimes observed (Fig. 3h, slight staining in far right embryo). Otx2 is normally expressed throughout the epiblast and gradually becomes restricted to the anterior half of the embryo (Simeone et al. 1993). In _Foxd3_−/− 6.5-dpc embryos, there was no Otx2 expression (Fig. 3i), but in embryos a few hours older (∼6.75 dpc) a few _Otx2_-expressing cells were present at the distal tip (Fig. 3i, far right embryo). To examine Nodal expression in mutant embryos, we generated _Foxd3_−/−; Nodal-lacZ+/− embryos. Using the Nodal-lacZ allele, β-galactosidase activity is normally detected throughout the epiblast (Varlet et al. 1997; Brennan et al. 2001), but no β-galactosidase activity was detected in most _Foxd3_−/− embryos, with an occasional embryo (2 of 15) showing β-galactosidase activity at the distal tip (arrow in Fig. 3j). No epiblast expression of the Nodal cofactor Cripto was detected (Fig. 3k; Ding et al. 1998).
In contrast to the loss of epiblast and mesodermal gene expression, distal expansion of the expression of extraembryonic endodermal markers Pem and Amn (Lin et al. 1994; Kalantry et al. 2001) was observed at 6.5 dpc (Fig. 3l,m), as was expression of the extraembryonic ectodermal markers Bmp8b, Fgfr2, and _Err_β (Fig. 3n–p). Formation of the anterior visceral endoderm (AVE), a signaling center required to pattern the anterior-posterior axis of the embryo, was also examined. Expression of the AVE markers Lim1 and Hex (Barnes et al. 1994; Thomas et al. 1998) was restricted to the distal tip of _Foxd3_−/− embryos (Fig. 3q,r). This result suggests that the occasional distal expression of Otx2 and Nodal at the tip of _Foxd3_−/− embryos (Fig. 3i,j) is most likely due to the ectopic position of the AVE. These observations demonstrate that in _Foxd3_−/− embryos, the AVE is specified but fails to move anteriorly in the absence of the epiblast and primitive streak. As summarized in Figure 3s, the data show that a loss of Foxd3 function results in a dramatic reduction in cells of the epiblast and their derivatives, a distal expansion of proximal extraembryonic tissues, and a mislocalization of the AVE. This suggests that Foxd3 plays a critical role in maintaining the number of epiblast cells and therefore in establishing the proximal-distal patterning in the early embryo.
Foxd3 is not required for maintenance of Oct4 or Fgf4 expression in blastocysts
The POU homeobox gene Pou5f1, encoding Oct4 protein, is required for the identity of pluripotent progenitor cells in the ICM of the blastocyst. Embryos lacking Oct4 die around 5.5 dpc, prior to the formation of the egg cylinder (Nichols et al. 1998). Oct4 mRNA is not expressed in _Foxd3_−/− embryos as early as 5.5–6.5 dpc (Fig. 3a), but because Oct4 function is required before this time, we examined _Foxd3_−/− embryos for expression of this key regulatory gene prior to implantation. The secreted protein Fgf4 is also required early in the peri-implantation mouse embryo, and Oct4 and Sox2 proteins have been shown to regulate Fgf4 expression directly (Yuan et al. 1995). We therefore also examined Sox2 and Fgf4 mRNAs in _Foxd3_−/− blastocysts.
Blastocysts were harvested from Foxd3+/− intercrosses and individually processed for RT-PCR. Expression of the hypoxanthine phosphoribosyl transferase (Hprt) gene was used as a positive control, and only _Hprt_-positive samples were used for subsequent analyses. Expression of Foxd3 in blastocysts was used to genotype the samples, and _Foxd3_-negative/_Hprt_-positive samples were scored as _Foxd3_−/− blastocysts. Oct4, Sox2, and Fgf4 mRNAs were detected in all blastocysts, regardless of Foxd3 expression (Fig. 4a). Oct4 mRNA was localized to the ICM in all blastocysts from intercrosses (Fig. 4b; n = 18). In addition, immunofluorescent localization of Oct4 in blastocysts harvested from intercrosses showed that 100% of the 23 blastocysts had robust nuclear staining of Oct4 protein (Fig. 4c). Therefore, expression of several genes known to be required for early ICM proliferation and expansion are initiated and maintained in the absence of Foxd3.
Figure 4.
Expression of Oct4, Sox2, and Fgf4 is maintained in _Foxd3_−/− blastocysts. (a) RT-PCR from single blastocysts shows that Oct4, Sox2, and Fgf4 mRNAs are expressed in _Foxd3_−/− blastocysts. Lanes 2 and 8 are from _Foxd3_−/− embryos, and all other lanes are from either Foxd3+/+ or +/− blastocysts. (b) Whole mount in situ hybridization for Oct4 mRNA demonstrated that blastocysts from intercrosses (18 of 18 total) have Oct4 mRNA localized to the ICM. (c) Immunofluorescent detection of Oct4 protein in blastocysts from an intercross show that all blastocysts (23 of 23 total) have robust nuclear staining of Oct4 protein.
Foxd3 function is required in the epiblast
To determine whether Foxd3 function is required in the epiblast or in extraembryonic trophoblast and endodermal tissues, we took advantage of the developmental restriction of ES cells. ES cells injected into a recipient blastocyst contribute almost exclusively to epiblast derivatives and not extraembryonic tissues (Beddington and Robertson 1989). Rosa26.1 ES cells were injected into blastocysts from Foxd3+/− intercrosses, and all chimeras were dissected between 8.5 and 9.5 dpc; none were allowed to progress further in development. Because ES cells do not contribute to the extraembryonic endoderm, we used this tissue to genotype recipient blastocysts retrospectively. We generated 140 embryos in these experiments, and retrospective genotyping indicated that of the recipient blastocysts, 43 were +/+, 81 were +/−, and 10 were −/− (six embryos could not be genotyped). Two examples of chimeras obtained from _Foxd3_−/− blastocysts are shown in Figure 5a–d. Of the 10 embryos obtained from _Foxd3_−/− blastocysts, eight were morphologically normal and two recapitulated the _Foxd3_−/− phenotype. Staining for β-galactosidase activity demonstrated that most rescued chimeras (5 of 8) contained a large portion of wild-type cells such as the chimera shown in Figure 5b. However, even a low (∼25%) contribution of wild-type ES cells (embryo shown in Fig. 5a) completely rescued the _Foxd3_−/− defects. The remaining two embryos derived from _Foxd3_−/− blastocysts were morphologically identical to _Foxd3_−/− embryos and had no contribution of wild-type ES cells, as detected by PCR for the lacZ gene. Therefore, every rescued _Foxd3_−/− embryo contained wild-type cells. _Foxd3_−/− chimeras might be underrepresented for two reasons. First, most chimeras were recovered at 9.5 dpc, and it is possible that nonrescued embryos would be resorbed by that time. Second, the yolk sac endoderm was successfully separated from the extraembryonic mesoderm in most, but not all, cases, so it is possible that some _Foxd3_−/− embryos were incorrectly scored as +/−. Histological analysis of the chimera shown in Figure 5a demonstrated that after rescue with wild-type cells, _Foxd3_−/− cells can differentiate into axial and paraxial mesoderm, as well as neural crest (Fig. 5c,d; data not shown). This result indicates that injected wild-type ES cells act non-cell autonomously to rescue development of _Foxd3_−/− embryos.
Figure 5.
Developmental and proliferative potential of _Foxd3_−/− embryos. (a,b) Two examples of chimeras obtained by injecting Rosa26.1 ES cells into blastocysts from Foxd3+/− intercrosses. Extraembryonic endoderm was used to genotype the recipient blastocysts, and chimeras were stained with Xgal to detect the wild-type Rosa26.1 cells. Wild-type ES cells rescued the mutant phenotype completely, indicating that Foxd3 function is required in the epiblast and that a low contribution of wild-type cells is sufficient to rescue the epiblast defect. (c,d) Histological analysis of chimera shown in panel a shows that _Foxd3_−/− cells (not stained blue) can differentiate into notochord, somites, and neural tube. (e,f) Teratocarcinomas derived from Foxd3+/− (e) and −/− (f) embryos. The portion of the embryo transferred is diagrammed. The heterozygous tumor extends far beyond the field shown and contains tissues derived from all three germ layers, including fat (f), skeletal muscle (m), neural tissue, and cartilage (data not shown). The _Foxd3_−/− embryonic cells do not produce any differentiated cell types and do not proliferate. The entire extent of the _Foxd3_−/− transplanted embryonic tissue is outlined with a white dashed line, and panels e and f are shown at the same magnification. Coagulated blood (orange) and calcium deposits (blue) are results of the surgery. f, fat; m, muscle; n, notochord; nt, neural tube; s, somite.
Foxd3 loss of function causes a loss of progenitor cells in the epiblast
To assess the developmental potential and proliferative capacity of _Foxd3_−/− cells, we attempted to generate teratocarcinomas by placing the embryonic portion of a 6.5-dpc embryo under the kidney capsule of a histocompatible mouse. Foxd3+/+ and +/− teratocarcinomas contained differentiated tissues representing all germ layers including muscle, adipose tissue, cartilage, neuroepithelial tissue, gut-like tubes, and dermoid cysts (Fig. 5e; data not shown). However, no tumors were obtained from 11 _Foxd3_−/− embryos transplanted, even when only the extreme distal tip was transplanted in an effort to avoid inhibition of teratoma production by extraembryonic ectoderm. Figure 5f shows the site of a _Foxd3_−/− transplant 8 wk after surgery as evidence that the _Foxd3_−/− cells failed to grow or differentiate. These data support the idea that the pluripotent cells of the epiblast are lost without Foxd3 function.
Foxd3 is required for ES cell derivation
To assess the requirement for Foxd3 in another pluripotent progenitor cell type, we attempted to generate _Foxd3_−/− ES cells by two independent methods. First, we tried to derive _Foxd3_−/− ES cells from blastocysts; Foxd3+/− mice were backcrossed onto a 129SvEvTac genetic background for five generations, and 153 blastocysts from intercross matings were cultured to derive ES cell lines using standard methods (Hogan et al. 1994). Despite obtaining 6 Foxd3+/+ and 10 Foxd3+/− cell lines, no _Foxd3_−/− ES cell lines were established.
Next, targeting of the wild-type allele in Foxd3+/− ES cells was attempted. The neo cassette was removed from the targeted locus by electroporation with a plasmid encoding Cre recombinase. Colonies were picked at random, expanded, and tested by Southern blot for the presence of the neo cassette. To verify removal of the neo cassette, cell lines were also cultured in the presence of G418, and it was confirmed that no cells could survive this selection. The original targeting vector, shown in Figure 1c, was then reelectroporated into Foxd3Ires-GfpΔneo+/−cells, and cells were selected in G418. In several independent experiments, a total of 147 G418-resistant Foxd3+/− cell lines were obtained, indicating that the null allele had been retargeted in those cells. However, no _Foxd3_−/− ES cell lines were obtained in these experiments, suggesting that _Foxd3_−/− ES cells do not survive in culture. In addition, electroporated cells were grown in the presence of 12.5ng/mL recombinant Fgf4, similar to experiments that successfully generated _Fgf4_−/− ES cells (Wilder et al. 1997). In multiple experiments, 146 G418-resistant Foxd3+/− cell lines were obtained, but no _Foxd3_−/− cell lines were identified. Taken together, these results suggest that Foxd3 function is essential for establishing pluripotent ES cells in vitro.
Foxd3 is required for maintenance of the inner cell mass
The inability to derive _Foxd3_−/− ES cells from blastocysts prompted an examination of the effect of prolonged in vitro culture on _Foxd3_−/− blastocysts. Blastocysts isolated from Foxd3+/− intercrosses were cultured in ES cell medium containing LIF. In culture, all blastocysts attached, hatched from their surrounding zona pellucida, and trophectoderm cells spread around a central ICM. For the initial 3–5 days of in vitro culture, _Foxd3_−/− outgrowths were indistinguishable from Foxd3+/+ or +/− (Fig. 6a). However, during days 5–9 in culture, the ICM of the _Foxd3_−/− embryos failed to expand, and trophoblast giant cells (arrows in Fig. 6a,b) underwent programmed cell death, as revealed by TUNEL staining (Fig. 6c). At the same time, Foxd3+/+ and +/− ICMs continued to expand and grow. Outgrowths were genotyped by PCR, and no outgrowths with a robust ICM were _Foxd3_−/−. In total, 16 of 69 (24%) blastocysts failed to survive in vitro. To test the possibility that a lack of Fgf4 from the ICM might be responsible for this phenotype, the culture medium was supplemented with 100ng/mL recombinant Fgf4 and 1μg/mL heparin. However, no difference was observed in the Fgf4-treated outgrowth cultures; 5 of 20 Fgf4-treated outgrowths failed to survive and expand an ICM, suggesting that Fgf4 cannot rescue the proliferation defect in _Foxd3_−/− ICM cells.
Figure 6.
Developmental potential of the _Foxd3_−/− ICM in vitro. (a) Blastocysts were harvested from Foxd3+/− intercrosses and placed on gelatin-coated dishes in ES cell medium. Blastocysts attached and formed robust outgrowths within the first 3 d of culture. After prolonged culture (b), the _Foxd3_−/− cultures failed to maintain an ICM. Arrows in a and b indicate trophoblast giant cells. (c) TUNEL staining demonstrates that in _Foxd3_−/− outgrowths, attached cells were undergoing programmed cell death by day 6 in culture. Arrows in c point to TUNEL-positive cells.
Discussion
The results presented here throw new light on the genetic regulation of early mouse development. The preimplantation mouse embryo contains only 20–40 cells in the ICM, and this pool of progenitor cells must proliferate and respond accurately to secreted signals to differentiate and undergo morphogenesis, forming both the embryo proper and supporting extraembryonic tissues. It is essential that the progenitor cells maintain their pluripotency throughout these critical stages and expand their cell numbers appropriately. We have now demonstrated that the winged helix gene Foxd3 plays a role in maintaining the epiblast and its derivatives and in establishing pluripotent ES cell lines.
_Foxd3_−/− embryos die around the time of gastrulation with a loss of epiblast and a concomitant expansion of the proximal extraembryonic ectoderm and endoderm. Our results suggest that the cells of the _Foxd3_−/− ICM and/or epiblast die. This conclusion is supported by in vitro blastocyst culture in which _Foxd3_−/− cells fail to proliferate and expand a normal ICM outgrowth. Mammalian embryogenesis requires a fine balance of extraembryonic and embryonic tissues. For example, when distal epiblast cells are transplanted to a proximal position adjacent to the extraembryonic ectoderm, these cells adopt a proximal cell fate, suggesting that signals from extraembryonic tissues influence the fate of epiblast cells (Tam and Zhou 1996). Thus far, only Bmp4 has been identified as an instructive signal produced by extraembryonic ectoderm; Bmp4 secretion from the extraembryonic ectoderm patterns the proximal portion of the epiblast and is required for generating extraembryonic mesoderm and precursors of primordial germ cells (Lawson et al. 1999). Such morphogenetic signaling occurs in both directions, and it has been suggested that Fgf4 secreted by the ICM and epiblast plays a role in patterning the extraembryonic ectodermal and trophoblast lineages (Tanaka et al. 1998). The apparent overgrowth of the extraembryonic ectoderm and loss of epiblast in _Foxd3_−/− embryos suggest that this balance is disrupted.
The chimeric rescue experiments show that wild-type ES cells completely rescue _Foxd3_−/− embryonic ectoderm through 9.5 dpc, indicating that Foxd3 function is required in the epiblast. In rescued mutants, _Foxd3_−/− cells can give rise to many different tissues and cell types, including mesoderm and neural crest. These results show that Foxd3 functions in a non-cell autonomous manner and suggest that Foxd3 may be required for the regulation of a cell surface or secreted signaling molecule. Additional evidence indicates that cell-cell interactions may be critical for Foxd3 function. It has not been possible to generate _Foxd3_−/− ES cells by targeting the second allele of Foxd3+/− cells, even though the putative _Foxd3_−/− ES cells would be generated in the presence of Foxd3+/− ES cells. We would argue that secretion of a factor from wild-type ES cells is not sufficient to rescue _Foxd3_−/− cells because either cell-cell contact is required for such a rescue or the secreted factor must be present above a threshold level that is not attained under the in vitro culture conditions used. An alternative explanation is that wild-type cells must be present in a particular location, perhaps at the junction between the extraembryonic and embryonic regions of the embryo, in order for rescue to be achieved. Finally, a third possibility is that Foxd3 is necessary for the production or function of a factor required transiently to maintain the small population of pluripotent stem cells in the ICM or epiblast, but not for the subsequent differentiation of these lineages. A tissue-specific deletion of Foxd3 will be required to address these issues.
It is known that the epiblast requires a sufficient number of cells to proceed past a critical stage in development (gastrulation), and one possibility is that following gastrulation, _Foxd3_−/− cells in chimeras can respond to other signals in the embryo and proliferate and/or differentiate accordingly. In support of this idea, mutations in the gene encoding the Bmpr/Alk3 receptor protein cause proliferation defects as early as 6.5 dpc with no production of embryonic mesoderm, although teratocarcinomas from _Bmpr_−/− cells, albeit smaller than wild type, are able to produce mesoderm (Mishina et al. 1995). The explanation for this difference is that the embryo requires a minimal number of epiblast cells to carry out the morphogenetic movements of gastrulation. Therefore, we would argue that in the Foxd3 chimera rescue experiments, the presence of a sufficient number of wild-type cells allows progress through gastrulation.
The results show that the early preimplantation embryo is unaffected by the loss of Foxd3. The ICM of _Foxd3_−/− blastocysts appears morphologically indistinguishable from that of wild-type blastocysts. In addition, expression of mRNA for the transcription factors Oct4 and Sox2 and the secreted ligand Ffg4 is detected in _Foxd3_−/− blastocysts, and Oct4 protein is localized to the nucleus. However, the ICM fails to properly expand in vitro, and cells from the ICM cannot respond to culture conditions designed to support ES cell derivation. Oct4 expression is initiated but then downregulated shortly after implantation in _Foxd3_−/− embryos (∼5.5 dpc, Fig. 3a), and this may explain why _Foxd3_−/− embryos survive one day longer than _Oct4_−/− embryos. In agreement with this idea, _Oct4_−/− cells differentiate into trophoblast cells, and _Foxd3_−/− embryos have an expansion of the extraembryonic ectoderm, the trophoblast progenitor cells (Nichols et al. 1998). _Fgf4_−/− blastocysts also fail to elaborate an ICM in vitro, but they can be rescued by addition of soluble Fgf4 in the culture medium (Feldman et al. 1995). In contrast, _Foxd3_−/− ICMs are not rescued by addition of Fgf4, which is perhaps not surprising because mutant blastocysts are making Fgf4 mRNA. This suggests that there may be a disruption in the ability of _Foxd3_−/− cells to receive or interpret the Fgf4 signal. It has been demonstrated in zebrafish that the Oct4 homolog, spiel-ohne-grenzen (spg), is required for the competence of neurectoderm cells in the mid-hindbrain to respond to Fgf8 (Reim and Brand 2002). In addition, in vitro work has demonstrated that Foxd3 and Oct4 interact to regulate gene expression (Guo et al. 2002). Taken together, the analysis of the _Foxd3_−/− phenotype suggests that Foxd3 may be acting with Oct4 to confer responsiveness to Fgf4 signals on the ICM and early epiblast.
It has not been possible thus far to establish _Foxd3_−/− ES cell lines by several independent approaches. This is also the case for _Oct4_−/− ES cell lines, and careful titration of Oct4 levels in ES cells demonstrates that the absolute level of Oct4 protein is critical for the differentiation of specific lineages in the preimplantation embryo (Niwa et al. 1998). These results are in agreement with the model outlined above, placing Foxd3 function downstream of Oct4, required for the ability of cells to respond to Fgf4. A confounding observation is that _Fgf4_−/− ES cells have been isolated (Wilder et al. 1997). However, these cells were derived with soluble Fgf4 added to the medium, and it is not clear whether this is required for the establishment of such lines. This result raises an alternative explanation, placing Foxd3 in a parallel regulatory pathway, independent of Oct4, Sox2, and Fgf4, that is required for early embryogenesis and establishment of multipotent ES cells.
The winged helix gene Foxd3 was first characterized by its expression in ES cells and multipotent cells of the neural crest. Ectopic expression of Foxd3 drives proliferation in cell culture and a change in fate of neural tube cells towards neural crest. Perhaps even more important for the potential control of stem cell fate, it was shown that continued expression of Foxd3 in migrating neural crest interferes with the subsequent differentiation of those cells (Dottori et al. 2001). The expression of Foxd3 in multipotent cells, the inhibition of differentiation by ectopic expression of Foxd3, and our studies of Foxd3 loss-of-function together suggest that Foxd3 may play a critical role in establishing or maintaining proliferating and self-renewing progenitor cell populations. Stem cells represent a unique tissue type with great potential for disease therapy. However, in order to take advantage of this important biological potential it is crucial that we understand the genetic pathways controlling the production, maintenance, and differentiation of these cells. Identification of key regulatory genes such as Foxd3 is an important step in exploiting the potential for manipulation of multipotency in vitro and eventually in vivo for therapeutic goals.
Materials and methods
Generation of Foxd3+/− mice and embryos
The Foxd3 locus was cloned by screening a 129SvEv mouse library (Research Genetics) using PCR and a probe from the cDNA (Labosky and Kaestner 1998). The targeting vector was constructed using a 4.0 kilobase (kb) _Sac_1 fragment as the 5′ homology arm and a 1.5kb _Xba_1 fragment as the 3′arm. A diptheria toxin (DTA) cassette was used to select against random integration, and positive selection was provided by the neo cassette, flanked by loxP sites for subsequent removal in culture. The Foxd3 targeted allele has the entire coding region deleted as well as ∼70 bp of the 3′ flanking DNA. Either a Gfp gene with an internal ribosomal entry site (Ires, for the Foxd3Ires-Gfp allele) or a Gfp cassette (for the Foxd3Gfp allele; Clontech) was inserted as diagrammed in Figure 1. With the Ires-Gfp cassette inserted, Gfp fluorescence was detected in Foxd3+/− ES cells, but this lineage reporter (Foxd3Ires-Gfp) did not function in the embryo (data not shown). When a Gfp gene without an Ires was used, generating a fusion mRNA that included the Foxd3 5′UTR (Foxd3Gfp), Gfp fluorescence was observed in the neural crest but never in embryos earlier than 8.5 dpc (data not shown).
TL1 ES cells were electroporated following standard protocols (Hogan et al. 1994), and 162 colonies were selected for resistance to G418 and screened by Southern blot as diagrammed in Figure 1. Thirty-six recombinants were obtained for a targeting frequency of 22%. Transient transfection with circularized cre plasmid was performed to remove the neo resistance cassette and generate the Foxd3Ires-GfpΔneo allele. Three different cell lines carrying the Foxd3Ires-Gfp allele were injected into C57BL/6 blastocysts, and male chimeras were bred to either C57BL/6 or Black Swiss females. Two independent cell lines transmitted through the germ line, and because both showed the identical phenotype, studies were restricted to one cell line. Two independent cell lines carrying either the Foxd3Ires-GfpΔneo or Foxd3Gfp allele were also used to generate those mouse lines.
Mice were genotyped either by Southern blot or by PCR with the following primers: wild-type allele, 5′-CGACGACGG GCTGGAGGAGAA-3′ and 5′-ATGAGCGCGATGTAAGGG TAG-3′; mutant allele, 5′-AAGGGCGAGGAGCTGTTCAC-3′ and 5′-TGCTGCTTCATGTGGTCGG-3′ (red boxes in Fig. 1c).
The mutation was maintained on several inbred and outbred genetic backgrounds (129SvEvTac, C57BL/6, Black Swiss, and CD-1) with no variation in phenotype. Removal of the neo cassette did not affect the phenotype (data not shown).
Chimeras
Chimeras were generated by injection of Rosa26.1 ES cells into blastocysts isolated from Foxd3+/− intercrosses. Varying numbers (from 5 to 15) of ES cells were injected. Embryos were dissected at 8.5 and 9.5 dpc, and extraembryonic tissues were protease-treated to separate the extraembryonic mesoderm from the extraembryonic endoderm as described (Hogan et al. 1994). No chimeras were analyzed later than 9.5 dpc. DNA was extracted from extraembryonic endoderm to genotype the host blastocysts retrospectively. Embryos were stained with Xgal using standard techniques (Hogan et al. 1994).
Histology and whole mount in situ hybridization
Histology was performed by standard approaches (Presnell and Schreibman 1997). BrdU incorporation was detected with an antibody from Zymed, and the TUNEL assays were performed as described (Conlon et al. 1995) with reagents from Roche Molecular Biochemicals and/or with the Apoptag kit from Intergen. Anti-phosphohistone H3 antibody was from Upstate Biotechnology, and Alexa Fluor® 488 goat-anti-rabbit secondary was from Molecular Probes. Oct4 antibody was from Becton Dickson. Whole mount in situ hybridization was performed using Costar 12-well inserts following standard protocols (Hogan et al. 1994). Digoxigenin-labeled RNA probes were prepared using reagents from Roche Molecular Biochemicals. To examine the expression of Nodal, mice carrying the Foxd3 null allele and the Nodal-lacZ allele (Varlet et al. 1997) were generated. These double heterozygous mice were mated to Foxd3+/− mice, and embryos were stained with Xgal as described (Hogan et al. 1994) to reveal Nodal gene expression.
Teratocarcinomas
Teratocarcinomas were generated by transplanting a portion of a gastrula-stage embryo under the kidney capsule of a histocompatible adult mouse as described (Damjanov et al. 1987). For these experiments, transferred embryos were derived from crosses of Foxd3+/− mice on a C57BL/6 genetic background, and host mice were F1 hybrids from a [C57BL/6 × 129/SvEvTac] cross.
Derivation of ES cell lines
Blastocysts were harvested from 129SvEvTac (N5 or higher) Foxd3+/− intercrosses and placed onto irradiated STO feeder layers in complete ES medium: DMEM with 4500mg/L glucose and without pyruvate, 2mM glutamine, 0.1mM nonessential amino acids, 0.1mM β-mercaptoethanol (Sigma), 15% fetal calf serum (tested for ES cells, Summit Biotechnology), 1000U/mL LIF (ESGRO), and 50μg/mL gentamycin. Reagents were from GIBCO/Invitrogen except where indicated. After 4–5 d in culture when the ICMs have reached an appropriate size, they were manually isolated, trypsinized, and replated onto fresh STO feeder layers. After a few days in culture, ES cell clumps appeared and the line was then cultured routinely.
In vitro culture of blastocysts
Blastocysts were harvested from Foxd3+/− intercrosses and placed onto gelatin-coated dishes in ES cell medium. Fgf4 (Sigma) was added at 100ng/mL along with 1μg/mL heparin. Outgrowths were photographed every other day, and tissue was harvested and processed for RT-PCR as described below. Cultures were genotyped for Foxd3 as indicated by Foxd3 mRNA. Samples not demonstrating hypoxanthine phosphoribosyl transferase (Hprt) expression were discarded.
RT-PCR
RNA for RT-PCR was extracted using the Dynabead mRNA Direct kit (Dynal), and PCR was performed using the wild-type primers described above. To monitor the integrity of the cDNA produced by reverse transcription, RT-PCR for Hprt was performed with the following primers: 5′-TTGTTGGATTT GAAATTCCAGACAAG-3′ and 5′-GCATTTAAAAGGAACT GTTGACAACG-3′. The Sox2, Oct4, and Fgf4 primers were as described (Nichols et al. 1998; Zappone et al. 2000).
Acknowledgments
We thank Diane Zhou, Jacquie Kloetzli-Campbell, Valerie Cluzet, Weitao Sun, and Iris Fontaine-Glover for technical assistance. Richard Behringer, Ray Dunn, Brigid Hogan, Elizabeth Lacy, Carol MacLeod, Jonathan Pearce, Janet Rossant, Michael Shen, and Guang Zhou generously supplied whole mount in situ probes. Special thanks to Brigid Hogan and Steve DiNardo for their thoughtful reading of this manuscript, Elizabeth Robertson for providing us with Rosa26.1 ES cells and Nodal-lacZ mice, Davor Solter for advice in generating teratocarcinomas, Austin Smith and Richard Behringer for helpful discussions, Melissa Mann for help with the single blastocyst RT-PCR, and Sigrid Eckardt for advice with immunostaining of blastocysts. L.A. Hanna was supported by NIH grant (F32HL10421). This work was supported by grants to P.A.L. from the American Heart Association, the Center for Research in Fibrodysplasia Ossificans Progressiva, and the NIH (R01HD36720), and by an NIH grant to D.S.K. (R01GM64768).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL plabosky@mail.med.upenn.edu; FAX (215) 898-9871.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1020502.
References
- Barnes JD, Crosby JL, Jones CM, Wright CV, Hogan BL. Embryonic expression of Lim-1, the mouse homolog of Xenopus Xlim-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev Biol. 1994;161:168–178. doi: 10.1006/dbio.1994.1018. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RSP, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Damjanov I, Damjanov A, Solter D. Production of teratocarcinomas from embryos transplanted to extra-uterine sites. In: Robertson EJ, editor. Teratocarcinomas and embryonic stem cells. Oxford: IRL Press; 1987. pp. 1–18. [Google Scholar]
- Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, Shen MM. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature. 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci. 2002;99:3663–3667. doi: 10.1073/pnas.062041099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann BG. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 1991;113:913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Hromas R, Ye H, Spinella M, Dmitrovsky E, Xu D, Costa RH. Genesis, a Winged Helix transcriptional repressor, has embryonic expression limited to the neural crest, and stimulates proliferation in vitro in a neural development model. Cell Tissue Res. 1999;297:371–382. doi: 10.1007/s004410051365. [DOI] [PubMed] [Google Scholar]
- Kalantry S, Manning S, Haub O, Tomihara-Newberger C, Lee HG, Fangman J, Disteche CM, Manova K, Lacy E. The amnionless gene, essential for mouse gastrulation, encodes a visceral-endoderm-specific protein with an extracellular cysteine-rich domain. Nat Genet. 2001;27:412–416. doi: 10.1038/86912. [DOI] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–190. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes & Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TP, Labosky PA, Grabel LB, Kozak CA, Pitman JL, Kleeman J, MacLeod CL. The Pem homeobox gene is X-linked and exclusively expressed in extraembryonic tissues during early murine development. Dev Biol. 1994;166:170–179. doi: 10.1006/dbio.1994.1305. [DOI] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes & Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce JJ, Evans MJ. Mml, a mouse Mix-like gene expressed in the primitive streak. Mech Dev. 1999;87:189–192. doi: 10.1016/s0925-4773(99)00135-5. [DOI] [PubMed] [Google Scholar]
- Presnell JK, Schreibman MP. Humason's animal tissue techniques. Baltimore, MD: The Johns Hopkins University Press; 1997. pp. 17–116. [Google Scholar]
- Reim G, Brand M. Spiel-ohne-grenzen/pou2 mediates regional competence to respond to Fgf8 during zebrafish early neural development. Development. 2002;129:917–933. doi: 10.1242/dev.129.4.917. [DOI] [PubMed] [Google Scholar]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. MesP1: A novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D'Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada DA, Fletcher CF, Jenkins NA, Copeland NG, Klemsz M, et al. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem. 1996;271:23126–23133. doi: 10.1074/jbc.271.38.23126. [DOI] [PubMed] [Google Scholar]
- Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RS. Hex: A homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ. Nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development. 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- Wilder PJ, Kelly D, Brigman K, Peterson CL, Nowling T, Gao QS, McComb RD, Capecchi MR, Rizzino A. Inactivation of the FGF-4 gene in embryonic stem cells alters the growth and/or the survival of their early differentiated progeny. Dev Biol. 1997;192:614–629. doi: 10.1006/dbio.1997.8777. [DOI] [PubMed] [Google Scholar]
- Xu D, Yoder M, Sutton J, Hromas R. Forced expression of Genesis, a winged helix transcriptional repressor isolated from embryonic stem cells, blocks granulocytic differentiation of 32D myeloid cells. Leukemia. 1998;12:207–212. doi: 10.1038/sj.leu.2400948. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes & Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, et al. Sox2 regulatory sequences direct expression of a β-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]