Cryptic speciation in a model invertebrate chordate (original) (raw)
Abstract
We applied independent species concepts to clarify the phylogeographic structure of the ascidian Ciona intestinalis, a powerful model system in chordate biology and for comparative genomic studies. Intensive research with this marine invertebrate is based on the assumption that natural populations globally belong to a single species. Therefore, understanding the true taxonomic classification may have implications for experimental design and data management. Phylogenies inferred from mitochondrial and nuclear DNA markers accredit the existence of two cryptic species: C. intestinalis sp. A, genetically homogeneous, distributed in the Mediterranean, northeast Atlantic, and Pacific, and C. intestinalis sp. B, geographically structured and encountered in the North Atlantic. Species-level divergence is further entailed by cross-breeding estimates. C. intestinalis A and B from allopatric populations cross-fertilize, but hybrids remain infertile because of defective gametogenesis. Although anatomy illustrates an overall interspecific similarity lacking in diagnostic features, we provide consistent tools for in-field and in-laboratory species discrimination. Finding of two cryptic taxa in C. intestinalis raises interest in a new tunicate genome as a gateway to studies in speciation and ecological adaptation of chordates.
Keywords: anatomy, Ciona intestinalis, cryptic species, gamete compatibility, phylogenetics
Correct classification of model species is a major component for corroborating conclusions drawn from comparative biology. When laboratory colonies are not correlated with species concepts, or experimental material is largely obtained from natural populations, preventing uncovered speciation requires the taxonomic assessment through a phylogeographic approach (1, 2). The tunicate Ciona intestinalis L. (Chordata: Ascidiacea) is a marine filter-feeding organism that commonly lives in shallow waters of temperate to boreal regions. Favored biotopes typically include natural and artificial substrates in moderately sheltered habitats with high nutrient content and low wave exposure, such as mussel farms, estuaries, bays, lagoons, and ports (3). C. intestinalis is raising ecological and economical concerns as an invasive pest that outcompetes for space and food, reduces native diversity, and influences entire ecosystems (4, 5). A chordate body plan, research amenability, worldwide distribution, and easy sampling are the main reasons for the success of C. intestinalis in biosciences (6, 7). In 2002, completion of the whole genome sequence marked an impressive transition in the history of C. intestinalis, transforming it into a model organism of prime importance in integrative genomics (8–10). Besides progress in culturing, line storage, and formal genetics, research with C. intestinalis is largely based on sampling in nature (like in the 18th century) under the view that natural populations globally belong to the linnean taxon (data not shown and refs. 11–15). Notably, the taxonomic identity of this ascidian has been exhaustively reviewed through synonymies, museum specimens, and geographical variation, highlighting the sensitivity of phenotypic characters to environmental conditions and intraspecific polymorphisms (i.e., tunic texture and color morphs) (16). However, recent reports of genetic divergence between Mediterranean–Pacific and Atlantic populations of C. intestinalis have provided clues about an ancient episode of speciation or subspeciation (17–19). Because deciphering the taxonomy of this organism is central to integrative research, we interpreted the identity of C. intestinalis in the framework of three species concepts, focusing on Mediterranean and North European populations (see Materials and Methods for collection sites). Initially, two types of intersterile populations were identified as part of an international screen for genetic polymorphisms, during crossing assays undertaken to create genetic linkage maps and laboratory strains (20).
Results
Gamete Compatibility.
According to Mayr's definition (21), species are “groups of interbreeding populations in nature, unable to exchange genes with other such groups living in the same area.” Finding of both cryptic species of C. intestinalis in a well known transition area (English Channel, Plymouth, U.K.; Brest, France) (22), provides a unique opportunity to examine patterns of sexual isolation in a simultaneous hermaphrodite and to relate gamete compatibility with geographical distance and genetic partitioning.
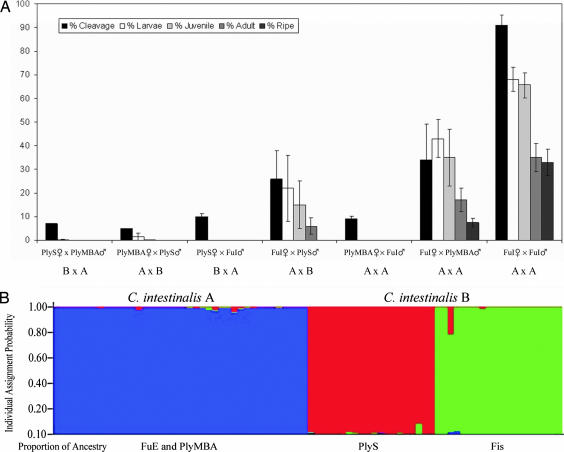
Reciprocal fertilization trials using individuals of the two species from Plymouth (PlyMBA sp. A and PlyS sp. B) always resulted in major dysfunctions within one to two cell cleavages, suggesting high levels of pre- and early postzygotic mechanisms against reproductive interactions (Fig. 1A). In contrast, heterospecific crossings between allopatric individuals (FuI sp. A × PlyS sp. B) displayed a higher degree of gamete compatibility, allowing development and growth of F1 generations. As shown in Fig. 1A, only Mediterranean female gametes could be fertilized and grown to adulthood in homo- and heterospecific combinations. Conversely, fertilization assays performed by using Plymouth spp. A and B oocytes resulted in lethal embryos that died before the larval stage. Hybrids suffered from reduced fitness, and, most notably, they did not produce functioning gametes. Indeed, although mature oocytes were never observed in the oviduct throughout the hybrid lifespan, spermatozoa collected from the male duct (vas deferens) were unable to inseminate C. intestinalis A eggs. The hypothesis of sexual isolation is reinforced by lack of microsatellite-based genetic recombination within C. intestinalis spp. A and B from the English Channel and from geographically distant populations (Fig. 1B and refs. 23 and 24). Progenies generated by homospecific crosses with FuI and PlyMBA sp. A specimens were capable of reaching sexual maturity, giving rise to F2 generations when backcrossed with field-collected Mediterranean individuals. If compared with sympatric individuals, fertilization success between allopatric C. intestinalis A was affected by early postzygotic barriers and low fitness, in support of genetic isolation by distance. Variability (see error bars in Fig. 1) reflected seasonal effects, with higher rates of fertilization and growth occurring in winter.
Fig. 1.
Sexual interactions and Bayesian clustering. (A) Heterotypic and homotypic crosses between allopatric and sympatric individuals of C. intestinalis A and B. Gamete compatibility is represented as key developmental steps from fertilization to sexual maturation. Each stage is expressed as percentage of the previous one (y axis). (B) Absence of natural hybridization between C. intestinalis A and B is uncovered by Bayesian-based clustering analysis of 12 microsatellite loci. Data were normalized. Error bars indicate standard deviations.
These results demonstrate the existence of two reproductively isolated entities within C. intestinalis according to the criteria of the biological species concept (21). Yet hybridization sensitivity related to behavioral differences in spawning and/or to ecophysiological limits will need to be addressed in detail.
Phylogenesis.
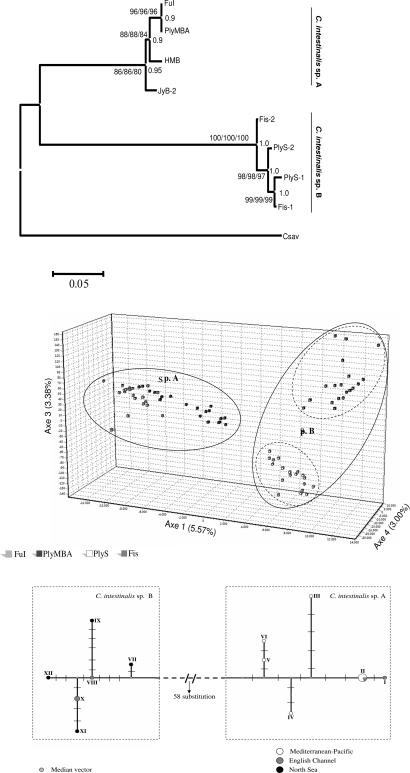
Genetic relationships inferred from 2,578 bp of concatenated nuclear (n; internal transcribed spacer 2, ITS-2; EPIC Hox5 and Gsx introns; Hox13 exon) and mitochondrial (mt; cytochrome oxidase I, COI) sequences under differential selective pressure (see Materials and Methods for details), using Ciona savignyi as the outgroup, generated two highly supported clades. One includes specimens from the English Channel, the Mediterranean Sea, and the Pacific Ocean, and the other includes those from North European coasts (Fig. 2A). Twenty individuals from four European localities were genotyped in 12 microsatellite loci, revealing a sharp break between phylogenetic subdivisions (see Materials and Methods for details). The two populations sampled within each clade of C. intestinalis were also distinct, unveiling two levels of genetic differentiation: (i) cryptic genetic diversity at the interspecies level and (ii) genetic structuring within each lineage denoted by the presence of two populations in each clade sharing distinct alleles (Figs. 1B and 2B). A low intralineage genetic diversity was observed between the two populations assayed [Table 1 and supporting information (SI) Table 3]. Genetic structure and distinct levels of gene flow between geographically predefined populations within lineages are further corroborated by the number of migrants among them (Table 2). The Bayesian-based clustering method demonstrated that the only model able to explain partitioning of genetic variability within the data set was that with K = 3 clusters. Each geographic population within C. intestinalis sp. B belongs only to one cluster, indicating distinct levels of population structure. Conversely, populations of C. intestinalis sp. A are equally distributed in a single cluster, indicating lower population distinction. Microsatellites clearly resolve C. intestinalis A and B populations and lineages, whereas sequence markers fail to discriminate populations at the intralineage level despite the geographical distance (Figs. 1B and 2B). We hypothesize that more subgroups may exist within C. intestinalis A and B at worldwide scale. Here they may have been missed owing to limited sampling coverage.
Fig. 2.
Genetic divergence. (A) A single maximum parsimony tree (1,148 steps of length, confidence interval = 0.96) inferred from 2,578-bp concatenated nuclear (Hox5, Gsx, Hox13, and ITS-2) and mitochondrial (COX1) sequences is shown (192 parsimony informative sites, 7.44%). Maximum likelihood/maximum parsimony/neighbor joining bootstrap support (1,000 iterations) and Bayesian posterior probability values are shown, respectively, on the left and right of each node. Numbers after locality abbreviations indicate distinct haplotypes. (B) Microsatellite genotype-based factorial correspondence analysis in three-dimensional representation. Color codes: black, FuI; light gray, PlyMBA; dark gray, Fis; white, PlyS. (C) Median joining haplotype network based on 515-bp COI alignment. Haplotypes are shaded according to minimum–maximum latitudes. I, PlyMBA; II, FuI, FuE, CdS, VC, Ve, Ta, LT, Al, and Br-1; III, HMB; IV, JyB-2; V, OnM; VI, JyB-1; VII, Fis-2; VIII, PlyS-2; IX, BH; X, PlyS-1 and Br-2; XI, Fis-1; XII, Ed.
Table 1.
Pairwise F statistics
| Population (type) | FuE (A) | PlyMBA (A) | PlyS (B) | Fis (B) |
|---|---|---|---|---|
| FuE (A) | — | 1.176 | 0.868 | 0.657 |
| PlyMBA (A) | 0.176 | — | 0.774 | 0.668 |
| PlyS (B) | 0.224 | 0.244 | — | 0.806 |
| Fis (B) | 0.276 | 0.272 | 0.237 | — |
Table 2.
Molecular variance
| Levels of variance | df | % |
|---|---|---|
| Among species | 1 | 6 |
| Among populations within species | 2 | 19 |
| Within populations | 156 | 75 |
| F_rt_ | 0.060 | |
| F_sr_ | 0.207 | |
| F_st_ | 0.254 |
The genetic divergence based on all mitochondrial and nuclear markers is strictly comparable to that observed between either C. intestinalis cryptic species and the congeneric C. savignyi (13% and 15%, respectively). This should not to be confused with individual (1.2%) (8) or interpopulation (3.0% in the Gulf of Naples) polymorphisms. The resolved network inferred from COI demonstrates the existence of two distinct sister haplogroups separated by 58 substitution steps. Each haplogroup includes several private alleles and some frequent ones, all of which have disjoint geographic origin. C. intestinalis A specimens form an assemblage consisting of a dominant Mediterranean/English Channel haplotype and two distinct Pacific ones. Specimens from the aforementioned localities are clearly separated by six equally frequent C. intestinalis B haplotypes (Fig. 2C).
Mobile elements are good candidates as molecular tools for mapping the geographic distribution of the two C. intestinalis species. For example, the LTR Gypsy Ty3 retrotransposon Cigr-1 and the MITE Cimi-1 retrotransposon are located, respectively, in intergenic and intronic positions of English but not Californian or Neapolitan individuals (18, 25). Herein we report a C. intestinalis A-specific 210-bp insertion in the second intron of the Hox5 gene, with 85% similarity to a Cili-2 non-LTR retrotransposon element (26) [SI Fig. 4]. This length polymorphism is easily recognizable on agarose gels and provides a fast molecular approach to distinguish between the two organisms (see Materials and Methods). In addition, a species-specific gene order was identified in the mitochondrial genome, and it can be also applied as a fast PCR screening method (58).
Morphology.
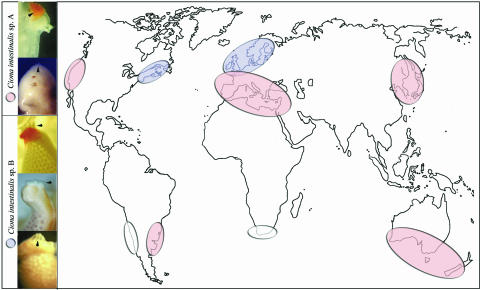
Taxonomy of Ciona spp. is currently formulated upon different anatomical characters, including presence of endostylar appendage, spermiduct papillae and postabdominal extension of mantle, position of pharyngeoepicardiac openings, and gill structure (3, 16, 27). An accurate analysis of external and internal anatomy in two populations per cryptic species confirmed the absence of diagnostic morphological characters (16) (SI Table 4). However, the two forms can be told apart by looking at spermiduct pigmentation. In C. intestinalis, the vas deferens opens well inside the atrial siphon, just below the anus, where it ends with a variable number of genital apertures in the shape of ellipsoidal papillae. With one exception (see below), characteristic bright orange pigmentation of these papillae is typical of C. intestinalis A populations analyzed in this study (149 individuals from 13 localities), while the duct itself is always uncolored. In contrast, pigmentation in C. intestinalis B (57 individuals from six localities) is confined to the duct, where it ranges from subdistal spots to more proximal extension. Remarkably, C. intestinalis A from South England lack orange pigmentation on both spermiduct and papillae, a condition observed also in sp. B populations from European and Canadian coasts of the North Atlantic Ocean (19) (Fig. 3). Although pigmentation of the genital openings is deprived of taxonomic value, this character may serve as an appropriate element for rapid discrimination by light microscopy. Pigmentation has been considered in the evaluation of speciation events in the colonial ascidians Pseudodistoma crucigaster and Cystoydes dellechiajei and in the assignment of C. intestinalis var. sydneiensis to the proper taxon (16, 28, 29), yet this phenotypic trait may be influenced by selective adaptation to environmental parameters (e.g., ref. 30). Herein the Mediterranean pattern is dominantly inherited in offspring of both homo- and heterospecific crosses (data not shown). We therefore conclude that in C. intestinalis this is a genetic and not environmentally controlled character with species-specific segregation.
Fig. 3.
Geographical distribution of C. intestinalis sp. A (pink) and sp. B (pale blue) and corresponding patterns of spermiduct pigmentation. See Morphology for detailed information on pattern distribution. Empty ovals indicate uncharacterized records of C. intestinalis populations. Arrowheads point to the ellipsoidal papillae at the end of the spermiduct.
Discussion
Is C. intestinalis a single cohesion taxon, or does it embody species- or subspecies-level divergence? According to reproductive and molecular data, the taxonomic classification of this invertebrate model chordate must be redrawn with the description of two morphologically cryptic species. Because of the poor detail of the original descriptions, the taxonomic status of the two cryptic species should be neatly dealt with through a PCR-assisted revision of anatomy, synonymies, and type specimens.
C. intestinalis spp. A and B inhabit largely disjoint geographical regions that overlap in the English Channel. We cannot exclude an accidental introduction of C. intestinalis sp. A along the southern coasts of England, where the only available record consists of volunteer individuals growing in the water facility of the Laboratory of the Marine Biological Association in Plymouth. However, an expansion of this taxon toward northeastern Atlantic is supported by the identification of populations in Brest (France) and Vigo (Spain). At this juncture we did not explore large sectors of the worldwide distribution of C. intestinalis sensu Linneus, including some areas of invasive introduction (e.g., refs. 31 and 32). In this respect, microsatellites lend themselves as informative markers for phylogeography, migration, and recruitment studies (23, 24).
Phylogenetic divergence and genetic homogeneity at population level are indicators of ancient and distinct evolutionary trajectories, as also supported by broad differences in the structure of the mitochondrial genome (F. Iannelli and C. Gissi, personal communication). Microsatellite and mtDNA analyses delineate separate scenarios: C. intestinalis sp. A is genetically homogeneous, suggestive of recent spread, whereas C. intestinalis sp. B is represented by fixed haplotypes and spatial genetic structure, compatible with an ancient origin. No episode of gene flow was detected where distribution of the two species intersects. The fact that C. intestinalis A and B lack significant and obvious differences in adult morphology, despite the low level of gene flow, implicates stabilizing selection on their phenotypes (33, 34). The two taxa colonize similar habitats; hence a careful approach to ecology, physiology, and life histories is needed to understand the circumstances that molded species divergence.
We analyzed sexual isolation by homo- and heterospecific cross-breeding of sympatric and allopatric individuals, following offspring viability until age of sexual maturation. The two cryptic species do not interbreed in the laboratory, probably as a result of effective pre- and postzygotic mechanisms of reproductive isolation. Adult hybrids generated by crossing gametes of allopatric specimens remained infertile until their senescence. The intensity of the above compatibility barriers appears to be related to the phylogenetic and geographic distances uncovered in this work. Altogether, sexual interaction assays point to selective reinforcement of gamete incompatibility in the absence of physical isolation between the two C. intestinalis cryptic species (2, 35), and they may explain why genetic hybrids were not detected. Our knowledge of the sexual behavior in C. intestinalis spp. does not allow inferences on the gamete bias observed between the two sibling taxa. Studies of one-way cross-infertility in C. intestinalis support the notion that a multilocus mechanism controls sexual interactions (18, 36–42).
Demonstration of a phylogenetic split in the invertebrate model chordate, C. intestinalis, clarifies a previous hypothesis of subspeciation beyond any possible ambiguities (17–19). This result provides new views for a large number of research fields and points to the importance of careful taxonomic definitions in the comparative approach. Furthermore, it widens the genomic toolbox for studying processes and functions in the evolutionary history of chordates.
Materials and Methods
Biological Material.
We analyzed specimens or sequences of C. intestinalis sp. A and sp. B from 21 geographical sites. Following is the locality list per species with abbreviations.
C. intestinalis sp. A: two populations in the Fusaro Lagoon, Naples, Italy (FuI and FuE); Villaggio Coppola, Naples, Italy (VC); Castellamare di Stabia, Naples, Italy (CdS); Venice, Italy (Ve); Taranto, Italy (Ta); Lago Faro, Messina, Italy (LF); Alicante, Spain (Al); Lake Timsah, Egypt (LT); Brest, France (Br); Marine Biological Association, Plymouth, England (PlyMBA); Half Moon Bay, California (HMB); Santa Barbara, California (SB); Yokohama Bay, Japan (JyB); Onagawa, Miyagi, Japan (OnM); and Chungmu Port, South Korea (KCP).
C. intestinalis sp. B: Plymouth Sound, England (PlyS); Edinburgh, Scotland (Ed); Breskens Harbor, The Netherlands (BH); Fiskebäckskil, Sweden (Fis); Brest, France (Br); and Mahone Bay, Canada (MB).
Animals from FuI, FuE, VC, CdS, PlyMBA, PlyS, and Fis were maintained in the animal facility at the Stazione Zoologica until ready for experimental use in fertilization assays. These were integrated with specimens kindly sent from several localities to our laboratory.
Culturing and Crossings.
Wild animals and hybrids were maintained according to standard procedures (11), with some modifications. To avoid cross-contaminations and for a correct classification, all parental and F1 animals have been genotyped and kept in separate tanks. Hetero- and homospecific crossing experiments were performed in different years (2003–2006). Experiments consisted of five trials per gamete combination and were conducted at 14°C, 16°C, and 18°C (11).
Morphology.
Five to 10 individuals from FuI, Fis, PlyMBA, and PlyS were anesthetized with menthol in filtered seawater, fixed in 4% formaldehyde in filtered seawater, and analyzed by using binocular stereomicroscopy before and after dissection. To examine spermiduct pigmentation, 5–10 individuals from each population were dissected, analyzed, and photographed.
DNA Extraction and Amplification.
Total DNA was extracted from fresh or 95% ethanol-preserved animal body wall. Fresh specimens were sustained for 2 days without feeding and with continuous change of 0.22 μM-filtered seawater to avoid stomach content contamination. DNA extraction was performed as previously described. A detailed list of primers and PCR conditions used to amplify mtDNA and nDNA markers is shown in SI Table 5 for Mt-haplotypes and sequence AA, see SI Table 6.
Genome Databases.
Californian sequences were downloaded from the Joint Genome Institute C. intestinalis database (http://genome.jgi-psf.org/Cioin2/Cioin2.home.html) (8), Japanese sequences were from the Ghost database (http://ghost.zool.kyoto-u.ac.jp/indexr1.html) (43), and C. savignyi sequences were from the Ensembl genome browser (www.ensembl.org/Ciona_savignyi/index.html) (44) after BlastN with C. intestinalis sequences.
Network.
Networks were generated by Network version 4.1.1.2 (www.fluxus-engineering.com/sharenet.htm) on ClustalW-aligned COI sequences using the Median Joining algorithm (45) and the parsimony postprocessing calculation option. Five sequences from each collection locality were used in the alignment.
Population Genetic Analysis.
Eighty Ciona spp. individuals (20 from each locality per population) were genotyped with four previously reported (23) and eight newly selected microsatellite markers (24). Allele detection was conducted by using an automated capillary sequencer (CEQ 2000XL DNA Analysis system; Beckman Coulter, Fullerton, CA), and electropherograms were analyzed with Beckman CEQ 2000 version 3.0 software. Quality of the allelic matrices and postgenotypic errors by population and locus were assessed in MICRO-CHECKER version 2.2.1 (46). Allelic richness per locus per population, and a factorial correspondence analysis of the microsatellite data, were calculated in GENETIX version 4.05.02 (47). Deviation from Hardy–Weinberg equilibrium was estimated in GENPOP version 3.4 (48). The partition of the genetic diversity, variation, gene flow, F statistics, number of migrants, and analysis of molecular variance were explored as implemented in GENALEX version 6 (49) from 12 microsatellite allelic frequency data among FuE, PlyMBA, PlyS, and Fis. Levels of significance were Bonferroni-corrected. A Bayesian-based clustering analysis, as implemented in STRUCTURE version 2.1 (50), was also applied to the microsatellite data set. This analysis computes the number of clusters of genotypes or subpopulations (K) that best fits the genetic variability encountered in the data set and interprets it without having prior information on the number of predefined sampled geographical populations. Under the admixture model, five independent runs were performed of K = 1–5 at 106 Markov chain–Monte Carlo repetitions and a burn-in period of 50,000 iterations without prior information and assuming correlated allele frequencies. The posterior probability was then calculated for each value of K, and the optimal number of clusters was chosen.
Sequence Analysis and Tree Reconstructions.
Sequences were aligned with BioEdit version 4.8.5 (51). The alignment was refined by eye. Maximum parsimony and maximum likelihood phylogenies were inferred in PAUP* 4.0b10 for Windows (52). Hierarchical likelihood ratio tests were performed by using MODELTEST version 3.06 (53) to find the best-fitting parameters for the maximum likelihood analysis given the alignment. Bootstrap support for individual clades (54) was calculated on 1,000 replicates by using the same methods, options, and constraints as in the tree inferences but with all identical sequences removed. Bayesian posterior probabilities of individual clades were calculated by using a variant of the Markov chain Monte Carlo algorithm as used in MrBayes version 3.1.2 (55). To increase the resolving power associated with additional informative sites, concatenated phylogenies were inferred from a final alignment of 2,578 bp and five genes. Before the combined analysis, the significance of the incongruence length difference test (56) was evaluated as evidence of congruence among all of the gene regions involved in the final alignment as partitions.
Cili-2 Retrotransposon.
The Neapolitan Hox5 sequence was screened against the reference collection of repeats and masks homologous portions by using CENSOR (57) (www.girinst.org/censor/index.php). Five to 10 individuals from each population were tested via PCR amplification for Cili-2 presence/absence.
Supplementary Material
Supporting Information
Acknowledgments
We thank L. Ballarin (University of Padova, Padova, Italy), J. Ballas (University of Vigo, Vigo, Spain), J. Bishop (Marine Biological Association of the U.K., Plymouth, U.K.), C. Dahlberg and M. Thorndyke (Kristinenberg Marine Research Station, Fiskebackskil, Sweden), S. Giacobbe (University of Messina, Messina, Italy), A. Gittenberger (National Museum of Natural History, Leiden, The Netherlands), K. Hotta (Keio University, Yokohama, Japan), C. Herbinger and S. Howes (Dalhousie University, Halifax, NS, Canada), I.-H. Kim (Kangnung National University, Kangnung, South Korea), L. Leveque (Roscoff Biological Station, Centre National de la Recherche Scientifique, Roscoff, France), S. Obenat (National University of Mar del Plata, Mar del Plata, Argentina), A. Ramos (University of Alicante, Alicante, Spain), G. Sharaf (Suez Canal University, Ismailia, Egypt), S. Shimeld (University of Reading, Reading, Berkshire, U.K.), M. Thorndyke, and the University of California Santa Barbara Ascidian Stock Center (NIH/R24GM075049) for kindly providing specimens; M. Nydam for sharing unpublished information; S. Groppelli and F. Di Bernardo for photographs; the Fishery, Marine Resources for Research, and Molecular Biology services at Stazione Zoologica Anton Dohrn for technical assistance; R. De Santis for laboratory support; J.-A. Sneli for translation of Gunnerus's description of Tethyum sociabile and illuminating discussions; and J. Bishop, D. Horner, J. Joss, M. Thorndyke, F. Toscano, three anonymous reviewers, and members of the laboratory for comments. This work was supported by grants from the European Union (Marine Genomics Europe, WP36, “The genetic identity of species,” to N.A.) and the Italian Ministry of University and Research (Grant 24/C08c to P.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF471239–EF471279).
References
- 1.Knowlton N. Annu Rev Ecol Syst. 1993;24:189–216. [Google Scholar]
- 2.Palumbi SR. Annu Rev Ecol Syst. 1994;25:547–572. [Google Scholar]
- 3.Millar RH. Ciona: Liverpool Marine Biological Committee Series No. 35. Liverpool, UK: Univ Press of Liverpool; 1953. [Google Scholar]
- 4.Carlton JT, Geller JB. Science. 1993;261:78–82. doi: 10.1126/science.261.5117.78. [DOI] [PubMed] [Google Scholar]
- 5.Stachowicz JJ, Whitlatch RB, Osman RW. Science. 1999;286:1577–1579. doi: 10.1126/science.286.5444.1577. [DOI] [PubMed] [Google Scholar]
- 6.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 7.Satoh N. Developmental Biology of Ascidians. Cambridge, UK: Cambridge Univ Press; 1994. [Google Scholar]
- 8.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 9.Imai KS, Levine M, Satoh N, Satou Y. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- 10.Mallon AM, Wilming L, Weekes J, Gilbert JGR, Ashurst J, Peyrefitte S, Matthews L, Cadman M, McKeone R, Sellick CA, et al. Genome Res. 2004;14:1888–1901. doi: 10.1101/gr.2478604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirino P, Toscano A, Caramiello D, Macina A, Miraglia V, Monte A. Mar Models Elec Rec. 2002 www.mbl.edu/BiologicalBulletin/MMER/cirino/CirCon.html.
- 12.Liu L-P, Xiang J-H, Dong B, Natarajan P, Yu K-J, Cai N-E. J Zhejiang Univ Sci B. 2006;7:467–474. doi: 10.1631/jzus.2006.B0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson C, Christiaen L, Deschet K, Jiang D, Joly JS, Legendre L, Nakatani Y, Tresser J, Smith WC. Methods Cell Biol. 2004;74:143–170. doi: 10.1016/s0091-679x(04)74007-8. [DOI] [PubMed] [Google Scholar]
- 14.Schmidtke J, Engel W. Biochem Genet. 1980;18:503–508. doi: 10.1007/BF00484397. [DOI] [PubMed] [Google Scholar]
- 15.Kano S, Chiba S, Satoh N. Mar Biotechnol (New York) 2001;3:58–67. doi: 10.1007/s101260000048. [DOI] [PubMed] [Google Scholar]
- 16.Hoshino Z, Nishikawa T. Publ Seto Mar Biol Lab. 1985;30:61–79. [Google Scholar]
- 17.Boffelli D, Weer CV, Weng L, Lewis KD, Shoukry MI, Pachter L, Keys DN, Rubin EM. Genome Res. 2004;14:2406–2411. doi: 10.1101/gr.3199704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki MM, Nishikawa T, Bird A. J Mol Evol. 2005;61:627–635. doi: 10.1007/s00239-005-0009-3. [DOI] [PubMed] [Google Scholar]
- 19.Nydam M, Harrison R. Mar Biol. 2007 0.1007/s00227-007-0617-0. [Google Scholar]
- 20.Kano S, Satoh N, Sordino P. Zool Sci. 2006;23:31–39. doi: 10.2108/zsj.23.31. [DOI] [PubMed] [Google Scholar]
- 21.Mayr E. Systematics and the Origin of Species. New York: Dover; 1942. [Google Scholar]
- 22.Hayward PJ, Ryland JS. Handbook of the Marine Fauna of North-West Europe. Oxford, UK: Oxford Univ Press; 1998. [Google Scholar]
- 23.Procaccini G, Pischetola M, Di Lauro R. Mol Ecol. 2000;9:1924. doi: 10.1046/j.1365-294x.2000.01071-4.x. [DOI] [PubMed] [Google Scholar]
- 24.Andreakis N, Caputi L, Sordino P. Mol Ecol Notes. 2007 10.1111/s.1471-8286.2006.0164g.x. [Google Scholar]
- 25.Ferrier DE, Holland PW. Mol Phylogenet Evol. 2002;24:412–417. doi: 10.1016/s1055-7903(02)00204-x. [DOI] [PubMed] [Google Scholar]
- 26.Simmen MW, Bird A. Mol Biol Evol. 2000;17:1685–1694. doi: 10.1093/oxfordjournals.molbev.a026267. [DOI] [PubMed] [Google Scholar]
- 27.Copello M, Devos L, Lafargue F. Vie Milieu. 1981;31:243–253. [Google Scholar]
- 28.Tarjuelo I, Posada D, Crandall KA, Pascual M, Turon X. Mol Ecol. 2004;13:3125–3136. doi: 10.1111/j.1365-294X.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Legentil S, Turon X. Biol J Linn Soc. 2006;88:203–214. [Google Scholar]
- 30.Sabbadin A, Graziani G. Nature. 1967;213:815–816. [Google Scholar]
- 31.Orensanz JM, Schwindt E, Pastorino G, Bortolus A, Casas G, Darrigran G, Elías R, López Gappa JJ, Obenat S, Pascual M, et al. Biol Invasions. 2002;4:115–143. [Google Scholar]
- 32.McDonald J. Mar Pollut Bull. 2004;49:854–874. doi: 10.1016/j.marpolbul.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 33.Stachowicz JJ, Fried H, Osman RW, Whitlatch RB. Ecology. 2002;83:2575–2590. [Google Scholar]
- 34.Petren K, Grant PR, Grant BR, Keller LF. Mol Ecol. 2005;14:2943–2957. doi: 10.1111/j.1365-294X.2005.02632.x. [DOI] [PubMed] [Google Scholar]
- 35.Snell TW, Hawkinson CA. Evolution (Lawrence, Kans) 1983;37:1294–1305. doi: 10.1111/j.1558-5646.1983.tb00245.x. [DOI] [PubMed] [Google Scholar]
- 36.Morgan TH. Proc Natl Acad Sci USA. 1923;9:170–171. doi: 10.1073/pnas.9.5.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murabe N, Hoshi M. Zool Sci. 2002;19:527–538. doi: 10.2108/zsj.19.527. [DOI] [PubMed] [Google Scholar]
- 38.Morgan TH. J Exp Zool. 1942;90:199–228. [Google Scholar]
- 39.Morgan TH. J Exp Zool. 1944;95:37–59. [Google Scholar]
- 40.Byrd J, Lambert CC. Mol Repr Dev. 2000;55:109–116. doi: 10.1002/(SICI)1098-2795(200001)55:1<109::AID-MRD15>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 41.Lambert CC, LaFargue F, Lambert G. Vie Milieu. 1990;40:293–295. [Google Scholar]
- 42.Kurn U, Sommer F, Hemmrich G, Bosch TC, Khalturin K. Dev Comp Immunol. 2007;31:360–371. doi: 10.1016/j.dci.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N. Zool Sci. 2005;22:837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- 44.Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, et al. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bandelt H-J, Forster P, Röhl A. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 46.Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- 47.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX, Logiciel Sous Windows Pour la Génétique des Populations. Montpellier, France: Université de Montpellier II; 2004. Version 4.05.02. [Google Scholar]
- 48.Raymond M, Rousset F. J Hered. 1995;86:248–249. [Google Scholar]
- 49.Peakall R, Smouse PE. Mol Ecol Notes. 2006;6:288–295. [Google Scholar]
- 50.Pritchard JK, Stephens M, Donnelly P. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall TA. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- 52.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2002. Version 4. [Google Scholar]
- 53.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 54.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 55.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 56.Farris JS, Kallersjo M, Kluge AG, Bult C. Cladistics. 1994;10:315–319. doi: 10.1111/j.1096-0031.1992.tb00071.x. [DOI] [PubMed] [Google Scholar]
- 57.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 58.Iannelli F, Pesole G, Sordino P, Gissi C. Trends Genet. 2007 doi: 10.1016/j.tig.2007.07.001. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information