Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9 (original) (raw)
Abstract
Transcriptional deregulation through the production of dominant-acting chimeric transcription factors derived from chromosomal translocations is a common theme in the pathogenesis of acute leukemias; however, the essential target genes for acute leukemogenesis are unknown. We demonstrate here that primary myeloid progenitors immortalized by various MLL oncoproteins exhibit a characteristic Hoxa gene cluster expression profile, which reflects that preferentially expressed in the myeloid clonogenic progenitor fraction of normal bone marrow. Continued maintenance of this MLL-dependent Hoxa gene expression profile is associated with conditional MLL-associated myeloid immortalization. Moreover, Hoxa7 and Hoxa9 were specifically required for efficient in vitro myeloid immortalization by an MLL fusion protein but not other leukemogenic fusion proteins. Finally, in a bone marrow transduction/transplantation model, Hoxa9 is essential for MLL-dependent leukemogenesis in vivo, a primary requirement detected at the earliest stages of disease initiation. Thus, a genetic reliance on Hoxa7 and Hoxa9 in _MLL_-mediated transformation demonstrates a gain-of-function mechanism for MLL oncoproteins as upstream constitutive activators that promote myeloid transformation via a _Hox_-dependent mechanism.
Keywords: MLL, Hox, myeloid, leukemia, transcription
Nonrandom chromosomal translocations are hallmark mutations frequently associated with human leukemias (Rabbitts 1994; Rowley 2001). Genes located at the breakpoints of leukemia-associated translocations are causally involved in disease pathogenesis and define a clear functional distinction between the pathways disrupted in chronic versus acute leukemias (Sawyers et al. 1991; Gilliland and Griffin 2002). Chronic leukemias are typically associated with mutations that constitutively activate either growth factor receptors or their downstream signaling components, leading to uncontrolled proliferation. In contrast, mutations of the transcriptional machinery are characteristic of acute leukemias, leading to a differentiation block and subsequent expansion of immature progenitors unable to generate mature effector cells.
The MLL gene codes for a novel transcriptional regulator that is frequently targeted by chromosomal translocations in clinically aggressive subtypes of acute lymphoid and myeloid leukemias (ALL and AML, respectively) of children and adults (Ayton and Cleary 2001). The MLL protein is a structural and functional homolog of Trithorax (Djabali et al. 1992; Gu et al. 1992; Tkachuk et al. 1992), a Drosophila homeotic regulator, and is required for mouse embryonic viability, efficient definitive hematopoiesis, and skeletal patterning along the anterior-posterior axis (Yu et al. 1995; 1998; Hess et al. 1997; Yagi et al. 1998; Ayton et al. 2001). Interestingly, MLL controls the maintenance, but not the initiation, of expression of Hox genes during embryogenesis (Yu et al. 1998). This distinctive role is likely to be mediated through effects on chromatin structure, as MLL is a histone methyl-transferase (HMT) and assembles a multiprotein complex with additional chromatin-modifying activities (Milne et al. 2002; Nakamura et al. 2002). The identities of MLL target genes that mediate its complex embryonic and hematopoietic functions are currently unknown. However, in a simple fibroblast model, MLL is required for multiple Hoxa and Hoxc gene expression via a mechanism that appears dependent on its HMT domain (Hanson et al. 1999; Milne et al. 2002). Within the hematopoietic system, Hox genes are expressed in the stem cell and immature progenitor compartments of normal bone marrow, but are transcriptionally down-regulated upon induction of terminal myeloid differentiation (Sauvageau et al. 1994; Lawrence et al. 1995; Pineault et al. 2002). Thus, normal stem cell and/or progenitor self-renewal may be _Hox_-dependent, and inappropriate Hox gene expression may underlie leukemic stem cell maintenance (Buske and Humphries 2000).
In acute leukemias, MLL participates in translocations with over 40 distinct partner chromosomal loci, resulting in the production of chimeric proteins containing a common N-terminal region of MLL fused to variable portions of each partner protein (Ayton and Cleary 2001). MLL partner proteins are structurally diverse, but possess motifs suggestive of normal functions associated with either transcriptional regulation within the nucleus or cytoplasmic signaling events. To date, structure/function analyses for multiple nuclear partner proteins suggest that transcriptional activator properties are essential for leukemogenesis (Lavau et al. 1997,2000a; Slany et al. 1998; DiMartino et al. 2000,2002; So and Cleary 2002,2003; Zeisig et al. 2003). Chimeric MLL proteins behave as dominant-acting oncogenes to promote leukemogenesis (Corral et al. 1996; Lavau et al. 1997,2000b; Dobson et al. 1999); however, it has been controversial as to whether this results in either MLL loss of function via trans-dominant inhibition or gain of MLL function. Furthermore, although MLL fusion proteins have been proposed to act as deregulated chimeric transcription factors, the target genes that mediate their oncogenic properties remain unknown. Here we used a genetic approach to demonstrate that Hoxa7 and Hoxa9 are required for the full oncogenic activity of an MLL oncoprotein. These results define an oncogenic transcriptional pathway in which MLL fusion proteins function as _Hox_-dependent gain-of-function mutants to induce acute myeloid leukemias.
Results
Myeloid transformation by MLL oncogenes is associated with expression of a specific subset of Hoxa genes
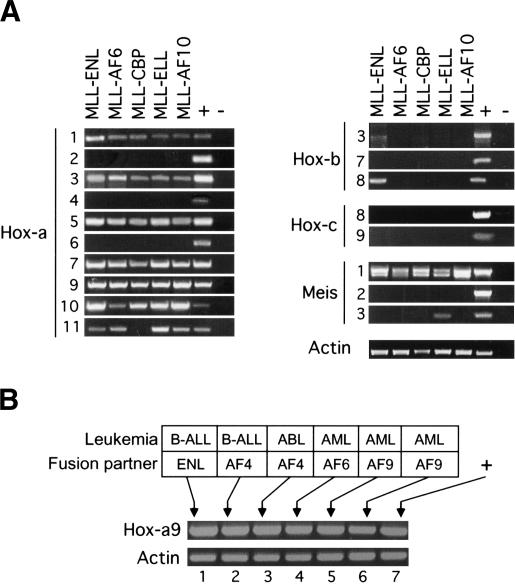
Given the conserved genetic requirement for MLL and its fly ortholog Trithorax in the maintenance of Hox gene expression, we used expression of Hox genes as surrogate gene expression markers for the status of MLL function in cells immortalized by MLL oncoproteins. Murine primary myeloid progenitor (MPMP) cell lines immortalized by MLL oncogenes were analyzed for the expression of various Hox genes. These lines were established from in vitro clonogenic assays and uniquely represent the earliest stages of MLL-associated acute leukemogenesis (Lavau et al. 1997; for review, see Ayton and Cleary 2001). Unlike human leukemias and leukemic cell lines, which have sustained multiple mutations associated with tumor initiation and progression, MPMP lines have sustained very few if any secondary mutations and retain cytokine dependence. Reverse transcriptase PCR (RT-PCR) analysis of MPMP lines immortalized by five different MLL fusion proteins revealed a characteristic Hoxa gene expression profile (Fig. 1A). All lines expressed Hoxa7, Hoxa9, Hoxa10, and Hoxa11 at the 5′ end of the Hoxa cluster. In contrast, 3′ Hoxa genes were variably expressed with periodicity, as evidenced by low levels of Hoxa1, higher levels of Hoxa3 and Hoxa5, and complete absence of Hoxa2, Hoxa4, and Hoxa6 expression.
Figure 1.
Consistent expression of select Hox genes in murine and human cells transformed by MLL fusion genes. (A) RT-PCR analysis was conducted on RNA isolated from MPMP cell lines immortalized by the MLL fusion genes indicated at the top of the lanes. Primers employed for RT-PCR were specific for various Hox or Meis transcripts indicated to the left of the panels. (B) RT-PCR was performed on human cell lines that express MLL fusions with various partner proteins, which are indicated along with the leukemia subtypes at the top of the gel lanes. Cell lines are as follows: HB1119 (lane 1), RS411 (lane 2), MV4-11 (lane 3), ML21 (lane 4), MonoMac6 (lane 5), and THP1 (lane 6).
Selective Hoxb and Hoxc genes, including those whose overexpression results in hematopoietic deregulation or those previously implicated as MLL targets during embryogenesis, were not consistently expressed in MPMP lines (Fig. 1A). Furthermore, expression of Hox genes was not a general property of myeloid transformation, because MPMP lines immortalized under identical in vitro conditions by E2A-HLF (Hunger et al. 1992) or E2A-PBX1 (Nourse et al. 1990) oncogenes did not express Hoxa, Hoxb, or Hoxc genes (data not shown). Given the known collaborative roles of Hoxa and Meis genes in myeloid oncogenesis (Nakamura et al. 1996), the expression of three members of the Meis family of Hox cofactors were also analyzed. Meis1, but not Meis2 or Meis3 was expressed in all MPMP lines immortalized by MLL fusion proteins. Similar to MPMP lines, HOXA9 was also expressed at high levels in all examined human cell lines derived from MLL-associated acute lymphoid, biphenotypic, or myeloid leukemias (Fig. 1B), consistent with previous observations in human ALL cells (Rozovskaia et al. 2001; Armstrong et al. 2002; Drabkin et al. 2002; Yeoh et al. 2002). Taken together, these data suggest that a specific subset of 5′ Hoxa genes is consistently coexpressed with Meis1 in cells transformed by MLL oncogenes, including cells representing the earliest stages of myeloid progenitor immortalization. These data provide further evidence to support a gain-of-function mechanism for MLL-associated transformation events.
_MLL fusion proteins are required for the initiation and maintenance of myeloid immortalization in vitro and 5_′ Hoxa gene expression
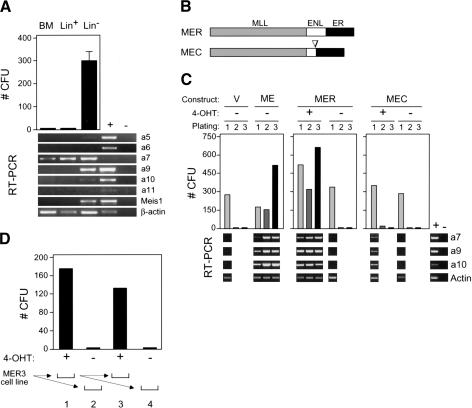
To determine the relationship between Hoxa gene expression and the myeloid clonogenic activity of hematopoietic progenitors, normal bone marrow (BM) cells were fractionated into populations enriched (lin+) or depleted (lin-) for the expression of lineage-associated antigens and then cultured for 7 d in methylcellulose supplemented with exogenous cytokines that promote myeloid differentiation. Myeloid clonogenic activity resided exclusively in the lin- fraction (Fig. 2A), known to be enriched in progenitors and stem cells. Robust expression of 5′ Hoxa genes including Hoxa7 and Hoxa9 was observed in the lin- clonogenic fraction; we therefore focused on the role of these Hoxa genes in MLL-associated myeloid transformation.
Figure 2.
Myeloid transformation requires sustained function of MLL-ENL and correlates with maintenance of Hox gene expression. (A) Myeloid clonogenic activity in normal bone marrow is associated with expression of Hoxa cluster genes. CFUs (per 10,000 plated cells) were determined after 7 d of culture in methylcellulose for whole bone marrow mononuclear cells (BM) or cellular fractions enriched based on the expression (Lin+) or absence (Lin-) of lineage markers. RT-PCR results are shown below for the Hox transcripts indicated to the right of each panel. (B) Conditional MLL-ENL proteins are schematically illustrated and consist of ER fusions at the C terminus of transformation-competent MLL-ENL (MER) or a mutant lacking the critical transformation domain of ENL (MEC). (C) Conditional immortalization of myeloid progenitors by MLL-ENL. Results of serial myeloid replating assays are shown for myeloid progenitors transduced with the constructs indicated at the top of the panels, cultured in the presence (+) or absence (-) of 4-OHT. Results of RT-PCR assays performed on cells harvested at day 7 of each plating are shown below for the transcripts indicated on the right. (D) MLL fusion protein activity is continuously required for the maintenance of transformation. A myeloid precursor cell line (MER3) immortalized by MLL/ENL-ER was plated in methylcellulose culture in the presence (+) or absence (-) of 4-OHT, and CFUs (per 10,000 plated cells) were determined on day 7 (lanes 1,2, respectively). Serial replating was performed as illustrated below, and CFUs again determined on day 7 (lanes 3,4, respectively).
The requirement for MLL fusion protein activity in the continued maintenance of Hoxa gene expression and myeloid transformation in vitro was assessed using conditional MLL fusion proteins. A mutant ligand-binding domain of the human estrogen receptor that is specifically responsive to 4-hydroxy-tamoxifen (4-OHT; Littlewood et al. 1995) was fused to the C terminus of MLL-ENL (MER), or a transformation-defective mutant (Slany et al. 1998) of MLL-ENL (MEC; Fig. 2B). The conditional constructs, as well as MLL-ENL and retroviral vector alone, were transduced into primary BM cells, which were then cultured in an in vitro immortalization assay in the presence or absence of 4-OHT (Fig. 2C). Under these conditions, transduction with vector alone was unable to sustain clonogenic activity through a second round of plating in methylcellulose and was associated with prior loss of 5′ Hoxa gene expression in first-round cells. Thus, in the absence of an immortalizing oncogene, the culture conditions promoted concomitant loss of both clonogenic activity and 5′ Hoxa gene expression. In contrast, MLL-ENL maintained clonogenic activity throughout serial replating and was associated with maintenance of 5′ Hoxa gene expression (Fig. 2C).
Similar to MLL-ENL, cells transduced with MER maintained clonogenic activity in vitro and 5′ Hoxa gene expression, but only in the presence of 4-OHT. Conversely, the immortalization-defective MEC protein was unable to maintain 4-OHT-dependent clonogenic activity through three rounds of serial replating. Low levels of Hox gene expression were detected in first-round cells transduced by MEC in the presence but not the absence of 4-OHT, reflecting transient residual clonogenic activity in second-round cells, which was eliminated by the third round of culture. Thus, 4-OHT-dependent regulation of an MLL fusion protein results in both conditional myeloid immortalization and associated 5′ Hoxa gene expression.
After three rounds of serial replating, MER-transformed cells were readily adapted to growth in liquid culture dependent upon the presence of cytokine (IL-3 or GM-CSF) and 4-OHT. In a representative MPMP cell line (Fig. 2D, line 3), 4-OHT was absolutely essential for continued clonogenic activity in vitro. Thus, MLL fusion protein activity is required for both initiation and maintenance of the immortalized phenotype, which correlates with 5′ Hoxa gene expression.
Hoxa7 and Hoxa9 independently contribute to efficient MLL_-associated myeloid immortalization in vitro_
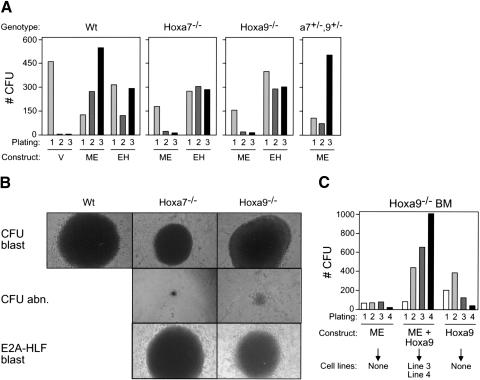
A genetic approach was employed to specifically address the requirement for Hoxa7 and Hoxa9 in myeloid transformation initiated by an MLL oncogene. In vitro serial replating assays were performed using hematopoietic progenitors harvested from wild-type mice or mice homozygous for inactivating mutations of Hoxa7 or Hoxa9, respectively (Chen et al. 1997, 1998). The enhanced replating potential typically induced by MLL-ENL in wild-type myeloid progenitors was significantly compromised in the absence of Hoxa7 or Hoxa9, despite comparable primary transduction efficiencies (Fig. 3A). In second- and third-round cultures, colony numbers were reduced, although not eliminated, in transduced _Hox_-deficient BM compared to wild-type BM. Most of the colonies obtained (52%-67%) were small and displayed atypical morphology (Fig. 3B) in contrast to the large, dense, blast-like colonies seen in cultures initiated with transduced wild-type BM (51%-82% blast-like wt colonies versus 12%-25% of _Hox_-deficient). The transformation potential of _Hox_-deficient progenitors was confirmed by transduction of the unrelated chimeric oncogene E2A-HLF, which enhanced the replating potential of _Hox_-deficient cells with efficiency and colony morphology equal to those of wild-type cells (Fig. 3A,B). Thus, MLL-ENL is selectively dependent on a subset of Hox genes to transform myeloid progenitors in vitro.
Figure 3.
Hoxa7 and Hoxa9 are independently required for efficient in vitro immortalization by MLL-ENL. (A) Representative results (CFUs/10,000 plated cells) of serial myeloid replating assays are shown for myeloid progenitors transduced with vector alone (V), MLL-ENL (ME), or E2A-HLF (EH) as indicated. Genotypes of BM cells used for transductions are indicated above the panels. (B) Morphologies for colony types observed in methylcellulose assays initiated with cells transduced by MLL-ENL (rows 1 and 2) or E2A-HLF (row 3). Genotypes of transduced cells are indicated above the panels. (C) Coexpression of Hoxa9 in combination with MLL-ENL rescues in vitro immortalization of _Hoxa9_-/- BM cells. Results are shown as CFU/10,000 plated cells. Transduced constructs are indicated below, along with indications of ability to establish IL-3-dependent cell lines in liquid media.
Hoxa7 and Hoxa9 are neighboring genes whose contributions to _MLL_-mediated transformation may depend on their total combined dosage. To assess whether they are required for overlapping or distinct parallel downstream transformation pathways, compound heterozygous Hoxa7+/-Hoxa9+/- progenitors were transduced with MLL-ENL and used to initiate serial replating assays. MLL-ENL efficiently maintained the clonogenic activity of myeloid progenitors derived from Hoxa7+/-Hoxa9+/- BM (Fig. 3A) and generated MPMP cell lines (data not shown). Thus, Hoxa7 and Hoxa9 do not exhibit nonallelic noncomplementation and may control independent pathways required for MLL-mediated transformation.
To confirm that _Hox_-deficient BM contains progenitors susceptible to MLL-mediated immortalization, MLL-ENL and Hoxa9 were cotransduced into _Hoxa9_-deficient progenitors in an attempt to rescue the immortalization defect. Coexpression of Hoxa9 and MLL-ENL efficiently maintained clonogenic activity of _Hoxa9_-deficient myeloid progenitors (Fig. 3C) and generated multiple MPMP cell lines. Similar to prior reports for wt BM (Schnabel et al. 2000), Hoxa9 alone also enhanced the replating of _Hoxa9_-deficient progenitors, albeit at reduced efficiency relative to MLL-ENL/Hoxa9 coexpression, but did not yield MPMP cell lines in liquid culture under our conditions. Thus, the _MLL_-targeted progenitor populations are present and functional in _Hoxa9_-deficient BM.
Hoxa9 is essential for induction of MLL_-associated myeloid leukemias_
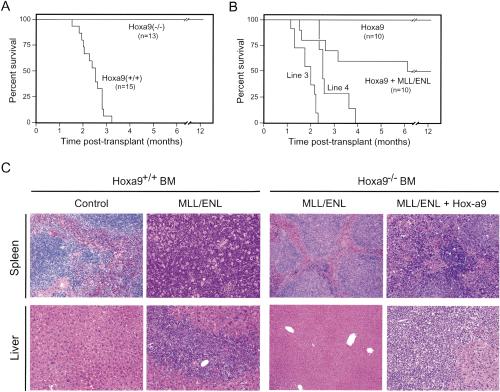
The genetic analysis was extended to determine the requirement for Hoxa9 during MLL-mediated induction of AML in vivo. All animals transplanted with _MLL-ENL_-transduced wild-type stem cells rapidly developed AML within 3 mo (Fig. 4A), similar to previous reports (Lavau et al. 1997). AML typically presented as massive hepatosplenomegaly resulting from extensive infiltration of the spleen and liver by immature myeloblasts (Fig. 4C), which displayed a Mac-1+/Gr-1+/-/c-Kitlo immunophenotype (data not shown). In contrast, no leukemias developed in mice transplanted with _MLL-ENL_-transduced _Hoxa9_-deficient BM during an observation period of >12 mo (Fig. 4A). However, transplantation of _Hoxa9_-/- BM cotransduced with MLL-ENL and Hoxa9, but not Hoxa9 alone, induced leukemias in 50% of recipient animals with latencies and histologic features (Fig. 4B,C) identical to those from wild-type BM, demonstrating that _Hoxa9_-/- cells were not inherently nonleukemogenic. The partial penetrance of the rescue transplants was likely due to inefficient cotransduction, because two independently derived _Hoxa9_-/- MPMP cell lines (Fig. 3C, lines 3 and 4) positively selected in vitro for MLL-ENL and Hoxa9 cotransduction induced fully penetrant rescue of the leukemogenic phenotype with regards to latency, histology, and immunophenotypic criteria (Fig. 4B; data not shown).
Figure 4.
Hoxa9 is essential for the leukemogenic properties of MLL-ENL. (A) Survival curves for cohorts of animals that were transplanted with MLL-ENL transduced progenitors from Hoxa9+/+ (n = 15) or _Hoxa9_-/- (n = 13) BM donors. (B) Coexpression of Hoxa9 in combination with MLL-ENL rescues in vivo leukemogenicity of _Hoxa9_-/- BM cells. Survival curves are shown for cohorts of animals that were transplanted with _Hoxa9_-/- progenitors transduced with Hoxa9 alone (n = 10), Hoxa9 + MLL-ENL (n = 10), or cell lines (lines 3 and 4) immortalized by coexpressed Hoxa9 and MLL-ENL. (C) Histology for spleens and livers from mice transplanted with transduced BM cells. Transduced genes and BM genotypes are indicated at the top.
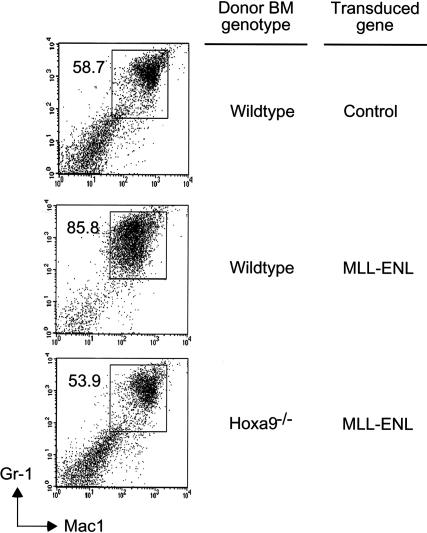
At 5 wk posttransplant, recipients of _MLL-ENL_-transduced wild-type BM displayed a significant preleukemic phenotype with moderate splenomegaly associated with a 2.7-fold increase in spleen weight and total splenocyte numbers relative to nontransplanted age-matched controls (data not shown). The BM was extremely pale due to myeloid progenitor expansion, as revealed by the abnormal appearance of a Mac-1+/Gr-1int population (Fig. 5). Comparable myeloid expansion was not present in recipients of _Hoxa9_-/- BM transduced with MLL-ENL, despite adequate engraftment, indicating that Hoxa9 was also essential for premalignant myeloid progenitor outgrowth in vivo.
Figure 5.
_Hoxa9_-dependent preleukemia in mice transplanted with MLL-ENL transduced BM. BM cells were analyzed for expression of Gr-1 and Mac-1 at 5 wk posttransplant. Numbers of Gr-1+/Mac-1+ cells were increased in recipients of wild-type BM transduced with MLL-ENL but not _Hoxa9_-/- BM, which showed levels of Gr-1+/Mac-1+ cells similar to those of control mice. Donor BM genotypes and transduced genes are indicated to the right. FACS data are representative of two animals in each cohort.
Discussion
Although many transcription factors have been directly implicated as dominant-acting oncoproteins, the target genes that mediate their oncogenic properties are largely unknown. Gene expression profiling of transformed cells has suggested a number of provisional candidate target genes. However, stringent genetic analyses in combination with biologically relevant functional assays for tissue-specific oncogenesis are essential for the correct assignment of bona fide downstream genes that make crucial contributions to the malignant phenotype. In this study we identified and validated subordinate genes that contribute to induction of AML by an MLL chimeric oncoprotein.
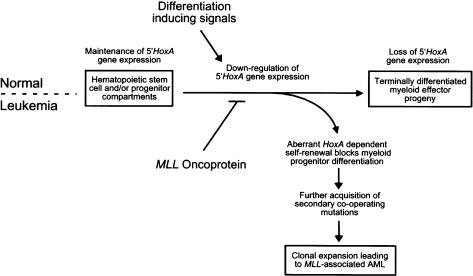
Our studies clearly define early, and apparently independent, roles for Hoxa7 and Hoxa9 in mediating the myeloid transforming properties of an MLL fusion protein. These results establish a linear pathway in which MLL oncoproteins function as upstream constitutive regulators to aberrantly maintain the expression of 5′ Hoxa genes required for transformation through inappropriate self-renewal of myeloid progenitors that cannot terminally differentiate (Fig. 6). Although our in vivo leukemogenesis studies only addressed the requirement for Hoxa9, it is possible that Hoxa7 may also be required for AML induction, given the compromised clonogenic properties of transduced _Hoxa7_-/- cells in vitro. Future experiments will determine whether other members of the Hoxa gene cluster also contribute to the MLL-associated malignant phenotype by these stringent criteria, and whether other MLL oncoproteins are similarly dependent on Hoxa7/Hoxa9. We note that maintenance of the characteristic Hoxa expression profile was consistently associated with myeloid immortalization by five distinct MLL fusion proteins, including nuclear partner proteins such as ENL, ELL, CBP, and AF10 or the cytoplasmic partner protein AF6, suggesting that deregulated Hoxa gene expression may be a common event in the initiation of MLL-associated leukemogenesis. It is likely that MLL oncoproteins directly deregulate Hox gene expression, as recent studies using chromatin immunoprecipitation have identified Hox gene promoters as direct targets for wild-type MLL in embryonic fibroblasts (Milne et al. 2002; Nakamura et al. 2002).
Figure 6.
Schematic model depicting Hoxa_-dependent myeloid transformation by MLL oncoproteins. Differentiation-inducing signals from the bone marrow microenvironment promote commitment of normal pluripotential HSCs toward terminal myeloid development that is associated with the loss of 5′_Hoxa gene expression. MLL oncoproteins disrupt this critical regulatory circuit, enhancing myeloid progenitor self-renewal via maintenance of 5′Hoxa gene expression, blocking differentiation events ultimately progressing to AML.
Multiple Hoxa cluster genes (Hoxa1, Hoxa3, Hoxa5, Hoxa7, Hoxa9, Hoxa10, and Hoxa11) were consistently expressed in MLL-immortalized MPMP cell lines. Conversely, several Hox genes (e.g., Hoxa4, Hoxc8, and Hoxc9) whose maintained expression requires wild-type MLL in midgestation embryos or embryonic fibroblasts (Yu et al. 1998; Hanson et al. 1999) were not expressed in MPMP cell lines immortalized by MLL oncogenes. Thus, transcriptional targets for oncogenic MLL mutants appear to represent only a subset of Hox genes regulated by wild-type MLL during embryogenesis. Differences in the spectrum of Hox genes regulated by either wild-type or mutant MLL proteins may be due in part to differential transcriptional effector properties of the novel partner proteins fused to the N terminus of MLL in leukemias.
Previous retroviral transduction/transplantation studies showed that forced expression of individual 5′ Hoxa genes such as Hoxa9 or Hoxa10 or the NUP98-HOXA9 fusion gene leads to the development of AML after particularly long latencies (Thorsteindottir et al. 1997; Kroon et al. 1998,2001). In our studies, Hoxa9 did not induce AML in _Hoxa9_-/- progenitors, consistent with the dosage-dependent nature of the _Hox_-associated leukemia phenotype. The long latencies in wild-type progenitors contrast with the more rapid induction of leukemias by MLL oncogenes. On this basis, we propose that sustained 5′ Hoxa gene expression is necessary, but not sufficient, for MLL-associated transformation. Interestingly, induction of AML is greatly accelerated if individual Hox genes such as Hoxa9 are coexpressed with the Hox DNA-binding cofactor Meis1 (Kroon et al. 1998). Indeed, Meis1 was originally isolated from retroviral genetic screens as a myeloid transforming oncogene only when aberrantly coexpressed with Hoxa7 or Hoxa9 (Nakamura et al. 1996). Because the latency period for AML induction by MLL-ENL is similar to that observed upon coexpression of Hoxa9 (or Hoxa10) with Meis1, and MPMP cell lines immortalized by numerous MLL fusion proteins consistently express Meis1, the potent transforming activity of MLL oncoproteins may reside in the consistent simultaneous expression of 5′ Hoxa genes in combination with Meis1. This predicts that MLL oncogenes may also be dependent on Meis1, a concept that we are currently evaluating using genetic approaches.
Although _Hox_-dependent lymphoid transformation was not directly addressed in the present study, we and others have observed a characteristic 5′ Hoxa gene expression profile in human lymphoid tumors and cell lines expressing translocation-derived MLL fusion proteins such as MLL-AF4 (Rozovskaia et al. 2001; Armstrong et al. 2002; Drabkin et al. 2002; Yeoh et al. 2002). These results suggest that _Hox_-dependent MLL-associated transformation is not myeloid-specific but may represent a general unifying feature of deregulated MLL activity regardless of the lineage affected. Furthermore, micro-array expression profiles revealed HOXA9 to be the most consistent expression marker for all human AML subtypes (Golub et al. 1999). In light of our current results, widespread HOXA9 expression in human AML may be due to aberrant MLL gain-of-function activity. This possibility warrants further analyses for MLL aberrations in human myeloid tumors that lack 11q23 chromosomal translocations but maintain HOXA9 expression.
During mouse embryonic development, normal MLL function is required for maintenance but not the initiation of expression of Hox genes such as Hoxa7 and Hoxc8 (Yu et al. 1998). These results imply that prior initiation of Hox expression may be an essential prerequisite for MLL-associated myeloid transformation to take place. It is currently unclear whether a similar mechanism operates during hematopoiesis to control MLL-dependent Hox transcriptional regulation, although some evidence lends support to this hypothesis. Firstly, we observed that cells enriched for the clonogenic progenitors that were used to initiate either in vitro immortalization or in vivo leukemogenesis assays preferentially expressed 5′ Hoxa genes. Secondly, the cellular targets for myeloid transformation by MLL-ENL are restricted to hematopoietic stem cells (HSC), CMP, and GMP subsets that normally express Hoxa9 (A. Cozzio, E. Passegue, P.M. Ayton, H. Karsunky, M.L. Cleary, and I.L. Weissman, in prep.). In contrast, progenitor populations such as the MEP that have down-regulated Hoxa9 expression are refractory to transformation by MLL-ENL. Thus, it is likely that a requirement for _MLL_-mediated transformation is pre-existent 5′ Hoxa gene expression in susceptible hematopoietic progenitors.
Genetic studies in the mouse indicate that MLL function is required for the correct execution of definitive hematopoiesis (Hess et al. 1997; Yagi et al. 1998). Loss of MLL function results in severe deficits in granulocyte and monocyte clonogenic (CFU-GM) progenitor activity associated with a dramatic reduction in colony number and size (Hess et al. 1997). Strikingly, it is the same CFUGM progenitor that is efficiently immortalized by MLL fusion proteins. Reminiscent of the MLL-deficient phenotype, we also observed the abnormal appearance of extremely small CFU-GM in serially replated cultures of _Hoxa7_-/- or _Hoxa9_-/- progenitors transduced by MLL-ENL. These observations suggest that 5′ Hoxa gene expression may also contribute to the essential role for normal MLL function during definitive hematopoiesis.
Retroviral transduction/transplantation studies have defined a critical role for multiple MLL fusion proteins in the initiation of in vitro myeloid immortalization and AML in vivo (Lavau et al. 1997,2000a,b; Slany et al. 1998; DiMartino et al. 2000,2002; So and Cleary 2002,2003; So et al. 2003; Zeisig et al. 2003). Our use of a conditional MLL fusion protein suggests that MLL oncoproteins may also be necessary for the continued maintenance of the immortalized phenotype. To our knowledge, this is the first demonstration that a dominant-acting chimeric transcription factor associated with acute leukemia is required for both initiation and maintenance of transformation. Continuous transcriptional deregulation of critical target genes by chimeric transcription factors may therefore be essential for the maintenance of the tumorigenic phenotype. These results compare well with other models using tetracycline-regulated transformation of pre-B cells or thymocytes with the BCR-ABL kinase or wild-type c-MYC, respectively (Felsher and Bishop 1999; Huettner et al. 2000).
In summary, the direct functional link between oncogenic MLL mutants and their specific Hox targets represents a unique entry point to delineate the molecular mechanisms of transcriptional deregulation in acute leukemias. Our genetic approach should serve as a useful paradigm for future functional analysis of oncogenic pathways.
Materials and methods
Hox gene expression analysis
Total RNA was isolated from MPMP cell lines or transduced myeloid cells using Trizol reagent (Invitrogen), and 1 μg was converted to random-primed cDNA using a preamplification kit (Invitrogen) according to the manufacturer's instructions. Resultant cDNA was diluted to a final volume of 100 μL, and 1 μL was used as the template for RT-PCR expression analysis. Nucleotide sequences of oligonucleotide primers used in this study are available upon request.
Retroviral vectors
A fragment of ENL cDNA (encoding amino acid residues 373-559) was ligated in-frame into the 3′ _Hpa_I site of MSCV/neo/5′MLL vector (DiMartino et al. 2002) to generate MSCV/MLL-ENLC. MSCV/EGFP was generated by replacement of the PGK-neo sequences with PGK-EGFP. MSCV/puro/MLL-ENLC and MSCV/EGFP/MLL-ENLC were constructed by ligation of a blunt-ended _Eco_RI-_Xho_I fragment of MLL-ENLC cDNA into the _Hpa_I site of either MSCV/puro or MSCV/EGFP. ENL sequences encoding residues 373-558 or an inactive mutant encoding ENL residues 373-543 (Slany et al. 1998) were cloned into pBS-ER (Littlewood et al. 1995) to generate pBS/ENL-ER fusions. Inserts were subsequently excised with _Eco_RI, blunted, and ligated in-frame into the 3′ _Hpa_I site in MSCV/neo/5′MLL vector to generate the corresponding transformation-competent or -incompetent MSCV/neo/MER or MSCV/neo/MEC constructs, respectively. MSCV/puro/E2A-HLF [a kind gift from K. Smith (Stanford University, Stanford, CA)] consisted of the E2A-HLF cDNA inserted into the _Eco_RI site of MSCV/puro.
In vitro myeloid immortalization assays
In vitro immortalization assays were performed as described (Lavau et al. 1997). For cotransduction experiments, clonogenic progenitors were further enriched by use of the lin- cell fraction of 5FU-treated BM as described below. 4-hydroxy-tamoxifen (4OHT; Sigma) was used at a final concentration of 1 μM. All experiments with conditional constructs were performed in the presence of either 4OHT or vehicle control (ethanol). Cytokine-dependent MPMP cell lines were generated by seeding cells from the third round of methylcellulose cultures in RPMI 1640 medium supplemented with 10% serum and 1 ng/mL of either murine IL-3 or GM-CSF (R&D Systems).
In vivo leukemogenesis assays
Donor BM cells were harvested from 4- to 8-week-old C57BL/6-Ly5.1 wild-type or Hoxa9-/- mice 5 d after intraperitoneal administration of 150 mg/kg 5FU. Mononuclear cells were stained with a cocktail of phycoerythrin (PE)-conjugated antibodies specific for lineage markers B220, CD3, CD4, CD8, Mac1, GR1, and Ter119 (Pharmingen). The lineage-negative (lin-) cellular fraction of BM was isolated using anti-PE microbeads (Miltenyi Biotech) and an Automacs cell separator as recommended by the manufacturer (Miltenyi Biotech). lin- cells were transduced with retroviruses as previously described (Lavau et al. 1997). Approximately 104 transduced lin- C57BL/6-Ly5.1 cells together with a radioprotective dose of 2 × 105 congenic C57BL/6-Ly5.2 recipient BM cells were transplanted into irradiated (960 rad) congenic C57BL/6-Ly5.2 mice via the retro-orbital plexus. For analysis of hematopoietic reconstitution or tumor immunophenotyping, cells were stained with anti-Ly5.1 antibody directly conjugated to fluorescein isothiocyanate in combination with PE-conjugated antibodies specific for lineage antigens, and then analyzed using a FACSCalibur flow cytometer (Becton Dickinson).
Histopathology
Tissues were fixed in 10% buffered formalin, embedded in paraffin, sectioned (5 μm), and stained with hematoxylin and eosin.
Acknowledgments
We thank Jorge DiMartino for constructs, cell lines, and advice in setting up serial replating assays. We acknowledge Kevin Smith for constructs and cell lines, and Cita Nicolas and Maria Ambrus for excellent technical assistance. We are indebted to Mario Capecchi for generously providing _Hoxa7_- and _Hoxa9_-deficient mice. This work was supported by funds from the National Cancer Institute (CA55029) and the Children's Health Initiative.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1111603.
References
- Armstrong S.A., Staunton, J.E., Silverman, L.B., Pieters, R., den Boer, M.L., Minden, M.D., Sallan, S.E., Lander, E.S., Golub, T.R., and Korsmeyer, S.J. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30**:** 41-47. [DOI] [PubMed] [Google Scholar]
- Ayton P.M. and Cleary, M.L. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20**:** 5695-5707. [DOI] [PubMed] [Google Scholar]
- Ayton P., Sneddon, S.F., Palmer, D.B., Rosewell, I.R., Owen, M.J., Young, B., Presley, R., and Subramanian, V. 2001. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis 30**:** 201-212. [DOI] [PubMed] [Google Scholar]
- Buske C. and Humphries, R.K. 2000. Homeobox genes in leukemogenesis. Int. J. Hematol. 71**:** 301-308. [PubMed] [Google Scholar]
- Chen F. and Capecchi, M.R. 1997. Targeted mutations in hoxa-9 and hoxb-9 reveal synergistic interactions. Dev. Biol. 181**:** 186-196. [DOI] [PubMed] [Google Scholar]
- Chen F., Greer, J., and Capecchi, M.R. 1998. Analysis of Hoxa7/Hoxb7 mutants suggests periodicity in the generation of the different sets of vertebrae. Mech. Dev. 77**:** 49-57. [DOI] [PubMed] [Google Scholar]
- Corral J., Lavenir, I., Impey, H., Warren, A.J., Forster, A., Larson, T.A., Bell, S., McKenzie, A.N., King, G., and Rabbitts, T.H. 1996. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: A method to create fusion oncogenes. Cell 85**:** 853-861. [DOI] [PubMed] [Google Scholar]
- DiMartino J.F., Miller, T., Ayton, P.M., Landewe, T., Hess, J.L., Cleary, M.L., and Shilatifard, A. 2000. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood 96**:** 3887-3893. [PubMed] [Google Scholar]
- DiMartino J.F., Ayton, P.M., Chen, E.H., Naftzger, C.C., Young, B.D., and Cleary, M.L. 2002. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood 99**:** 3780-3785. [DOI] [PubMed] [Google Scholar]
- Djabali M., Selleri, L., Parry, P., Bower, M., Young, B.D., and Evans, G.A. 1992. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2**:** 113-118. [DOI] [PubMed] [Google Scholar]
- Dobson C.L., Warren, A.J., Pannell, R., Forster, A., Lavenir, I., Corral, J., Smith, A.J., and Rabbitts, T.H. 1999. The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 18**:** 3564-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H.A., Parsy, C., Ferguson, K., Guilhot, F., Lacotte, L., Roy, L., Zeng, C., Baron, A., Hunger, S.P., Varella-Garcia, M., et al. 2002. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia 16**:** 186-195. [DOI] [PubMed] [Google Scholar]
- Felsher D.W. and Bishop, J.M. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4**:** 199-207. [DOI] [PubMed] [Google Scholar]
- Gilliland D.G. and Griffin, J.D. 2002. The roles of FLT3 in hematopoiesis and leukemia. Blood 100**:** 1532-1542. [DOI] [PubMed] [Google Scholar]
- Golub T.R., Slonim, D.K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J.P., Coller, H., Loh, M.L., Downing, J.R., Caligiuri, M.A., et al. 1999. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 286**:** 531-537. [DOI] [PubMed] [Google Scholar]
- Gu Y., Nakamura, T., Alder, H., Prasad, R., Canaani, O., Cimino, G., Croce, C.M., and Canaani, E. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71**:** 701-708. [DOI] [PubMed] [Google Scholar]
- Hanson R.D., Hess, J.L., Yu, B.D., Ernst, P., van Lohuizen, M., Berns, A., van der Lugt, N.M., Shashikant, C.S., Ruddle, F.H., Seto, M., et al. 1999. Mammalian Trithorax and poly-comb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. 96**:** 14372-14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J.L., Yu, B.D., Li, B., Hanson, R., and Korsmeyer, S.J. 1997. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90**:** 1799-1806. [PubMed] [Google Scholar]
- Huettner C.S., Zhang, P., Van Etten, R.A., and Tenen, D.G. 2000. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat. Genet. 24**:** 57-60. [DOI] [PubMed] [Google Scholar]
- Hunger S.P., Ohyashiki, K., Toyama, K., and Cleary, M.L. 1992. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes & Dev. 6**:** 1608-1620. [DOI] [PubMed] [Google Scholar]
- Kroon E., Krosl, J., Thorsteinsdottir, U., Baban, S., Buchberg, A.M., and Sauvageau, G. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17**:** 3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Thorsteinsdottir, U., Mayotte, N., Nakamura, T., and Sauvageau, G. 2001. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 20**:** 350-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Szilvassy, S.J., Slany, R., and Cleary, M.L. 1997. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 16**:** 4226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Du, C., Thirman, M., and Zeleznik-Le, N. 2000a. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 19**:** 4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Luo, R.T., Du, C., and Thirman, M.J. 2000b. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc. Natl. Acad. Sci. 97**:** 10984-10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence H.J., Sauvageau, G., Ahmadi, N., Lopez, A.R., LeBeau, M.M., Link, M., Humphries, K., and Largman, C. 1995. Stage- and lineage-specific expression of the HOXA10 homeobox gene in normal and leukemic hematopoietic cells. Exp. Hematol. 23**:** 1160-1166. [PubMed] [Google Scholar]
- Littlewood T.D., Hancock, D.C., Danielian, P.S., Parker, M.G., and Evan, G.I. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23**:** 1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne T.A., Briggs, S.D., Brock, H.W., Martin, M.E., Gibbs, D., Allis, C.D., and Hess, J.L. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10**:** 1107-1117. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Largaespada, D.A., Shaughnessy Jr., J.D., Jenkins, N.A., and Copeland, N.G. 1996. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat. Genet. 12**:** 149-153. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C.M., and Canaani, E. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10**:** 1119-1128. [DOI] [PubMed] [Google Scholar]
- Nourse J., Mellentin, J.D., Galili, N., Wilkinson, J., Stanbridge, E., Smith, S.D., and Cleary, M.L. 1990. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell 60**:** 535-545. [DOI] [PubMed] [Google Scholar]
- Pineault N., Helgason, C.D., Lawrence, H.J., and Humphries, R.K. 2002. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 30**:** 49-57. [DOI] [PubMed] [Google Scholar]
- Rabbitts T.H. 1994. Chromosomal translocations in human cancer. Nature 372**:** 143-149. [DOI] [PubMed] [Google Scholar]
- Rowley J.D. 2001. Chromosome translocations: Dangerous liaisons revisited. Nat. Rev. Cancer 1**:** 245-250. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T., Feinstein, E., Mor, O., Foa, R., Blechman, J., Nakamura, T., Croce, C.M., Cimino, G., and Canaani, E. 2001. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4: 11) abnormality. Oncogene 20**:** 874-878. [DOI] [PubMed] [Google Scholar]
- Sauvageau G., Lansdorp, P.M., Eaves, C.J., Hogge, D.E., Dragowska, W.H., Reid, D.S., Largman, C., Lawrence, H.J., and Humphries, R.K. 1994. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. 91**:** 12223-12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers C.L., Denny, C.T., and Witte, O.N. 1991. Leukemia and the disruption of normal hematopoiesis. Cell 64**:** 337-350. [DOI] [PubMed] [Google Scholar]
- Schnabel C.A., Jacobs, Y., and Cleary, M.L. 2000. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 19**:** 608-616. [DOI] [PubMed] [Google Scholar]
- Slany R.K., Lavau, C., and Cleary, M.L. 1998. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol. Cell Biol. 18**:** 122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C.W. and Cleary, M.L. 2002. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol. Cell Biol. 22**:** 6542-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood 101**:** 633-639. [DOI] [PubMed] [Google Scholar]
- So C.W., Karsunky, H., Passegue, E., Cozzio, A., Weissman, I.L., and Cleary, M.L. 2003. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell 3**:** 161-171. [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Sauvageau, G., Hough, M.R., Dragowska, W., Lansdorp, P.M., Lawrence, H.J., Largman, C., and Humphries, R.K. 1997. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol. Cell Biol. 17**:** 495-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachuk D.C., Kohler, S., and Cleary, M.L. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71**:** 691-700. [DOI] [PubMed] [Google Scholar]
- Yagi H., Deguchi, K., Aono, A., Tani, Y., Kishimoto, T., and Komori, T. 1998. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92**:** 108-117. [PubMed] [Google Scholar]
- Yeoh E.J., Ross, M.E., Shurtleff, S.A., Williams, W.K., Patel, D., Mahfouz, R., Behm, F.G., Raimondi, S.C., Relling, M.V., Patel, A., et al. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1**:** 133-143. [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hess, J.L., Horning, S.E., Brown, G.A., and Korsmeyer, S.J. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378**:** 505-508. [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hanson, R.D., Hess, J.L., Horning, S.E., and Korsmeyer, S.J. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. 95**:** 10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig B.B., Schreiner, S., Garcia-Cuellar, M.P., and Slany, R.K. 2003. Transcriptional activation is a key function encoded by MLL fusion partners. Leukemia 17**:** 359-365. [DOI] [PubMed] [Google Scholar]