Genetic dissection of the budding yeast Arp2/3 complex: A comparison of the in vivo and structural roles of individual subunits (original) (raw)
Abstract
In previous work, we identified the yeast Arp2/3 complex, which localizes to cortical actin patches and is required for their motility and integrity in vivo. This complex contains proteins homologous to each subunit of the Acanthamoeba and human Arp2/3 complex except for a 40-kDa subunit (p40), which was missing from the purified yeast complex. Here, we demonstrate by using immunoprecipitation and gel-filtration analysis that Arc40p, the homolog of p40 identified from the yeast genome database, associates with the yeast Arp2/3 complex. We have carried out gene disruptions of each subunit of the yeast Arp2/3 complex to study each subunit’s role in the function of the complex. Surprisingly, we find that only ARC40 is fully essential for cell viability. Strains lacking each of the other subunits exhibit varying degrees of defects in cell growth and viability and in assembly and polarization of cortical actin patches. We have also examined each subunit’s role in maintaining the structural integrity of the Arp2/3 complex. Arp2p, Arp3p, and Arc40p fall into the monomer pool in Δarc19 and Δarc35 cells, suggesting that Arc19p and Arc35p are the central scaffolding components of the complex. Arp2p and Arp3p do not have major roles in maintaining complex integrity, and Arc15p is required for association of Arp2p and Arc40p, but not other subunits, with the complex. These results provide evidence that each subunit contributes differently to the assembly and function of the Arp2/3 complex.
A complex containing two actin-related proteins, Arp2p and Arp3p, has recently emerged as a strong candidate for nucleating actin assembly that drives the motility of the pathogenic bacterium Listeria monocytogenes (1, 2). This complex, termed the Arp2/3 complex, contains seven subunits conserved among eukaryotes and localizes to regions of actin-based motility, such as the actin comet tails of Listeria (1), and the leading edges of Acanthamoeba and fibroblasts (3–5). Biochemical studies have shown it to bind both pointed ends and sides of actin filaments to create T structures resembling the brush-like actin structures seen at the leading edges of fish keratocytes (6, 7). Furthermore, the complex has a weak intrinsic actin nucleation activity that is significantly stimulated by the ActA protein of Listeria, and by Wiskott–Aldrich syndrome protein (WASP) family proteins (2, 8–11).
Much of the understanding of the complex has been focused on the actin-related proteins Arp2p and Arp3p. Because actin trimer formation is rate-limiting in actin polymerization (12, 13), and the Arp2/3 complex contains two molecules structurally similar to actin, this complex is structurally well suited to serve as an actin filament nucleator (4). Through structure modeling, Kelleher et al. predicted that the barbed end of an Arp2p/Arp3p heterodimer interacts with the pointed end of actin molecules to nucleate filament growth toward the barbed end (4). The biochemical findings that the complex stimulates filament nucleation and interacts with the pointed ends of actin filaments support this model (2, 6). However, there is still no evidence that the nucleation and the pointed-end binding activities directly involve the two actin-related proteins in the complex. Furthermore, the roles of the non-actin related subunits in these biochemical activities remain undetermined.
Yeast serves as an excellent genetic system to study the in vivo function of Arp2/3 complex. In fission yeast, Arp3p is an essential actin-patch component that functions to promote cell cycle-specific actin rearrangements (14). Sop2p, the fission yeast homolog of the 40-kDa subunit (p40) of the Arp2/3 complex is an essential protein that interacts with Arp3p but localizes to filamentous structures distinct from actin patches (15). In budding yeast, Arp2p and Arp3p have been shown to be components of actin patches (16, 17), the highly motile actin-rich structures that accumulate at sites of polarized growth during the yeast cell cycle. Both Arp2p and Arp3p function to maintain the proper organization of actin patches, and Arp3p is required for the motility of actin patches in vivo (16, 17). An Arp2p- and Arp3p-containing complex purified from budding yeast contained six equal stoichiometric subunits (17). Sequence identification of these subunits showed that they are highly conserved with the subunits of the human Arp2/3 complex (5). The only subunit missing in the purified budding yeast complex was p40. A homolog of p40, termed ARC40, has been found in the budding yeast genome database (15), but it is not clear whether this protein directly contributes to Arp2/3 complex function.
In this study, we demonstrate that Arc40p associates with the budding yeast Arp2/3 complex. To study the contributions of each subunit to complex function, we have disrupted the genes encoding each subunit of the yeast Arp2/3 complex. We find only ARC40 to be fully essential in our strain background. Deletion of genes encoding the other subunits gave rise to viable strains with varying degrees of growth defects, permitting us to analyze their relative roles in maintaining actin organization and the integrity of the Arp2/3 complex.
MATERIALS AND METHODS
Gene Disruption of Arp2/3 Complex Subunits.
A heterozygous ARP3 gene disruption strain (RLY180) was generated as described (17).
The ARP2 gene was PCR-amplified from genomic DNA by using primers SRp1 (5′-GCG CGC AAG CTT CTG TGA TAT GTA TAT TTG TT-3′) and SRp2 (5′-GCG CGC GGA TCC CTA TCC TCT AAC GGC GCT CA-3′) and cloned into pBluescript II SK(+) (Stratagene) by using _Hin_dIII and _Bam_HI sites to generate pDW1. _Bcl_I and _Nde_I sites were used to delete 87% of the ARP2 ORF. These sites were blunted, and the TRP1 gene from YDp-W (18), was inserted to generate the ARP2 gene disruption plasmid pDW3. pDW3 was cut with _Sac_I and _Bam_HI and transformed into the Trp− diploid strain RLY141 to generate RLY184. ARP2 gene disruption was confirmed by using PCR and restriction digest analysis (data not shown).
The ARC40 gene was amplified from genomic DNA by using primers DWp1 (5′-GCG CG_G GAT CC_A TCA GGA TCA TAC TTA GAG G-3′) and DWp2 (5′-GCG CG GGT ACC CTGCTA GTC AAT AAA AAC AC-3′) and cloned into pSK+ by using _Bam_HI and _Kpn_I sites. _Acc_I and _Bgl_II sites were used to delete 96% of the ARC40 ORF, which was replaced by the TRP1 gene from YDp-W as above to generate pDW22. To generate the Δarc40 strain, pDW22 was cut with _Stu_I and _Kpn_I and transformed into RLY175. ARC40 gene disruption was confirmed by using PCR (data not shown).
ARC15, ARC18, ARC19, and ARC35 genes were disrupted by using the one-step PCR-based method described (16). For ARC15 deletion, a PCR fragment carrying the TRP1 marker was amplified from pRS304 (19) with primers DWp19 (5′-CAG AGA AGA CTC AAC ACA ACA CAC GCG AAC GAT CAA GCA AGA TTG TAC TGA GCG TGC AC-3′) and DWp20 (5′-TTA CGT ATA TAT ATG TAT ATT TCT TTA TAC TAA GTT TTA CTG TGC GGT ATT TCA CAC CG-3′). The PCR product was transformed into the diploid strain RLY345. Correct integration was confirmed by using PCR and restriction digest analysis (data not shown).
For ARC18 deletion, a PCR fragment carrying the HIS3 marker from YDpH (18) was amplified with primers DWp21 (5′-TTG GTA TTA TCA GTC TCT CCA CTC CCA GTA TAT TAA TAA TCCC GGGGATCCGGTG ATT G-3′) and DWp22 (5′-TTC TAG TTT TTA TTA TTT TCC TTG CAC CGT ATA CCT TTA CTG CAG GTC GAC GGA TCC GG-3′). The PCR product was transformed into RLY138. Correct integration was confirmed by PCR and restriction digest analysis (data not shown).
For ARC19 deletion, a PCR fragment carrying the TRP1 marker from pRS304 (19) was amplified with primers ECp10 (5′-TT GAC AAA AGA TAA AAG AG GAA GCA ACT GAT AGT AGG AA AGA TTG TAC TGA GAG TGC AC-3′) and ECp11 (5′-GC CCT GAC GGA GAA AGA TTC CCT TTC CTA TAT TAG GAA TCT GTG CGG TAT TTC ACA CCG-3′). The PCR product was transformed into RLY344. Correct integration was confirmed by using PCR and restriction digest analysis (data not shown).
For ARC35 deletion, a PCR fragment carrying the HIS3 gene from pRS303 (19) by using primers ECp14 (5′-AAG ACT AGC AAT CTT CTT GCC TCA AAA CGG TAA TCC GCA AGA TTG TAC TGA GAG TGC AC-3′) and ECp15 (5′-AAC CCT TTT TAC GGA TTC TTA CGT ACT TAT TTA ATC TTT CTG TGC GGT ATT TCA CAC CG-3′) and transforming it into RLY138. Correct gene replacement was confirmed by using PCR and restriction digest analysis (data not shown).
Phalloidin Staining and Immnunofluorescence.
Phalloidin staining and immunofluorescence was carried out as described (20). FITC-conjugated donkey anti-rabbit and donkey anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch.
Analysis of Cytokinesis Defects.
Cells were grown to logarithmic phase in yeast extract/peptone/dextrose (YPD) media, fixed with 5% formaldehyde for 1 hour, and observed under a Labophot-2 microscope (Nikon) with a Plan Apo 100× DICH objective. Zymolyase treatment was carried out as described (21).
Generation of Polyclonal Arp3p and Arc40p Antibodies.
The C-terminal 115 aa of ARP3 were cloned into vector pGEX4T-1 (Amersham Pharmacia) and expressed as insoluble glutathione _S_-transferase fusion polypeptide in Escherichia coli. Polyacrylamide gel slices containing this polypeptide were used to raise antisera in rabbit (Cocalico Biologicals, Reamstown, PA). To generate an affinity matrix for purification, the antigen was solubilized in 8 M urea and conjugated to CNBr-activated Sepharose 4B (Pharmacia). Antibody affinity purification was performed by using standard techniques (22).
The C-terminal 195 aa of Arc40p were cloned into pGEX4T-3 (Amersham Pharmacia) and expressed as soluble glutathione _S_-transferase fusion polypeptide in E. coli. This polypeptide was purified on glutathione-agarose beads (Sigma) and used to raise antisera in rabbit (Cocalico Biologicals). The antigen was coupled to Bio-Rad Affi-Gel 10, and antibody affinity purification was performed by using standard techniques (22).
Gel Filtration Chromatography.
One-liter cultures of each strain were grown in YPD media to OD600 = 10–15 and frozen in liquid N2 to yield 5–10 g of cells. One gram of cell pellets was lysed by liquid N2 grinding method (20), resuspended in 0.7 ml of UBA buffer (50 mM Hepes, pH 7.5/100 mM KCl/3 mM MgCl2/0.2 mM ATP) + protease inhibitors mixture (17), centrifuged 45 min at 90,000 rpm in an RP120AT rotor (Sorvall) to yield 15 mg/ml extracts. Extracts (125 μl) were run on a Bio-Rad SE 1000/17 gel-filtration column at a flow rate of 0.5 ml/min. Alternate 0.5-ml fractions were collected, trichloroacetic acid-precipitated, run on SDS/PAGE gels, and blotted with respective antibodies. Catalase (232 kDa) and yeast actin (42 kDa) were used as standards.
Purification of Arp3p-Associated Complex from Wild-Type and Δ_arc15_ Strains.
Extracts from wild-type (RLY188) and Δarc15 (RLY763) strains bearing (myc)5(his)6-tagged Arp3p as their sole copy of Arp3p were prepared and fractionated on Q Sepharose column as described (17). Arp3p eluted between 170 and 240 mM KCl and was precipitated with 40% ammonium sulfate. The precipitate was resuspended in 50 mM Hepes (pH 7.5), 100 mM KCl, 3 mM MgCl2, and 20 mM immidazole and was bound to 1 ml of Ni-NTA aggarose column (Qiagen, Chatsworth, CA). After the column was washed with the same buffer containing 0.5 M KCl, Arp3p-associated complex was eluted in batch with 200 mM imidazole and dialyzed into UBA. In each case, 50 g of cells yielded 0.25–0.5 mg of final complex.
Immunoprecipitation Analysis.
To construct a stain that expresses hemagglutinin (HA)-tagged Arc40p under the GAL1 promoter, the ARC40 ORF was cloned between the _Eco_RI and _Xho_I sites of pRL62 (21)to generate pDW59, which was then linearized with _Eco_RV and integrated into the URA3 locus of RLY722 to generate RLY723. RLY723 cells were induced with 2% galactose for 6 hr and frozen in liquid N2. A 15 mg/ml liquid N2 grinding extract was prepared from these cells and incubated with protein A-Sepharose (Amersham Pharmacia) beads preloaded with mouse anti-HA (Babco, Richmond, CA) or anti-myc (23) antibody (as a control) for 1 hr. The beads were washed and boiled in sample buffer before SDS/PAGE and immunoblot analysis.
Arc40p interaction with (myc)5(his)6-tagged Arp3p in wild-type and Δarc15 cells was tested by preparing 15 mg/ml liquid N2 grinding extracts from RLY188 and RLY763 strains, and incubating them for 1 hr with protein A-Sepharose beads preloaded with anti-myc (23) or anti-HA (control) antibodies. The beads were washed and boiled in sample buffer before SDS/PAGE and immunoblot analysis.
RESULTS
Subunit Analysis of the Arp2/3 Complex by Gene Disruption.
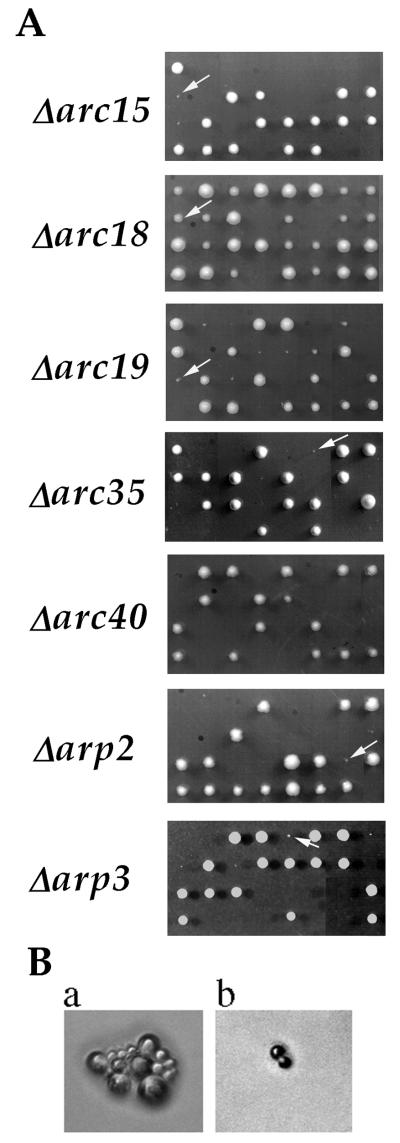
We have used a gene-disruption approach to characterize the contribution of each subunit to Arp2/3 complex function. Disruption of the genes encoding each subunit, including ARC40, was carried out in a diploid yeast strain of the S288c background. After confirmation of gene disruption, the heterozygous diploids were sporulated and subjected to tetrad analysis. As shown in Fig. 1A, gene disruption of each of the subunits impairs cell growth and viability by varying degrees. The frequency of viability for spores lacking ARP2, ARP3, ARC40, ARC35, ARC19, ARC18, or ARC15 was 11%, 16%, 0%, 2.5%, 52%, 88%, or 11%, respectively (>20 tetrads analyzed for each mutant). All mutant colonies that were visible after 5 days of growth at room temperature were viable when restreaked onto YPD plates or when grown in YPD liquid culture. Room temperature YPD liquid cultures of wild-type and Δ_arc18_ cells grew with a doubling time of 2–3 hr (as measured by OD600), and all other subunit-null strains had doubling times of 7–9 hr. Although 52% of Δ_arc19_ spores were viable, they still manifested strong growth defects, whereas all of the viable Δ_arc18_ spores formed colonies with sizes more similar to those of the wild-type (Fig. 1A). In contrast to the null mutant of each of the original six subunits, which either grew up to visible colonies or died as microcolonies, all Δarc40 spores died as a single cell with a large bud (>50 cells analyzed) (Fig. 1B). These results indicate that although all Arp2/3 complex subunits are conserved across species, they do not all contribute equally to complex function. Specifically, Arc18p seems to be important for a more subtle aspect of complex function than the other subunits, and ARC40 is genetically distinct from the original six subunits.
Figure 1.
Growth phenotype of gene-disruption mutants of Arp2/3 complex subunits. (A) Tetrads from heterozygous diploid strains Δarc15 (RLY514), Δarc18 (RLY515), Δarc19 (RLY344), Δarc35 (RLY343), Δarc40 (RLY722), Δarp2 (RLY184), and Δarp3(RLY180) after 5 days of growth on YPD at room temperature. The two large colonies in each tetrad always corresponded to wild-type spores, and the two small or missing colonies corresponded to disruption spores. Arrows point to viable gene-disruption colonies. (B) Terminal colony morphologies of a representative Δarc35 spore (a) and a representative Δarc40 spore (b).
Previously, it was reported that gene disruption of ARP2 and ARP3 is lethal in budding yeast (17, 24, 25). This discrepancy can be attributed to a difference in strain background for ARP2, because the previously published ARP2 gene disruption was in a YB18-derived background and ours is in S288c. This is not the case for ARP3, however, because we had previously found that ARP3 gene disruption was lethal even in the S288c background (17). This difference can be attributed to media conditions, because previous tetrad analysis was carried out on slightly caramelized plates, and we have since noticed that caramelization results in lethality of all Δarp3 spores.
Effects of Subunit Gene Disruption on Actin Organization.
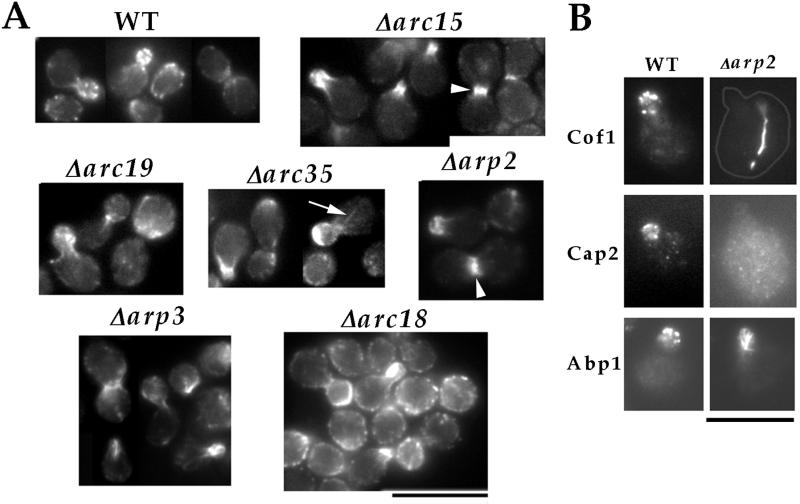
We examined the effects of the null mutation in each nonessential Arp2/3 complex subunit on the organization of actin by rhodamine–phalloidin staining. Even in the null strains with the most severe growth defects, such as Δarp2, Δarp3, and Δarc35, F actin polarized to the site of cell surface growth—the growing buds (Fig. 2A). However, the most prominent actin structures that formed in these buds were no longer actin patches but large filamentous aggregates, similar to those previously observed in _arp3_-2 mutant cells at the nonpermissive temperature (17). We also examined the localization of actin-binding proteins that normally associate with actin patches but not with actin cables (Fig. 2B). In Δarp2 cells, Cof1p (26) did not exist in patch-like structures but formed bars in 70% of the mother cells. Cap2p (27), meanwhile, did not appear to be localized in Δarp2 cells. Abp1p, which normally associates with both actin patches and cables (28), also localized to structures resembling the F actin aggregates in the buds of Δarp2 cells. Therefore, the biochemical composition of the aberrant actin structures that formed in Δarp2 cells is different from that of actin patches. This result, together with the fact that actin cables (arrow in Fig. 2A) and the cytokinetic actin ring (arrowheads in Fig. 2A) can form in the mutants, suggest that the Arp2/3 complex is required specifically for the formation of actin patches. Δarc18 mutant cells, the most healthy among the subunit-null mutants, retained a significant number of actin patches (Fig. 2A). However, these patches are distributed abundantly in the mother, indicating a loss of polarity. This suggests that the Arp2/3 complex may also play a role in maintaining the polarized distribution of actin patches.
Figure 2.
Characterization of actin defects in cells lacking each core Arp2/3 complex subunit. (A) Phalloidin staining of cells lacking each Arp2/3 complex subunit: wild-type (RLY1), Δarc15 (RLY570), Δarc18 (RLY571), Δarc19 (RLY572), Δarc35 (RLY573), Δarp2 (RLY574), Δarp3 (RLY739). Arrow points to actin cables and arrowheads to cytokinetic actin rings. (B) Localization of Cof1p, Cap2p, or Abp1p in wild type (RLY1) or Δarp2 (RLY574) cells. An outline has been drawn to show the profile of the Δarp2 cell stained with anti-Cof1p. (Bar = 10 μm.)
Because cytokinetic actin rings were more prominently visible in Arp2/3 subunit-null strains than in wild-type cells, it was possible that these mutations cause cytokinesis defects. We examined cytokinesis efficiency by quantifying the percentage of cells in chains of three or more buds. No such chains were observed in wild-type or Δarc18 mutant cells, but Δarc15, -19, and -35 and Δarp2 and -3 cell populations contained 6%, 5%, 9%, 4%, and 1% cells that formed chains, respectively. To distinguish whether these chains were formed because of cytokinesis or cell-separation defects, cells were fixed with formaldehyde, and the percentage of cells in chains was quantified after zymolyase treatment (21). The percentage of cells in chains in Δarc15, -19, and -35 and Δarp2 and -3 cells dropped to 1%, 3%, 0%, 1%, and 0%, respectively, suggesting that the defect was in cell separation rather than cytokinesis. This result suggests that Arp2/3 complex function is not required for cell division in budding yeast.
Roles of Individual Subunits in Maintaining the Structural Integrity of the Arp2/3 Complex.
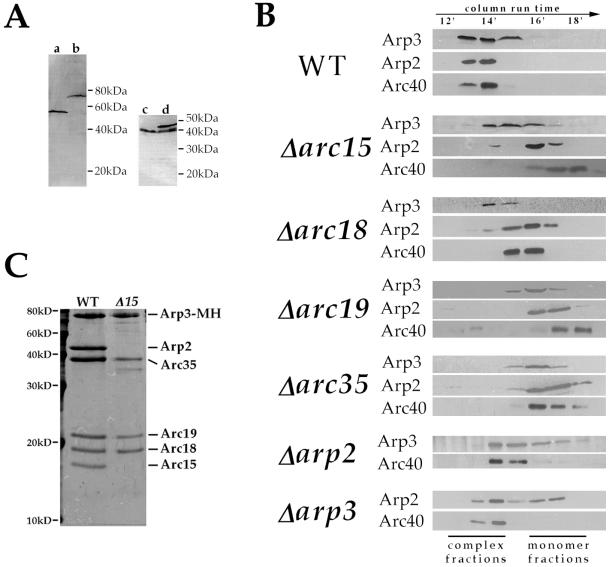
Potential roles of individual Arp2/3 complex subunits include (i) maintaining the structural integrity of the complex; (ii) directly participating in stimulation of actin polymerization; (iii) mediating the localization or activation of the complex. As the first step toward understanding the role of each subunit, we studied the requirement of each subunit in maintaining the structural integrity of the complex by gel filtration chromatography analysis of extracts prepared from each viable null strain. For this study, rabbit polyclonal antibodies were raised against the C termini of Arp3p and Arc40p. The affinity-purified anti-Arp3p antibody recognizes a single band corresponding to Arp3p in a total yeast extract, and affinity purified anti-Arc40p antibody recognized the Arc40p (Fig. 3A). In wild-type extracts, the majority of Arp2p and Arp3p cofractionated as a 230-kDa complex, suggesting that most of these proteins exist in the Arp2/3 complex rather than as monomers (Fig. 3B). Arc40p also cofractionated with Arp2p and Arp3p, providing the first indication that Arc40p may in fact be a component of the yeast Arp2/3 complex. In Δarc19 and Δarc35 extracts, Arp2p, Arp3p, and Arc40p migrated as monomers, indicating that Arc19p and Arc35p play a major role in maintaining complex integrity. This result also further supports that Arc40p is a component of the budding yeast Arp2/3 complex. Arp3p and Arc40p ran in the complex fractions in Δarp2 extract, and similarly, Arc40p and a significant portion of Arp2p remained associated with the complex in Δarp3 extract, suggesting that Arp2p and Arp3p are not required to maintain the integrity of the complex. In Δarc15 extracts, Arc40p and the majority of Arp2p ran as monomers, but Arp3p still appeared in a partial complex. Purification of the Arp3p-containing complex from Δarc15 cells further confirmed that this partial complex contains polypeptides corresponding to all of the original subunits except Arp2p and Arc15p (Fig. 3C). This result indicates that Arc15p is required for the interactions of Arp2p and Arc40p with the rest of the complex. Because Arp2p and Arc40p do not depend on each other to associate with the complex (Fig. 3B), it is possible that Arc15p interacts directly with both of these subunits. Similar to Δarc15, the Δarc18 mutation caused a significant fraction of Arp2p, but not Arp3p, to fall into monomer fractions, indicating that Arc18 is required for the association of Arp2p, but not Arp3p, with the complex. In Δarc18 cells, however, Arc40p migrates at a size intermediate between those of the complex and Arc40p monomer.
Figure 3.
Roles of individual subunits in maintaining complex integrity. (A) Characterization of anti-Arp3p and anti-Arc40p antibodies. a and b are immunoblots of extracts prepared from a wild-type strain (RLY1) and a strain expressing (myc)5-(his)6-tagged Arp3p as its sole copy of Arp3p (RLY188), respectively, by using affinity-purified anti-Arp3p antibody. c and d are immunoblots of extracts prepared from a wild-type strain (RLY1) and a strain expressing both wild-type and green fluorescent protein-tagged Arc40p (RLY821), respectively, by using affinity-purified anti-Arc40p antibody. (B) Extracts from wild-type and subunit-deletion strains were prepared as described in Materials and Methods and run on an S1000 gel filtration column at a flow rate of 0.5 ml/min. The fractions were trichloroacetic acid-precipitated and analyzed by using immunoblotting with antibodies against Arp2p (16), Arp3p, and Arc40p. (C) Subunit composition of Arp3p-associated complexes purified from the wild-type (RLY188) and Δarc15 (RLY763) strains as described in Materials and Methods. Complexes were run on SDS/15% polyacrylamide gels and visualized by using Coomassie blue staining.
Arc40p Coimmunoprcipitates with the Arp2/3 Complex.
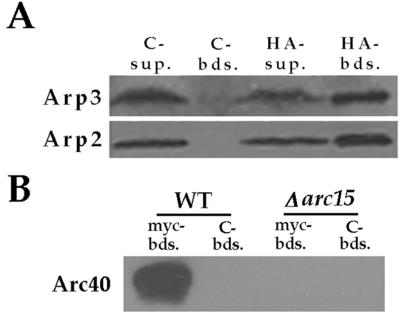
We performed coimmunoprecipitation analysis to further address whether Arc40p interacts with the yeast Arp2/3 complex. We found that an HA epitope-tagged Arc40p, expressed from the Gal1 promoter, coimmunoprecipitated with Arp2p and Arp3p, indicating that although this protein does not copurify with the core Arp2/3 complex subunits by our purification procedure, it can associate with the complex (Fig. 4A). Furthermore, at endogenous protein levels, myc-tagged Arp3p from wild-type but not Δarc15 cells immunoprecipitates Arc40p (Fig. 4B), indicating that the myc tag on Arp3p does not drastically disrupt interaction with Arc40p and confirming that Arc15p is required for binding of Arc40p to the Arp2/3 complex.
Figure 4.
Interaction of Arc40p with Arp2/3 complex. (A) HA-tagged Arc40p was immunoprecipitated with anti-HA or control (anti-myc) beads from an extract prepared from RLY723 strain. The associated proteins were analyzed by using immunoblotting with antibodies against Arp2p and Arp3p. Sup, supernatant; Bds, beads. (B) Myc-tagged Arp3p was immunoprecipitated from wild-type (RLY188) or Δarc15 (RLY763) extracts by using anit-myc or control (anti-HA) beads. The associated proteins were analyzed by using immunoblotting with the Arc40p antibody.
DISCUSSION
The Arp2/3 complex is composed of seven proteins, each of which defines a gene family conserved among eukaryotes. The biochemical activities of this complex include a regulated actin nucleation activity (2, 9–11), as well as pointed-end and side-binding interactions with actin that create T bar structures resembling the brush-like actin structures seen at the leading edge of fish keratocytes (6, 7). This complex is therefore likely to be critical for organizing actin into specific structures and to promote actin nucleation during polymerization-driven cell motility. In this work, we have carried out genetic characterization of each Arp2/3 complex subunit as a first step toward understanding the specific contributions of individual subunits to complex function.
It was surprising that none of the null mutations in the six core subunits was completely lethal, because it was reported previously that ARP2 and ARP3 were essential genes. This discrepancy can be attributed to a combination of differences in strain background and media conditions.
Because there is no detectable monomer pool of Arp2p, Arp3p, or Arc40p in wild-type cells and currently no evidence for Arp2/3 complex subunits functioning separately from the complex, it is reasonable to interpret the subunit knockout phenotypes in terms of Arp2/3 complex function. The severe growth defects seen in Δarc15, Δarc19, Δarc35, Δarp2, and Δarp3 strains are likely to be caused by a substantial loss of complex function. The less severe defects observed in Δarc18 cells indicate that the complex is able to retain partial function in the absence of this subunit. All viable null strains, with the exception of Δarc18, exhibit similar defects in actin organization, including a loss of actin patches and the appearance of aberrant bundles of F actin at sites of polarization, suggesting that the Arp2/3 complex is critical for the formation of cortical actin patches. The mutant cells are still able to accumulate F actin, albeit in aberrant structures, to the growing bud, suggesting that actin assembly activities independent of the Arp2/3 complex remain polarized toward the bud.
The Δarc18 mutation is less detrimental to cell growth and viability than other subunit knockouts. Δarc18 cells retain a significant number of actin patches, correlating with the higher spore viability, but the actin patches distribute to both the bud and the mother cells. Such a polarization defect has also been described for a temperature-sensitive arp2 mutation (16). Therefore, it is possible that although a partially functional Arp2/3 complex may be sufficient for the generation of actin patches, a fully functional Arp2/3 complex is required to polarize these actin structures to the buds.
The viable strains bearing gene disruption of each of the Arp2/3 complex subunits provide valuable starting reagents for studying the role of individual subunits in the assembly and biochemical activity of Arp2/3 complex. Gel-filtration chromatography analysis showed that disruption of ARC19 or ARC35 causes Arp2p, Arp3p, and Arc40p to shift to monomer size, indicating that Arc19p and Arc35p are likely to be the central scaffolding proteins of the complex. Chemical crosslinking data on the Acanthamoeba Arp2/3 complex support a scaffolding role for Arc35p, because this protein has been crosslinked to Arp2p, Arp3p, and p40 (29). Interestingly, Arp2p and Arp3p are not required for holding each other or Arc40p in the complex, indicating that these subunits are probably not central structural components and may be exposed at the surface, consistent with a role in mediating the interaction with actin. The Δarc15 mutation causes the majority of Arp2p and Arc40p, but not Arp3p, to fall to the monomer pool. Because a partial complex containing all subunits except Arp2p, Arc15p, and Arc40p can be purified from this strain and because Arc40p and Arp2p associate with the complex in the absence of each other, Arc15p seems to play a specific role in linking these two subunits to the complex. However, because neither Arp2p or Arc40p were directly crosslinked to Arc15p (29), it is unclear whether Arc15p physically links Arp2p and Arc40p to the complex or whether the Δarc15 mutation leads to a structural disruption that causes Arp2p and Arc40p to dissociate. Like the Δarc15 mutation, the Δarc18 mutation causes a majority of Arp2p, but not Arp3p, to dissociate from the complex. However, a difference between the two mutant strains is that Arc40p and perhaps a slightly greater amount of Arp2p remains complex-associated in the Δarc18 strain.
Although our data do not rule out the possibility that individual subunits have functions outside of the complex, the differences in Arp2/3 complex subunit-null phenotypes reported here can be interpreted in the context of our complex integrity analysis. Null mutations of ARC15, -19, and -35 and ARP2 and -3 subunits of the Arp2/3 complex lead to similarly severe phenotypes in respect to viability loss, poor colony growth, and loss of actin patches. The absence of Arp2p and/or Arp3p, the two subunits presumed to directly contact actin during nucleation, leads to loss of complex function comparable to Δarc19 and Δarc35 mutations, which make the complex fall apart. Although a partial complex lacking Arp2p and Arc40p is maintained in the Δarc15 extract, this mutant strain grows just as poorly as Δarc19 and Δarc35 strains. This result is consistent with a critical role for Arp2p in the complex. The Δarc18 mutation also affects the association of Arp2p and Arc40p with the complex. However, the Δarc18 phenotype is much less severe than that of Δarc15. This difference may be explained if a greater residual population of intact complex is present in vivo in Δarc18 cells than in Δarc15 cells, and/or if Arc15p is also involved in the activation or localization of the complex in addition to its structural role. We have in fact found that the Δarc15, but not the Δarc18, mutation disrupts the interaction of the Arp2/3 complex with Bee1p, the yeast WASP-like protein that activates the actin nucleation activity of the Arp2/3 complex in vitro (11).
Both the gel-filtration data and coimmunoprecipitation experiments strongly suggest that Arc40p is a component of the Arp2/3 complex. Its absence in the purified complex may be explained by a looser association with the complex, possibly because of the myc-His tag on Arp3p in the strain used for purification. The lack of Arc40p in the purified complex, however, suggests that it does not have a role in holding the rest of the complex together, and yet Arc40p appears to be most important for cell viability. Because we do not detect a monomer pool of Arc40p in wild-type cells, it seems unlikely that this subunit functions separately from the complex, suggesting that Arc40p mediates a unique and critical aspect of complex function.
Table 1.
Yeast strains
| Strain | Genotype | Ref. |
|---|---|---|
| RLY1 | MATa ura3-52 his3-Δ200 leu2-3 lys2-801 | 20 |
| RLY138 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3/leu2-3 lys2-801/lys2-801 | 20 |
| RLY141 | MATa/MATα URA3/ura3-52 his3-Δ200/HIS3 trp1-1/trp1-1 leu2-3/leu2-3 lys2-801/LYS2 | R.L., unpublished data |
| RLY175 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 lys2-801/lys2-801 | R.L., unpublished data |
| RLY180 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3/leu2-3 lys2-801/lys2-801 Δarp3∷HIS/ARP3 | 17 |
| RLY184 | MATa/MATα URA3/ura3-52 HIS3/his3-Δ200 leu2-3/leu2-3 trp1-1/trp1-1 lys2-801/lys2-801 Δarp2∷TRP1/ARP2 | This study |
| RLY186 | MATa ura3-52 his3-Δ200 leu2-3 trp1-1 Δarp2∷TRP1 pDW6 | This study |
| RLY188 | MATa ura3-52 his3-Δ200 leu2-3 lys2-801 Δarp3∷HIS3 pDW20 | 17 |
| RLY343 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3/leu2-3 lys2-801/lys2-801 Δarc35∷HIS3/ARC35 | This study |
| RLY344 | MATa/MATα URA3/ura3-52 his3-Δ200/HIS3 leu2-3/leu2-3 trp1-1/trp1-1 LYS2/lys2-801 Δarc19∷TRP1/ARC19 | This study |
| RLY345 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 leu2-3/leu2-3 Lys2/lys2-801 | |
| RLY514 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3/leu2-3 trp1-1/trp1-1 LYS2/lys2-801 Δarc15∷TRP1/ARC15 | This study |
| RLY515 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3/leu2-3 lys2-801/lys2-801 Δarc18∷HIS3/ARC18 | This study |
| RLY570 | MATa ura3-52 his3-Δ200 trp1-1 leu2-3 lys2-801 Δarc15∷TRP1 | This study |
| RLY571 | MATa ura3-52 his3-Δ200 leu2-3 lys2-801 Δarc18∷HIS3 | This study |
| RLY572 | MATa ura3-52 his3-Δ200 leu2-3 trp1-1 Δarc19∷TRP1 | This study |
| RLY573 | MATa ura3-52 leu2-3 lys2-3 his3-Δ200 Δarc35∷HIS3 | This study |
| RLY574 | MATα leu2-3 trp1-1 his3-Δ200 Δarp2∷TRP1 | This study |
| RLY722 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 lys2-801/lys2-801 Δarc40∷TRP1/ARC40 | This study |
| RLY723 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 lys2-801/lys2-801 Δarc40∷TRP1/ARC40 Gal-HAP40∷URA3 (pDW59) | This study |
| RLY739 | MATα ura3-52 his3-Δ200 leu2-3 lys2-801 Δarp3∷HIS3 | This study |
| RLY763 | MATa ura3-52 leu2-3 lys2-801 his3-Δ200 ARP3-5myc6his∷URA3 Δarc15∷TRP1 | This study |
| RLY722 | MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 lys2-801/lys2-801 Δarc40∷TRP1/ARC40 GFP-ARC40∷HIS3 (pDW72) | This study |
Acknowledgments
We thank Barbara Winsor, David Drubin, and John Cooper for providing the antibodies against Abp1p, Arp2p, Cof1p, and Cap2p. We thank Scott Schuyler, Katie Shannon, Terry Lechler, and Mimi Shirazu for critical reading of the manuscript and members of the Li laboratory for stimulating discussions and encouragement. This work was supported by Grant GM57063 from the National Institutes of Health to R.L.
ABBREVIATION
HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Welch M D, Iwamatsu A, Mitchison T J. Nature (London) 1997;385:265–268. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 2.Welch M D, Rosenblatt J, Skoble J, Portnoy D A, Mitchison T J. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 3.Machesky L M, Atkinson S J, Ampe C, Vandekerckhov J, Pollard T D. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelleher J F, Atkinson S J, Pollard T D. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch M D, DePace A H, Verma S, Iwamatsu A, Mitchison T J. J Cell Biology. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullins R D, Heuser J A, Pollard T D. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svitkina T M, Verkhovsky A B, McQuade K M, Borisy G G. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Machesky L M, Insall R H. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 9.Machesky L, Mullins R, Higgs H, Kaiser D, Blanchoin L, May R, Hall M, Pollard T. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner M W. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 11.Winter D, Lechler T, Li R. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- 12.Cooper J A, Buhle E L J, Walker S B, Tsong T Y, Pollard T D. Biochemistry. 1983;22:2193–2202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- 13.Tobacman L S, Korn E D. J Biol Chem. 1983;258:3207–3214. [PubMed] [Google Scholar]
- 14.McCollum D, Feokistova A, Morphew M, Balasubramanian M, Gould K L. EMBO J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- 15.Balasubramanian M K, Feoktistova A, McCollum D, Gould K L. EMBO J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- 16.Moreau V, Madania A, Martin R P, Winsor B. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter D, Podtelejnikov A V, Mann M, Li R. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- 18.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 20.Li R. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lippincott J, Li R. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 313–315. [Google Scholar]
- 23.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwob E, Martin R P. Nature (London) 1992;355:179–182. doi: 10.1038/355179a0. [DOI] [PubMed] [Google Scholar]
- 25.Huang M, Souciet J, Chua J, Galibert F. Yeast. 1996;12:839–848. doi: 10.1002/(SICI)1097-0061(199607)12:9%3C839::AID-YEA982%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 26.Moon A L, Janmey P A, Louie K A, Drubin D G. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amatruda J F, Cooper J A. J Cell Biol. 1992;117:1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drubin D G, Miller K G, Botstein D. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullins R D, Stafford W F, Pollard T D. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]