Quality control of mitochondria: protection against neurodegeneration and ageing (original) (raw)
Abstract
Dysfunction of mitochondria has severe cellular consequences and is linked to ageing and neurodegeneration in human. Several surveillance strategies have evolved that limit mitochondrial damage and ensure cellular integrity. Intraorganellar proteases conduct protein quality control and exert regulatory functions, membrane fusion and fission allow mitochondrial content mixing within a cell, and the autophagic degradation of severely damaged mitochondria protects against apoptosis. Here, we will summarize the current knowledge on these surveillance strategies and their role in human disease.
Keywords: mitochondria, mitochondrial dynamics;, mitochondrial proteases, mitophagy, neurodegeneration
Mitochondria: essential and delicate organelles
After the symbiotic engulfment of aerobic α-proteobacteria by pre-eukaryotic cells more than 1.5 billion years ago, mitochondria evolved as specialized organelles with a plethora of cellular functions. They not only house the respiratory chain (RC) and provide cellular energy but are also the site of essential biosynthetic pathways. Mitochondria serve as calcium stores and are integrated in a number of signalling pathways, including cell death cascades, thus controlling cellular homoeostasis in multiple ways (McBride et al, 2006). Given the multitude of functions, it does not surprise that mitochondrial dysfunction has severe cellular consequences, and is linked to ageing and neurological disorders in human (Lin and Beal, 2006).
Partly as a consequence of their endosymbiotic origin, the maintenance of mitochondrial activities imposes considerable challenges on eukaryotic cells. While the majority is nuclearly encoded, a few subunits of the RC are encoded by the mitochondrial genome, which requires a coordinated expression of nuclear and mitochondrial genomes. Moreover, mitochondria are highly dynamic organelles. Constant fusion and fission events, mediated by conserved cellular machineries, lead to the formation of a reticulated mitochondrial network (Cerveny et al, 2007; Detmer and Chan, 2007; Hoppins et al, 2007). This allows adapting mitochondrial activities to different physiological demands, but makes the assembly of functional mitochondria a difficult task to achieve.
Additional challenges for mitochondrial integrity come from reactive oxygen species (ROS), which induces protein modifications, lipid peroxidation and DNA damage and are an inevitable by-product of oxidative phosphorylation. Dysfunctional mitochondria bear the risk of futile ATP hydrolysis and increased oxidative stress. Even more, extensive mitochondrial damage may lead to the dissipation of the membrane potential across the inner membrane and induce cell death by the release of pro-apoptotic proteins (Kroemer et al, 2007). It is therefore crucial to tightly control oxidative phosphorylation and monitor the functionality of the RC to maintain mtDNA integrity and limit mitochondrial damage.
Cellular defence against mitochondrial damage
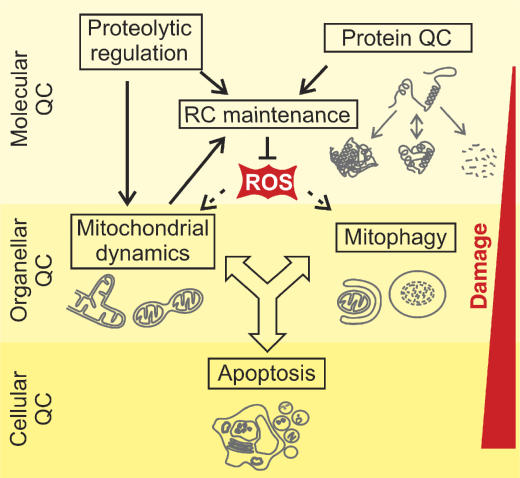
Cells have evolved elaborate systems to cope up with the diverse challenges imposed on mitochondrial integrity (Figure 1). The first line of defence is provided by a highly conserved, intraorganellar proteolytic system that conducts the surveillance of protein quality control (QC) within mitochondria (Koppen and Langer, 2007). Molecular chaperones and energy-dependent proteases monitor the folding and assembly of mitochondrial proteins and selectively remove excess and damaged proteins from the organelle. A second line of defence is provided at the organellar level by the dynamic nature of the mitochondrial population of a cell. The functionality of damaged mitochondria can be restored by fusion with neighbouring, intact mitochondria, assigning an important role for QC to components regulating mitochondrial dynamics (Detmer and Chan, 2007). Severe damage of mitochondria, however, impairs fusion and results in fragmentation of mitochondria, which are then selectively removed by an autophagic process, termed mitophagy (Kim et al, 2007). Mitophagy prevents the release of pro-apoptotic proteins from damaged mitochondria. Consistent with a cytoprotective function of autophagy, apoptosis is suppressed upon induction but induced upon inhibition of autophagy (Maiuri et al, 2007).
Figure 1.
Quality control (QC) surveillance of mitochondria. Intraorganellar proteases exert QC and regulatory functions to maintain respiratory chain (RC) activity. The functionality of damaged mitochondria can be restored by fusion and content mixing within the mitochondrial network. Severely damaged mitochondria fragment and are removed by mitophagy or induce apoptosis by the release of pro-apoptotic proteins.
Selective degradation of mitochondrial proteins: the first line of defence
The mitochondrial QC system recognizes and removes non-assembled and misfolded proteins selectively. Key components are ATP-dependent proteases, which are derived from bacterial proteases and highly conserved in eukaryotes (Koppen and Langer, 2007). These proteases degrade damaged proteins to peptides, which are subsequently either exported from the organelle or degraded further to amino acids by various oligopeptidases (Figure 2). As many substrates are part of multimeric protein complexes within mitochondria, degradation of damaged proteins may involve substrate extraction from large assemblies and is therefore likely closely linked to assembly and disassembly processes. ATP-dependent proteases sense the folding state of substrates by exerting chaperone-like properties and trigger the proteolysis of non-native proteins. A central role in this process is exerted by conserved ATPase modules, which are characteristic of the AAA+ family of ATPases and present in all ATP-dependent proteases.
Figure 2.
Quality control (QC) of mitochondrial proteins. ATP-dependent proteases present in various subcompartments of mitochondria recognize non-native polypeptides and trigger their proteolysis to peptides that are further degraded by oligopeptidases. At the same time, energy-dependent proteases can act as processing enzymes ensuring assembly and integrity of RC. Recent evidence links the UPS to mitochondrial QC and the regulation of mitochondrial dynamics. OM, outer membrane; IMS, intermembrane space; IM, inner membrane; M, matrix.
QC of matrix proteins
Up to two ATP-dependent proteases, the Lon and the ClpXP proteases, are present in the mitochondrial matrix in various organisms. While next to nothing is known about the physiological function of ClpXP in mitochondria, Lon protease (also termed PIM1 protease in yeast) has been demonstrated to degrade various misfolded and non-assembled polypeptides in the matrix. These include thermally denatured proteins as well as oxidatively damaged (carbonylated) proteins, such as the matrix-localized iron–sulphur protein aconitase (Bota and Davies, 2002; Major et al, 2006). In most cases, impaired folding appears to trigger protein degradation by Lon. Molecular chaperone proteins of the Hsp70 and Hsp100 family stabilize misfolded proteins against aggregation or mediate the dissolution of protein aggregates and thereby ensure proteolysis (Wagner et al, 1994; Bateman et al, 2002; Röttgers et al, 2002).
The limited capacity of the QC system in the matrix is illustrated by the accumulation of oxidatively modified and aggregated aconitase in aged cells or in age-related human disorders and may contribute to the age-related mitochondrial decline (Bota et al, 2002). Consistently, Lon-deficient cells accumulate protein aggregates in mitochondria and show deterioration of mitochondrial functions both in human and yeast (Suzuki et al, 1994; Bota et al, 2005). It is presently unclear whether these cellular defects are simply caused by the accumulation of protein aggregates within mitochondria. Alternatively, protein aggregation could be cytoprotective and may represent dumping of non-native, otherwise deleterious polypeptides. Cellular defects in the absence of Lon, such as the loss of the mitochondrial genome, may instead arise from the impaired proteolysis of specific regulatory proteins. Such a regulatory role could explain the presence of Lon protease in mitochondrial nucleoids and its affinity for DNA (Liu et al, 2004; Cheng et al, 2005).
QC by AAA proteases in the inner membrane
The mitochondrial inner membrane harbours the RC making inner membrane proteins a prime target of mitochondrial ROS. At least two membrane-integrated ATP-dependent proteolytic complexes, termed AAA proteases, conduct the QC surveillance in the inner membrane and the selective degradation of non-assembled and damaged proteins (Koppen and Langer, 2007). Whereas the _i_-AAA protease is active on the intermembrane side, _m_-AAA proteases expose their catalytic domains to the matrix side of the inner membrane. _m_- and _i_-AAA proteases exert overlapping substrate specificities in yeast and it is apparently solely the membrane topology of substrates that determines which protease is involved in degradation. AAA proteases recognize misfolded, solvent-exposed domains of substrates, loops of multispanning membrane proteins, or short N- or C-terminal tails protruding from the lipid bilayer (Leonhard et al, 2000). The presence of at least two AAA proteases inversely inserted into the inner membrane therefore ensures degradation of membrane proteins irrespective of their topology. AAA proteases mediate the ATP-dependent dislocation of substrate proteins from the membrane allowing their degradation in a hydrophilic environment (Leonhard et al, 2000). This is reminiscent of the turnover of ER membrane proteins by 26S proteasomes during ER-associated protein degradation (ERAD) (Meusser et al, 2005). Therefore, it appears that AAA proteases represent a distinct proteolytic system in a membrane, which is not accessible for the ubiquitin–proteasome system (UPS) from the cytosol.
Assembly and maintenance of the RC by AAA proteases
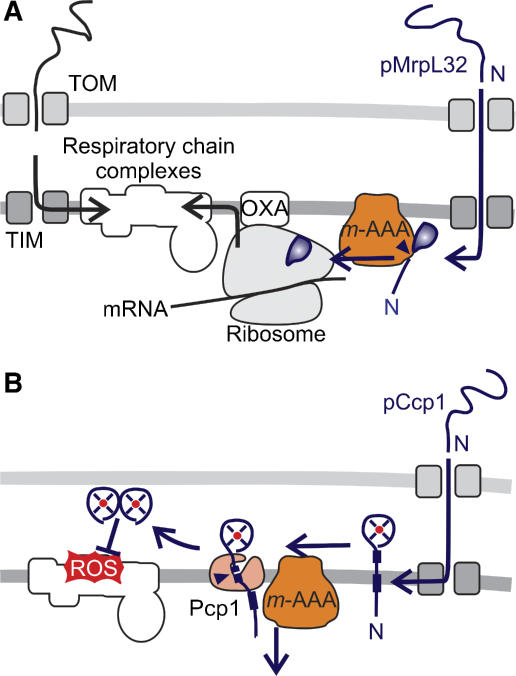
In addition to its function in QC, _m_-AAA proteases control directly the biogenesis of the RC. A nuclearly encoded subunit of mitochondrial ribosomes, MrpL32, is matured by yeast and murine _m_-AAA proteases, which act as a processing and not as a QC enzyme in this context (Figure 3A) (Nolden et al, 2005). Since MrpL32 processing is a prerequisite for its assembly into ribosomes and activation of mitochondrial translation, the _m_-AAA protease directly regulates the synthesis of RC subunits within mitochondria. Complementation experiments provide direct evidence that the dysfunction of mitochondria in the absence of _m_-AAA protease is mainly caused by an impaired processing of MrpL32 rather than by misfolded polypeptides (Nolden et al, 2005). It should be noted, however, that both regulatory and QC functions of the _m_-AAA protease are interdependent and may interfere with each other. For instance, QC substrates accumulating in aged cells or under stress conditions may compete with MrpL32 for binding to the _m_-AAA protease, resulting in an inhibition of translation.
Figure 3.
Regulation of RC assembly and maintenance by proteases. (A) The assembly of mitochondrial ribosomes and synthesis of mitochondrially encoded RC subunits require maturation of newly imported MrpL32 by the _m_-AAA protease. (B) Biogenesis of the ROS scavenger Ccp1 in yeast mitochondria depends on ATP-dependent membrane dislocation of the precursor protein by the _m_-AAA protease and maturation by the rhomboid protease Pcp1.
The _m_-AAA protease also exerts a protective function for assembled RC complexes, strikingly independent of its proteolytic activity. It mediates maturation of the ROS scavenger cytochrome c peroxidase (Ccp1) in the intermembrane space of yeast mitochondria and therefore limits ROS damage (Figure 3B). Ccp1 receives two-step processing by the _m_-AAA protease and the rhomboid protease Pcp1, an intramembrane-cleaving peptidase in the inner membrane (Esser et al, 2002). Rhomboid cleavage depends on the ATP-dependent membrane dislocation of Ccp1 by the _m_-AAA protease but not on its proteolytic activity (Tatsuta et al, 2007). A similar role has been proposed for the _i_-AAA protease assisting the import of mammalian polynucleotide phosphorylase (PNPase) into yeast mitochondria (Rainey et al, 2006). It therefore appears that the ATP-dependent membrane dislocation represents a novel function of AAA proteases of general relevance.
QC of mitochondrial proteins by the UPS
Increasing evidence suggests that the cytosolic UPS affects the QC of mitochondrial proteins in two ways (Figure 2): first, in the cytosol prior to import of nuclearly encoded proteins into mitochondria and, second, in the mitochondrial outer membrane. A proteomic survey of ubiquitinated proteins identified many proteins that finally reside in different mitochondrial subcompartments, suggesting that non-imported preproteins are degraded by the cytosolic UPS (Peng et al, 2003). Direct evidence for UPS-mediated degradation in the cytosol has been provided for apo-cytochrome c or the E2 component of 2-oxoglutarate dehydrogenase complex (Pearce and Sherman, 1997; Habelhah et al, 2004). Notably, although imported post-translationally, some preproteins are synthesized from messenger RNAs in close proximity to mitochondria and are found associated with the outer membrane (Zahedi et al, 2006). It is therefore conceivable that preproteins are ubiquitinated at the outer membrane and subject to proteolysis by the UPS. This is of interest as a RING-like ubiquitin ligase is present in the outer membrane of mammalian mitochondria that was demonstrated to affect mitochondrial dynamics (Nakamura et al, 2006; Yonashiro et al, 2006; Karbowski et al, 2007). However, it is presently unclear whether this ligase primarily exerts a regulatory or a QC control function. The latter would imply that mitochondrial outer membrane proteins are subject to proteolysis by 26S proteasomes in the cytosol and therefore point to similarities to ERAD.
Mitochondrial dynamics and QC
The dynamic nature of the mitochondrial network provides additional protection against mitochondrial damage. Ongoing cycles of fusion and fission of mitochondrial membranes are mediated by conserved protein machineries (Cerveny et al, 2007; Detmer and Chan, 2007; Hoppins et al, 2007). At least four dynamin-related GTPases mediate fission and fusion of mitochondrial membranes and determine the shape of the mitochondrial reticulum: mitofusins MFN1 and MFN2 in the outer and OPA1 in the inner membrane control mitochondrial membrane fusion, while DRP1 triggers fission events. Fusion, on the one hand, allows content mixing between intact and dysfunctional mitochondria. Replacement of damaged material, such as mutant mtDNA, contributes to the integrity and homogeneity of the mitochondrial population in a cell. In agreement with a protective function of mitochondrial fusion, a decline in respiratory activities has been observed in fusion-deficient murine fibroblasts lacking OPA1 or mitofusins (Chen et al, 2005, 2007). Fission, on the other hand, allows sequestration of irreversibly damaged, fusion-incompetent mitochondria and their subsequent elimination by autophagy (see below). Fusion and fission are highly regulated and again a central regulatory role of QC enzymes is emerging (Figure 4).
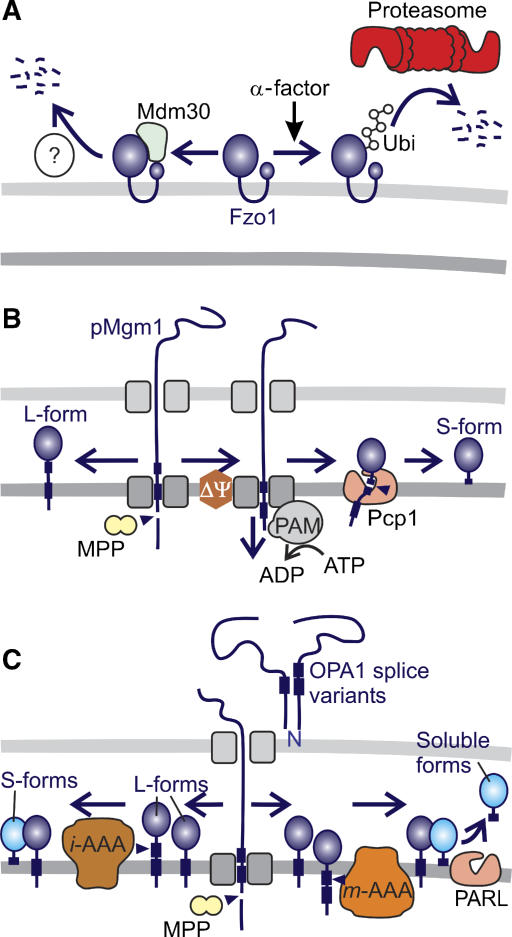
Figure 4.
Regulation of mitochondrial dynamics by proteolysis. (A) Degradation of the yeast mitofusin Fzo1 can occur along two independent pathways. Constitutive turnover is dependent on the F-box protein Mdm30. Fzo1 turnover induced by cell cycle arrest (in the presence of α-factor) is mediated by UPS but does not require Mdm30. (B) Alternative topogenesis of Mgm1. Proteolytic conversion of L-Mgm1 to S-Mgm1 by the rhomboid protease Pcp1 depends on the mitochondrial membrane potential (ΔΨ), matrix ATP, and the mitochondrial import motor (PAM). (C) Model for the processing of OPA1 in mammalian mitochondria. After processing of newly imported OPA1 by MPP, long and short isoforms of OPA1 are generated by constitutive (left pathway) and inducible (right pathway) cleavage at sites 2 and site 1, respectively. _m_- and _i_-AAA proteases and PARL have been linked to OPA1 processing, but the exact proteolytic pathways remained speculative.
Degradation of yeast mitofusin Fzo1
Fusion of yeast mitochondria depends on the constitutive turnover of the mitofusin Fzo1, an essential component of the fusion machinery in the outer membrane (Figure 4A). Proteolysis of Fzo1 was found to depend on the F-box protein Mdm30 (Neutzner and Youle, 2005; Escobar-Henriques et al, 2006). F-box proteins have been identified as components of SCF-E3 ubiquitin ligase complexes mediating proteasomal degradation of specific substrates (Petroski and Deshaies, 2005). Surprisingly, the Mdm30-dependent proteolysis of Fzo1 did not require other SCF complex components or 26S proteasomes, suggesting the existence of a yet unidentified proteolytic system for the turnover of outer membrane proteins (Escobar-Henriques et al, 2006). Notably, mating factor-induced cell cycle arrest induces rapid degradation of Fzo1 (Neutzner and Youle, 2005). However, under these conditions, proteolysis does not depend on Mdm30 but requires a functional UPS system. Thus, the level of yeast mitofusin Fzo1 in the mitochondrial outer membrane is controlled by two independent proteolytic pathways. It will be interesting to examine whether mammalian mitofusins are regulated in a similar manner.
Processing of the dynamin-like GTPase OPA1
Most compelling evidence for a regulatory role of mitochondrial peptidases exists for inner membrane fusion and cristae morphogenesis mediated by the dynamin-like GTPase OPA1 (Mgm1 in yeast) (Figure 4B and C). Fusion depends on proteolytic processing and the balanced formation of short and long isoform(s) (Herlan et al, 2003; Song et al, 2007). In yeast, the long isoform of Mgm1 (L-Mgm1) inserts into the mitochondrial inner membrane upon import and is cleaved by the mitochondrial-processing peptidase MPP (Herlan et al, 2003). A subset of the long isoforms undergoes a second cleavage event, which is mediated by the rhomboid protease Pcp1 in the inner membrane and results in the formation of the short Mgm1 isoform (S-Mgm1) (Herlan et al, 2003; McQuibban et al, 2003; Sesaki et al, 2003). The efficiency of Pcp1 processing was found to depend on the ATP-dependent protein import motor in the matrix space and thereby on the energy status of mitochondria (Herlan et al, 2004). Similarly, the balance between long and short isoforms of OPA1 is determined by matrix ATP also in mammalian mitochondria. However, in contrast to yeast mitochondria, low ATP levels trigger OPA1 cleavage (Duvezin-Caubet et al, 2006; Baricault et al, 2007). S-OPA1 accumulates and mitochondria fragment in cells harbouring a dysfunctional RC as well as in cells from patients suffering from mitochondrial myopathies (Duvezin-Caubet et al, 2006). These observations suggest that mitochondrial functionality and morphology are tightly coupled by the energy-sensitive processing of OPA1.
Eight splice variants of OPA1 are synthesized which are processed at two different sites, sites 1 and 2, resulting in the accumulation of at least five different OPA1 isoforms (Ishihara et al, 2006; Duvezin-Caubet et al, 2007; Olichon et al, 2007). OPA1 processing occurs constitutively at site 2 but cleavage can be induced at site 1 at low membrane potential, by ROS treatment or by apoptotic stimuli (Ishihara et al, 2006; Baricault et al, 2007; Griparic et al, 2007; Song et al, 2007). Currently, three peptidases have been linked to the processing of OPA1. Overexpression of S-OPA1 protects cells lacking the rhomboid protease PARL against apoptosis, demonstrating that they act along the same apoptotic pathway (Cipolat et al, 2006). However, _Parl_−/− cells do not show an impaired processing of OPA1 or mitochondrial morphology defects (Cipolat et al, 2006; Duvezin-Caubet et al, 2007). Rather, PARL appears to facilitate the release of S-OPA1 from the inner membrane (Cipolat et al, 2006). Moreover, PARL can substitute for the yeast rhomboid protease Pcp1 in processing Mgm1 but does not process OPA1 when co-expressed in yeast (Duvezin-Caubet et al, 2007). The _m_-AAA protease subunit paraplegin has also been proposed to mediate inducible cleavage of OPA1 at site 1 (Ishihara et al, 2006). But again, OPA1 processing is not impaired in paraplegin-deficient fibroblasts (Duvezin-Caubet et al, 2007). This might be explained by the partial redundancy of various _m_-AAA protease subunits and the formation of homo- and hetero-oligomeric isoenzymes (Koppen et al, 2007). Co-expression of OPA1 with _m_-AAA isoenzymes in yeast indeed revealed that OPA1 can be cleaved by mammalian _m_-AAA proteases in the heterologous yeast system (Duvezin-Caubet et al, 2007). Knockdown experiments identified the _i_-AAA protease Yme1L as being responsible for the constitutive cleavage of OPA1 at site 2 (Griparic et al, 2007; Song et al, 2007). The physiological consequence of this cleavage is still unclear as Yme1L affects mitochondrial morphology also in an OPA1-independent manner (Griparic et al, 2007). Therefore, it appears that various proteases regulate, directly or indirectly, OPA1 processing and adjust OPA1 activity and mitochondrial morphology to different physiological demands.
Elimination of damaged mitochondria by mitophagy
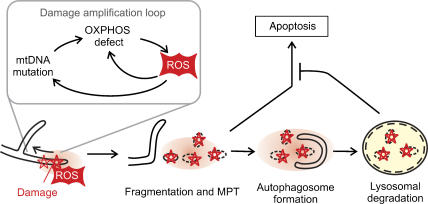
Mitochondrial dysfunction impairs OPA1 processing and triggers fragmentation sequestering damaged mitochondria from the intact mitochondrial network. Fragmented mitochondria can be removed by mitophagy before apoptosis is induced by the release of pro-apoptotic proteins from mitochondria (Figure 5) (Kim et al, 2007). Substantial insight into general mechanisms regulating autophagy came from studies in yeast which identified ∼30 proteins (Atg) involved in autophagic processes, many of them conserved in higher eukaroytes (Yorimitsu and Klionsky, 2005). Deletion of yeast ATG genes causes mitochondria-related phenotypes, such as growth defects on non-fermentable medium, suggesting a pivotal role of autophagy for QC of mitochondria (Zhang et al, 2007). Two mitochondrial proteins, termed Uth1 and Aup1, have been linked specifically to mitophagy and are not part of the general autophagic machinery: Uth1 in the mitochondrial outer membrane and the putative phosphatase Aup1 in the intermembrane space (Kissova et al, 2004; Tal et al, 2007).
Figure 5.
Quality control of mitochondria by mitophagy. ROS produced by damaged mitochondria induces mitochondrial fragmentation and mitochondrial permeability transition (MPT). Damaged mitochondria are engulfed by autophagosomes selectively and eliminated, preventing the release of pro-apoptotic proteins and apoptosis.
It is currently not understood how damaged mitochondria are specifically selected, but accumulating evidence suggests that mitochondrial dysfunction by itself triggers mitophagy (Kim et al, 2007). Mitochondria-derived ROS, at low concentrations, may act as signalling molecules and trigger mitophagy through redox regulation of Atg4, an essential cysteine protease in the autophagic pathway (Scherz-Shouval et al, 2007). The formation of autophagosomes depends on Atg4-mediated cleavage of Atg8 and its subsequent conjugation to phosphatidyl ethanolamine at autophagosomal membranes (Yorimitsu and Klionsky, 2005). Antioxidant treatment suppresses lipidation of Atg8 and autophagosome formation, suggesting that lipid conjugation is a direct consequence of ROS production (Scherz-Shouval et al, 2007).
Mitophagy was found to depend on mitochondrial fission. In primary neuronal cells, mitophagy can be induced by NO, which results in ROS production and mitochondrial fragmentation (Barsoum et al, 2006). Inhibition of mitochondrial fission or induction of mitochondrial fusion inhibits NO-induced mitophagy (Barsoum et al, 2006). Similarly, autophagic degradation of yeast mitochondria, induced by depletion of the inner membrane protein Mdm38, depends on fission mediated by the Drp1 homologue Dnm1 (Nowikovsky et al, 2007). After mitochondrial fission, progression of mitophagy appears to depend on the mitochondrial permeability transition (MPT) and can be inhibited by MPT inhibitors like cyclosporine A or overexpression of the anti-apoptotic protein Bcl2 (Xue et al, 2001; Rodriguez-Enriquez et al, 2006). MPT finally leads to the rupture of the outer membrane, the release of pro-apoptotic proteins from the intermembrane space and apoptosis, if mitochondria are not selectively removed by mitophagy. Thus, mitophagy and apoptosis appear to share common steps, although the physiological outcome is strikingly different (Maiuri et al, 2007).
Role of mitochondrial QC in disease and ageing
Numerous studies illustrate the relevance of disturbed mitochondrial activities for ageing and degenerative processes (Lin and Beal, 2006). An impaired energy supply or Ca2+ buffering, increased ROS production, or control of apoptosis by mitochondria may contribute to the progressive decline of long-lived, postmitotic cells. Mutations in mitochondrial proteins frequently result in neurological symptoms illustrating the susceptibility of neurons for mitochondrial dysfunctions. Given the protective functions of mitochondrial QC, it is not surprising that pathogenic mutations for an increasing number of neurodegenerative disorders are identified in key components of the intraorganellar QC system and the mitochondrial fusion machinery (Table I).
Table 1.
Neurodegenerative diseases linked to mitochondrial QC
| Associated disease | Gene | Function |
|---|---|---|
| Parkinson's disease (PD) | HTRA2 | Serine protease in mitochondrial intermembrane space |
| PINK1 | Serine/threonine kinase in mitochondrial intermembrane space | |
| PARKIN | E3 ubiquitin ligase | |
| Charcot–Marie–Tooth disease | ||
| Subtype 2A (CMT2A) | MFN2 | Dynamin-like GTPase in outer membrane mediating fusion |
| Subtype 4A (CMT4A) | GDAP1 | Outer membrane protein promoting fission |
| Autosomal dominant optic atrophy (DOA) | OPA1 | Dynamin-like GTPase required for inner membrane fusion and cristae morphology |
| Hereditary spastic paraplegia (HSP) | Paraplegin | Subunit of hetero-oligomeric _m_-AAA protease in inner membrane |
| HSP60 | Matrix-localized chaperonin |
Parkinson's disease
Parkinson's disease (PD), a common age-associated neurodegenerative disorder, is characterized by the preferential loss of dopamine-secreting neurons in the substantia nigra and the accumulation of intraneuronal inclusions (Lewy bodies). Although the aetiology of PD is heterogeneous and pathogenic mutations have been identified in various genes, mitochondrial dysfunction appears to have a prevalent role in the pathogenesis of the disease (Mandemakers et al, 2007). A role of protein QC within mitochondria is suggested by the finding of heterozygous missense mutations in HTRA2/OMI in sporadic cases of PD. HtrA2, homologous to bacterial Deg proteases, is localized in the mitochondrial intermembrane space and protects against mitochondrial stress. HtrA2 may act as a QC enzyme and degrade misfolded polypeptides in the mitochondrial intermembrane space or, in analogy to bacterial DegS, be part of an adaptive stress signalling cascade. HtrA2 is released from the intermembrane space of mitochondria during apoptosis (Suzuki et al, 2001). However, HtrA2-deficient mice show neurodegeneration and Parkinson-like phenotypes, but did not provide evidence for a pro-apoptotic function of HtrA2 (Jones et al, 2003; Martins et al, 2004).
Intriguingly, HtrA2 associates with PINK1, a serine/threonine kinase, which has been found to be mutated in PD patients (Plun-Favreau et al, 2007). Both PINK1 and HtrA2 appear to act along the same stress-protective pathway. PINK1 is required for phosphorylation of HtrA2, which increases its proteolytic activity in vitro (Plun-Favreau et al, 2007). While it remains to be clarified whether HtrA2 is a PINK1 substrate, PINK1-mediated phosphorylation has been demonstrated for TRAP1, a putative molecular chaperone with significant homology to the HSP90AA1 family (Pridgeon et al, 2007). PINK1-mediated phosphorylation of TRAP1 is induced by oxidative stress. Overexpression of PINK1, on the other hand, protects cells from apoptosis induced by oxidative stress, suggesting that PINK1 and TRAP1 are part of an anti-apoptotic signalling cascade (Pridgeon et al, 2007). Thus, although neither the physiological role of HtrA2 nor of TRAP1 has been defined, an intimate link among mitochondrial QC, stress signalling and PINK1-associated PD pathogenesis is emerging. Interestingly, Parkin has been genetically linked to PINK and functions downstream as an E3 ubiquitin ligase (Clark et al, 2006; Park et al, 2006; Exner et al, 2007). Therefore, although substrates are largely unknown, Parkin is functionally linked to mitochondria (Dodson and Guo, 2007).
Hereditary spastic paraplegia
Mutations in a subunit of the _m_-AAA protease, termed paraplegin, cause an autosomal recessive form of hereditary spastic paraplegia (HSP) (Casari et al, 1998). HSP is a genetically heterogeneous group of neurological disorders that is characterized by progressive and cell-specific axononal degeneration of cortical motor neurons starting from their distal extremities (Soderblom and Blackstone, 2006). The pathogenic mechanism is presently not understood but the retrograde mode of degeneration suggests that mitochondria at synaptic endings are affected initially. Paraplegin constitutes a subunit of a hetero-oligomeric _m_-AAA isoenzyme, which can substitute for QC and regulatory functions of the yeast _m_-AAA protease (Atorino et al, 2003; Koppen et al, 2007). Accordingly, mitochondrial dysfunction and axonal degeneration in the absence of paraplegin may result from the accumulation of non-degraded, misfolded inner membrane proteins or impaired regulatory steps during mitochondrial biogenesis, or both. It is of interest in this context that mutations in the chaperone HSP60, localized in the mitochondrial matrix, have been described to cause an autosomal dominant form of HSP (Hansen et al, 2002), suggesting that an impaired mitochondrial QC can indeed cause axonal degeneration.
Insights into the pathogenesis of HSP came from a paraplegin-deficient murine model that recapitulates main clinical features of HSP (Ferreirinha et al, 2004). While RC deficiencies were not apparent, enlarged and structurally abnormal mitochondria accumulated in synaptic terminals of motor neurons at early stages, correlating with the onset of motor impairment. These abnormalities become more prominent with age and involve also proximal regions. It is conceivable that the accumulation of aberrant mitochondria leads to clogging and subsequent swelling of axons, as fission and dynamic changes in mitochondrial morphology affect axonal transport (Rugarli and Langer, 2006). Moreover, an impaired axonal transport of enlarged mitochondria may also affect their autophagic degradation, which requires retrograde transport because of the paucity of lysosomes at synaptic endings (Hollenbeck, 1993).
While these findings could explain the accumulation of dysfunctional mitochondria at synaptic endings, they are difficult to reconcile with the proposed role of paraplegin for OPA1 processing. Impaired OPA1 cleavage in the absence of the paraplegin-containing _m_-AAA protease would impair fusion and cause fragmentation of mitochondria. This could facilitate their retrograde transport and autophagic degradation. Although functional mitochondria would be depleted at synapses, the presence of enlarged mitochondrial structures in the absence of paraplegin remains to be explained. Therefore, it appears that additional effects of paraplegin on mitochondrial fission or mitophagy must be envisioned.
Mitochondrial dynamics and neurodegeneration
The importance of mitochondrial dynamics for neuronal survival is illustrated directly by the identification of pathogenic mutations in components regulating mitochondrial dynamics. Mutations in OPA1 lead to dominant optic atrophy (Alexander et al, 2000; Delettre et al, 2000), the most commonly inherited optic neuropathy, characterized by the specific loss of retinal ganglion cells (Delettre et al, 2002). Moreover, Charcot–Marie–Tooth disease, a frequent peripheral neuropathy affecting both motor and sensory nerves (Züchner and Vance, 2005), is caused by mutations in MFN2 and GDAP1 (ganglioside-induced differentiation protein 1) localized in the mitochondrial outer membrane (Züchner et al, 2004; Niemann et al, 2005). While mutations of MFN2 inhibit fusion, inactivation of GDAP1 promotes fission of mitochondria.
A conditional mouse model allowing inactivation of MFN2 in the cerebellum of adult mice revealed that Purkinje cells are highly susceptible to mitochondrial fusion defects (Chen et al, 2007). In the absence of MFN2 and mitochondrial fusion, they accumulate fragmented mitochondria with an impaired respiratory activity, likely caused by the loss of mtDNA nucleoids. Interestingly, aberrant mitochondria are predominantly detectable in neuronal terminals of MFN2-deficient murine Purkinje cells (Chen et al, 2007). This is reminiscent of observations in paraplegin-deficient neurons and consistent with axonal transport deficiencies in the absence of MFN2 (Baloh et al, 2007).
The problem of tissue specificity
Although we are beginning to understand the selective vulnerability of neurons for mitochondrial damage, the striking tissue-specific consequences of pathogenic mutations in QC components including the mitochondrial fusion machinery remain difficult to explain. Multiple mechanisms can be envisioned that are not mutually exclusive. It is conceivable that certain cell types are more dependent on mitochondrial activities than others and therefore more susceptible to mitochondrial damage. This may include neurons with a high demand for mitochondrial transport, such as corticospinal motor neurons with long axons or Purkinje cells with highly branched dendrites. Others are inherently more exposed to stress and mitochondrial damage, such as dopamine-producing neurons affected in PD (Mandemakers et al, 2007). Moreover, functional redundancy and tissue-specific expression of different mitochondrial QC components could affect QC surveillance and the resistance of mitochondria against damage. For instance, the mitofusins MFN1 and MFN2 were detected in functionally redundant, homo- and hetero-oligomeric complexes (Chen et al, 2003). In murine Purkinje cells, MFN2 is more highly expressed than MFN1, providing an explanation for their selective vulnerability in CMT2A (Chen et al, 2007). Similarly, overlapping substrate specificity was demonstrated for homo- and hetero-oligomeric isoenzymes of the _m_-AAA protease in murine and human mitochondria (Koppen et al, 2007). AFG3L2, homologous to paraplegin, assembles with paraplegin into a hetero-oligomeric _m_-AAA protease, but can also form homo-oligomeric, proteolytically active complexes. Accordingly, homo-oligomeric AFG3L2 complexes may lessen phenotypic consequences of the loss of paraplegin in HSP. At the same time, differences in the relative abundance of AFG3L2 and paraplegin in different tissues, as observed in mouse, may relate to the tissue-specific defects observed in HSP patients (Koppen et al, 2007). If differences in the enzymatic properties or substrate specificity of homo- and hetero-oligomeric _m_-AAA isoenzymes exist, the situation may become even more complex. Thus, although recent years have seen considerable progress in our understanding of QC surveillance mechanisms, much remains to be learned before a molecular understanding of pathogenic consequences of mitochondrial damage can be reached.
References
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B (2000) OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet 26: 211–215 [DOI] [PubMed] [Google Scholar]
- Atorino L, Silvestri L, Koppen M, Cassina L, Ballabio A, Marconi R, Langer T, Casari G (2003) Loss of _m_-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J Cell Biol 163: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh RH, Schmidt RE, Pestronk A, Milbrandt J (2007) Altered axonal mitochondrial transport in the pathogenesis of Charcot–Marie–Tooth disease from mitofusin 2 mutations. J Neurosci 27: 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baricault L, Segui B, Guegand L, Olichon A, Valette A, Larminat F, Lenaers G (2007) OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res 313: 3800–3808 [DOI] [PubMed] [Google Scholar]
- Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E (2006) Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J 25: 3900–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JM, Iacovino M, Perlman PS, Butow RA (2002) Mitochondrial DNA instability mutants of the bifunctional protein Ilv5p have altered organization in mitochondria and are targeted for degradation by Hsp78 and the Pim1p protease. J Biol Chem 277: 47946–47953 [DOI] [PubMed] [Google Scholar]
- Bota DA, Davies KJ (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol 4: 674–680 [DOI] [PubMed] [Google Scholar]
- Bota DA, Ngo JK, Davies KJ (2005) Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic Biol Med 38: 665–677 [DOI] [PubMed] [Google Scholar]
- Bota DA, Van Remmen H, Davies KJ (2002) Modulation of Lon protease activity and aconitase turnover during aging and oxidative stress. FEBS Lett 532: 103–106 [DOI] [PubMed] [Google Scholar]
- Casari G, De-Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, DeMichele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93: 973–983 [DOI] [PubMed] [Google Scholar]
- Cerveny KL, Tamura Y, Zhang Z, Jensen RE, Sesaki H (2007) Regulation of mitochondrial fusion and division. Trends Cell Biol 17: 563–569 [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 280: 26185–26192 [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160: 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC (2007) Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130: 548–562 [DOI] [PubMed] [Google Scholar]
- Cheng X, Kanki T, Fukuoh A, Ohgaki K, Takeya R, Aoki Y, Hamasaki N, Kang D (2005) PDIP38 associates with proteins constituting the mitochondrial DNA nucleoid. J Biochem (Tokyo) 138: 673–678 [DOI] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D'Adamio L, Derks C, Dejaegere T, Pellegrini L, D'Hooge R, Scorrano L, De Strooper B (2006) Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126: 163–175 [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 26: 207–210 [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Pelloquin L, Belenguer P, Hamel CP (2002) OPA1 (Kjer type) dominant optic atrophy: a novel mitochondrial disease. Mol Genet Metab 75: 97–107 [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC (2007) Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol 8: 870–879 [DOI] [PubMed] [Google Scholar]
- Dodson MW, Guo M (2007) Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson's disease. Curr Opin Neurobiol 17: 331–337 [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Jagasia R, Wagener J, Hofmann S, Trifunovic A, Hansson A, Chomyn A, Bauer MF, Attardi G, Larsson NG, Neupert W, Reichert AS (2006) Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J Biol Chem 281: 37972–37979 [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Koppen M, Wagener J, Zick M, Israel L, Bernacchia A, Jagasia R, Rugarli EI, Imhof A, Neupert W, Langer T, Reichert AS (2007) Processing of OPA1 reconstituted in yeast depends on the subunit composition of the _m_-AAA protease. Mol Biol Cell 18: 3582–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M, Westermann B, Langer T (2006) Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J Cell Biol 173: 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E (2002) A novel two-step mechanism for removal of a mitochondrial signal sequence involves the _m_-AAA complex and the putative rhomboid protease Pcp1. J Mol Biol 323: 835–843 [DOI] [PubMed] [Google Scholar]
- Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, Kruger R, Winklhofer KF, Vogel F, Reichert AS, Auburger G, Kahle PJ, Schmid B, Haass C (2007) Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci 27: 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreirinha F, Quattrini A, Priozzi M, Valsecchi V, Dina G, Broccoli V, Auricchio A, Piemonte F, Tozzi G, Gaeta L, Casari G, Ballabio A, Rugarli EI (2004) Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J Clin Invest 113: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L, Kanazawa T, Van der Bliek AM (2007) Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol 178: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H, Laine A, Erdjument-Bromage H, Tempst P, Gershwin ME, Bowtell DD, Ronai Z (2004) Regulation of 2-oxoglutarate (alpha-ketoglutarate) dehydrogenase stability by the RING finger ubiquitin ligase Siah. J Biol Chem 279: 53782–53788 [DOI] [PubMed] [Google Scholar]
- Hansen JJ, Dürr A, Isabelle C-R, Georgopoulos C, Ang D, Nielson MN, Davoine CS, Brice A, Fontaine B, Gregersen N, Bross P (2002) Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 70: 1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M, Bornhovd C, Hell K, Neupert W, Reichert AS (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J Cell Biol 165: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M, Vogel F, Bornhövd C, Neupert W, Reichert AS (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem 278: 27781–27788 [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ (1993) Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol 121: 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J (2007) The machines that divide and fuse mitochondria. Annu Rev Biochem 76: 751–780 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K (2006) Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 25: 2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Van Keuren ML, Fernandes-Alnemri T, Meisler MH, Alnemri ES (2003) Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 425: 721–727 [DOI] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ (2007) The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol 178: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ (2007) Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N (2004) Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem 279: 39068–39074 [DOI] [PubMed] [Google Scholar]
- Koppen M, Langer T (2007) Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol 42: 1–22 [DOI] [PubMed] [Google Scholar]
- Koppen M, Metodiev MD, Casari G, Rugarli EI, Langer T (2007) Variable and tissue-specific subunit composition of mitochondrial _m_-AAA protease complexes linked to hereditary spastic paraplegia. Mol Cell Biol 27: 758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C (2007) Mitochondrial membrane permeabilization in cell death. Physiol Rev 87: 99–163 [DOI] [PubMed] [Google Scholar]
- Leonhard K, Guiard B, Pellechia G, Tzagoloff A, Neupert W, Langer T (2000) Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol Cell 5: 629–638 [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443: 787–795 [DOI] [PubMed] [Google Scholar]
- Liu T, Lu B, Lee I, Ondrovicova G, Kutejova E, Suzuki CK (2004) DNA and RNA binding by the mitochondrial Lon protease is regulated by nucleotide and protein substrate. J Biol Chem 279: 13902–13910 [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G (2007) Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 8: 741–752 [DOI] [PubMed] [Google Scholar]
- Major T, von Janowsky B, Ruppert T, Mogk A, Voos W (2006) Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease Pim1. Mol Cell Biol 26: 762–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandemakers W, Morais VA, De Strooper B (2007) A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. J Cell Sci 120: 1707–1716 [DOI] [PubMed] [Google Scholar]
- Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, Abuin A, Grau E, Geppert M, Livi GP, Creasy CL, Martin A, Hargreaves I, Heales SJ, Okada H, Brandner S, Schulz JB, Mak T, Downward J (2004) Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol 24: 9848–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S (2006) Mitochondria: more than just a powerhouse. Curr Biol 16: 551–560 [DOI] [PubMed] [Google Scholar]
- McQuibban GA, Saurya S, Freeman M (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423: 537–541 [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T (2005) ERAD: the long road to destruction. Nat Cell Biol 7: 766–772 [DOI] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S (2006) MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep 7: 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A, Youle RJ (2005) Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J Biol Chem 280: 18598–18603 [DOI] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U (2005) Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot–Marie–Tooth disease. J Cell Biol 170: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T (2005) The _m_-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123: 277–289 [DOI] [PubMed] [Google Scholar]
- Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ (2007) Mdm38 protein depletion causes loss of mitochondrial K(+)/H(+) exchange activity, osmotic swelling and mitophagy. Cell Death Differ 14: 1647–1656 [DOI] [PubMed] [Google Scholar]
- Olichon A, Elachouri G, Baricault L, Delettre C, Belenguer P, Lenaers G (2007) OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ 14: 682–692 [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Pearce DA, Sherman F (1997) Differential ubiquitin-dependent degradation of the yeast apo-cytochrome c isozymes. J Biol Chem 272: 31829–31836 [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, Harvey K, Deas E, Harvey RJ, McDonald N, Wood NW, Martins LM, Downward J (2007) The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat Cell Biol 9: 1243–1252 [DOI] [PubMed] [Google Scholar]
- Pridgeon JW, Olzmann JA, Chin LS, Li L (2007) PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol 5: e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM (2006) A new function in translocation for the mitochondrial _i_-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol 26: 8488–8497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ (2006) Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy 2: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttgers K, Zufall N, Guiard B, Voos W (2002) The ClpB homolog Hsp78 is required for the efficient degradation of proteins in the mitochondrial matrix. J Biol Chem 277: 45829–45837 [DOI] [PubMed] [Google Scholar]
- Rugarli EI, Langer T (2006) Translating _m_-AAA protease function in mitochondria to hereditary spastic paraplegia. Trends Mol Med 12: 262–269 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Southard SM, Hobbs AE, Jensen RE (2003) Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem Biophys Res Commun 308: 276–283 [DOI] [PubMed] [Google Scholar]
- Soderblom C, Blackstone C (2006) Traffic accidents: molecular genetic insights into the pathogenesis of the hereditary spastic paraplegias. Pharmacol Ther 109: 42–56 [DOI] [PubMed] [Google Scholar]
- Song Z, Chen H, fiket M, Alexander C, Chan DC (2007) OPA1 processing control mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178: 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki CK, Suda K, Wang N, Schatz G (1994) Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science 264: 273–276 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R (2001) A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell 8: 613–621 [DOI] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H (2007) Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem 282: 5617–5624 [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Augustin S, Nolden M, Friedrichs B, Langer T (2007) _m_-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J 26: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I, Arlt H, van Dyck L, Langer T, Neupert W (1994) Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J 13: 5135–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Fletcher GC, Tolkovsky AM (2001) Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr Biol 11: 361–365 [DOI] [PubMed] [Google Scholar]
- Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, Inatome R, Yanagi S (2006) A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J 25: 3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ (2005) Autophagy: molecular machinery for self-eating. Cell Death Differ 12: 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schonfisch B, Guiard B, Pfanner N, Meisinger C (2006) Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell 17: 1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S (2007) The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 3: 337–346 [DOI] [PubMed] [Google Scholar]
- Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V et al. (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot–Marie–Tooth neuropathy type 2A. Nat Genet 36: 449–451 [DOI] [PubMed] [Google Scholar]
- Züchner S, Vance JM (2005) Emerging pathways for hereditary axonopathies. J Mol Med 83: 935–943 [DOI] [PubMed] [Google Scholar]