A Novel Site of Action for α-SNAP in the SNARE Conformational Cycle Controlling Membrane Fusion (original) (raw)
Abstract
Regulated exocytosis in neurons and neuroendocrine cells requires the formation of a stable soluble _N_-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex consisting of synaptobrevin-2/vesicle-associated membrane protein 2, synaptosome-associated protein of 25 kDa (SNAP-25), and syntaxin 1. This complex is subsequently disassembled by the concerted action of α-SNAP and the ATPases associated with different cellular activities-ATPase _N_-ethylmaleimide-sensitive factor (NSF). We report that NSF inhibition causes accumulation of α-SNAP in clusters on plasma membranes. Clustering is mediated by the binding of α-SNAP to uncomplexed syntaxin, because cleavage of syntaxin with botulinum neurotoxin C1 or competition by using antibodies against syntaxin SNARE motif abolishes clustering. Binding of α-SNAP potently inhibits Ca2+-dependent exocytosis of secretory granules and SNARE-mediated liposome fusion. Membrane clustering and inhibition of both exocytosis and liposome fusion are counteracted by NSF but not when an α-SNAP mutant defective in NSF activation is used. We conclude that α-SNAP inhibits exocytosis by binding to the syntaxin SNARE motif and in turn prevents SNARE assembly, revealing an unexpected site of action for α-SNAP in the SNARE cycle that drives exocytotic membrane fusion.
INTRODUCTION
Soluble _N_-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) comprise a superfamily of small, mostly membrane-anchored proteins that mediate membrane fusion in the secretory pathway of eukaryotic cells. They are characterized by the presence of SNARE motifs, homologous stretches of 60–70 amino acids located next to the membrane anchor domains. Key to the understanding of SNARE function in membrane fusion was the discovery of an assembly-disassembly cycle that is associated with major conformational changes. SNARE motifs of appropriate sets of SNAREs are unstructured, but they spontaneously assemble into tight complexes of extraordinary stability, forming elongated coiled-coils. When residing in different membranes, SNARE assembly leads to the formation of metastable “_trans_”-complexes in which the N-terminal parts of the SNARE motifs are associated, whereas the C-terminal membrane anchors are still residing in separate membranes. Progression of assembly toward the C-terminal membrane anchors is thought to proceed down a steep energy gradient and force the membranes together, resulting in fusion, with the SNAREs being converted from trans to “_cis_” complexes (for reviews, see Söllner, 2004; Brunger, 2005; Hong, 2005; Jahn and Scheller, 2006).
To be reused in another round of fusion, SNAREs need to be reactivated by disassembly of _cis_-complexes, which is mediated by the hexameric ATPase _N_-ethylmaleimide-sensitive factor (NSF), a member of the ATPases associated with different cellular activities protein superfamily. NSF operates on all SNARE complexes and prevents accumulation of “spent” _cis_-complexes, thus ensuring that sufficient concentrations of free SNAREs are available for the maintenance of intracellular membrane traffic. NSF does not interact directly with SNARE complexes, but rather it requires cofactors termed soluble NSF attachment proteins (SNAPs). In mammals, SNAPs form a small protein family with three isoforms, termed α-, β-, and γ-SNAP, respectively, with α-SNAP being the most abundant and ubiquitous of the isoforms (Clary et al., 1990). SNAPs are recruited from the cytoplasm to SNARE complexes in the membrane, and SNARE-bound SNAPs in turn recruit NSF, resulting in a “20S-complex.” ATP hydrolysis by NSF results in the disassembly of the SNARE complex, with NSF and SNAPs dissociating from the membrane (Söllner et al., 1993) (for review, see Whiteheart and Matveeva, 2004).
The SNARE assembly–disassembly cycle thus consists of two parts: the first part leading from free and energized SNAREs toward assembly and fusion, resulting in _cis_-complexes representing the low point in potential energy; and the second part involving disassembly and re-“charging” of SNAREs, mediated by NSF and SNAPs (Fasshauer, 2003). Indeed, numerous lines of evidence support the view that the function of NSF and SNAP is confined to SNARE disassembly, with no role in the assembly–fusion part of the cycle. Impairment or loss of either protein leads to a general inhibition of membrane traffic, which is associated with an accumulation of SNARE complexes and ultimately leads to cell death (Novick et al., 1980; Barnard et al., 1997; Ungermann et al., 1998; Littleton et al., 2001; Hanley et al., 2002). Conversely, an increase of the intracellular concentration of NSF and of α-SNAP was shown to result in an enhancement of fusion (DeBello et al., 1995; Kibble et al., 1996; Xu et al., 1999, 2002), suggesting that—at least in certain fusion reactions—disassembly/reactivation of SNAREs is rate limiting. This feature is best documented for exocytosis in neurons and neuroendocrine cells where the turnaround of vesicles and thus the need for regenerating SNAREs is higher than in any other system.
Intriguingly, however, several lines of evidence have emerged that α-SNAP on its own, i.e., independent of NSF, may also be inhibitory rather than stimulatory for fusion. Increasing α-SNAP dosage in Drosophila resulted in diminished neuronal exocytosis, accumulation of SNARE complexes, and other morphological defects (Babcock et al., 2004). Furthermore, addition of α-SNAP was shown to inhibit cell-free fusion reactions, including sperm acrosome exocytosis (Tomes et al., 2005) and yeast vacuole fusion (Wang et al., 2000). In all cases, concomitant increase in NSF levels was able to overcome the inhibition, but the mechanism of α-SNAP–mediated inhibition was not identified. Because α-SNAP and NSF do not bind to each other unless α-SNAP is bound to its substrate (Weidman et al., 1989; Whiteheart et al., 1992; Wilson et al., 1992), these effects cannot be due to α-SNAP scavenging the free pool of NSF. Rather, they indicate a second site of α-SNAP action in membrane fusion that is different from the established function of α-SNAP as cofactor in NSF-mediated reactivation of SNARE complexes.
In the present study, we have investigated whether α-SNAP can operate in the fusion pathway downstream of NSF-mediated disassembly of SNARE complexes. As model reaction, we used exocytosis in neuroendocrine PC12 cells that is mediated by the SNAREs syntaxin 1, SNAP-25, and synaptobrevin-2. Unlike other SNARE-mediated fusion reactions, the pathway leading to membrane fusion is arrested at a late stage, requiring calcium influx for completion that is probably mediated by an interaction of the calcium sensor synaptotagmin with the fusion machinery. Thus, regulated exocytosis is ideally suited to study individual steps in the fusion pathway, allowing for discriminating reactions such as vesicle docking, priming, triggering, and fusion (Sudhof, 2004). Our findings show that α-SNAP in the absence of NSF activity potently inhibits fusion, and this inhibition is exerted by binding to free syntaxin 1, thus directly inhibiting its SNARE function in membrane fusion.
MATERIALS AND METHODS
Cell Culture, Transfection, and Plasmid Constructs
PC-12 cells (clone 251) (Heumann et al., 1983) were maintained and propagated as described previously (Lang et al., 1997). Cells were transfected with human proneuropeptide Y fused N-terminally to enhanced green fluorescent protein (NPY-GFP) (Holroyd et al., 2002) described previously (Lang et al., 1997). For experiments, cells were plated onto 20-mm glass coverslips coated with poly-l-lysine as described previously (Avery et al., 2000). Details of plasmid constructs used in this study are contained in Supplemental Material.
Total Internal Reflection Fluorescence Microscopy (TIRFM)
For TIRFM, we used an Axiovert 200 M microscope (Carl Zeiss, Jena, Germany) equipped with an α-Plan-Fluar 100×/1.45 numerical aperture oil immersion objective and a single line GFP filter set (beam splitter 495LP, emission filter 525/50; both from Chroma Technology, Brattleboro, VT). Objective-based TIRFM was achieved by passing the 488-nm spectral line from an argon-ion laser through a TIRF condenser (TILL Photonics, Gräfelfing, Germany). Images were acquired by a frame-transfer, back-illuminated 16-bit cooled charge-coupled device camera (DV435; Andor Technology, Belfast, Ireland) by using ImSpector software (Garching Innovation, Munich, Germany). An Optovar (1.6×) was used to enlarge the image on the camera chip to avoid spatial undersampling. Cells expressing membrane-bound GFP-tagged α-SNAP were used 2 d after transfection. They were mounted in the microscopy chamber in Ringer's solution (130 mM NaCl, 4 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 48 mM glucose, and 10 mM HEPES-NaOH, pH 7.4) and imaged for 15 min at a rate of 1 image/min in Ringer's solution with or without 1 mM _N_-ethylmaleimide (NEM). Analyses of TIRFM images are described in Supplemental Material.
Microscopy
Fluorescence microscopy was performed essentially as described previously (Lang et al., 2002), with the following modifications. For imaging, we used cooled, back-illuminated frame transfer charge-coupled device cameras with either a 2 × 512 × 512 EEV chip with 13- × 13-μm pixel size, or a 512 × 512 NTE chip with 24- × 24-μm pixel size (Princeton Instruments, Trenton, NJ). To avoid spatial undersampling by the large pixels magnifying lenses (1.6× for the EEV chip and 2.5× Optovar for the NTE chip) were used during imaging. For image acquisition MetaMorph (Molecular Devices, Sunnyvale, CA) was used. 1-(4-Trimethylammonium)-6-phenyl-1,3,5-hexatriene (TMA-DPH; Invitrogen, Karlsruhe, Germany) fluorescence was detected in the blue channel (excitation filter D 360/50, dichroic mirror 400DLP, and emission filter E 420 LP), GFP fluorescence in the green channel (excitation filter 480/40, dichroic mirror 505LP, and emission filter 527/30), and cyanine (Cy)3 and Cy5 were detected in the red (excitation filter 560/55, dichroic mirror 595LP, and emission filter HQ 645/75) and the long-red channel (excitation filter 620/60, dichroic mirror 660LP, and emission filter 700/75).
Cell-free Assay for Exocytosis
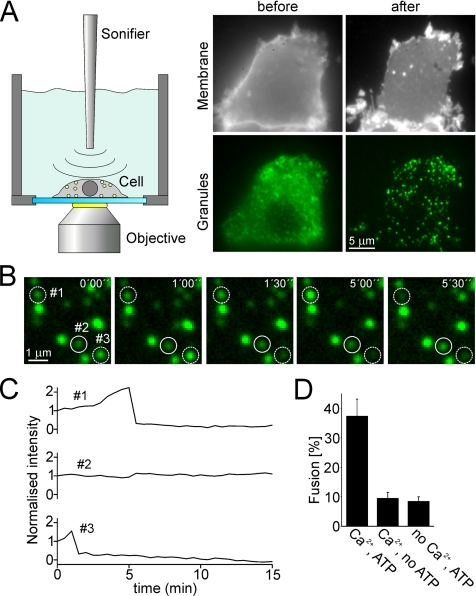
For on-stage preparation of membrane sheets, coverslips were mounted in a chamber filled with ice-cold K-Glu buffer containing 10 mM 1,3-diamino-2-propanol-N,N,N′,_N′_-tetraacetic acid (DPTA), 2 mM ATP, 4 mM MgCl2, and 0.5 mM dithiothreitol (DTT). The chamber was mounted onto the microscope stage, and a 2.5-mm tip of a sonication device (Sonifier B12; Branson Ultrasonics, Danbury, CT) was dipped into the chamber (Figure 2A). The distance between tip and glass coverslip was adjusted to ∼10 mm. A cell expressing NPY-GFP was disrupted using several sonication pulses (power setting of 5.5, corresponding to 100-W ultrasonic power delivered to horn tip). When a membrane sheet with numerous docked fluorescing secretory granules had formed, an image was taken, and the solution was exchanged with K-Glu buffer warmed to room temperature. Where indicated, the buffer was supplemented with rat brain cytosol (prepared as described previously; Avery et al., 2000), α-SNAP (wild type or the L294A variant), NEM, or NSF. After 5 min of preincubation another image was taken, and the solution was exchanged with K-Glu buffer. For stimulation, the buffer was supplemented with 3 mM CaCl2 resulting in ∼35 μM free Ca2+ (calculated assuming a Ca2+ dissociation constant of 81 μM for DPTA (Heinemann et al., 1994). Immediately after solution exchange, a 15-min imaging sequence was started with an image being taken every 30 s. At the end of the imaging sequence a camera dark frame was taken; in addition, phospholipids were visualized by adding 10–30 μl of a saturated solution of the lipophilic dye TMA-DPH in phosphate-buffered saline (PBS) containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4 pH 7.2. The continuity of the membrane sheets, as judged from their TMA-DPH staining, was documented by taking an image in the blue channel. Detailed description of the procedure used for quantification of exocytotic activity is documented in the Supplemental Materials.
Figure 2.
Ca2+-dependent exocytosis of secretory granules by using a membrane sheet-based cell-free assay. (A) Membrane sheets generated by sonication of PC12 cells retain docked secretory granules. Cells expressing the secretory granule marker NPY-GFP were grown on glass-coverslips and mounted on the microscope stage. GFP-labeled cells were selected and ruptured by brief pulses of ultrasound, resulting in a flat plasma membrane sheets with numerous green dots. Top, staining of the plasma membrane of a PC-12 cell before and after rupture with the lipophilic dye TMA-DPH. Bottom, GFP-channel showing secretory granules. (B) Granules docked to a membrane sheet undergo Ca2+-dependent exocytosis. Membrane sheet was preincubated for 5 min in an ATP-containing and calcium-free solution followed by the addition of ∼35 μM free calcium to trigger exocytosis (start at t = 0). Images were acquired every 30 s for 15 min. Exemplary images (time as indicated) show that the fluorescence intensity either changed (dashed circles) or remained constant (continuous circle). (C) Intensity traces of the granules encircled in B. Granules were scored as having undergone exocytosis when the drop of fluorescence intensity between two consecutive images exceeded 25%. (D) Exocytosis is dependent on the presence of Ca2+ and ATP in the triggering phase. Exocytotic membrane fusion was calculated by relating the number of granules scored positive for exocytosis during the 15 min stimulation phase to the number of granules present in the first image. Values are given as mean ± SEM (n = 9–20 membrane sheets for each condition recorded in at least three independent experiments). SEM denotes the SE of measurement.
Immunofluorescence
Immunostaining on membrane sheets was performed essentially as described previously (Lang et al., 2001), with the following modifications. Fixation was extended to 60–90 min, and incubations with primary and secondary antibodies were performed for 60 min. For binding of α-SNAP before immunostaining, membrane sheets were incubated immediately after preparation for 5 min at 37°C with 2 μM recombinant α-SNAP (wild type [wt] or L294A) in a humid chamber in K-Glu buffer containing 10 mM DPTA, 2 mM ATP, 4 mM MgCl2, and 0.5 mM DTT supplemented (where indicated) with recombinant light chains of clostridial neurotoxins, NEM, purified NSF, or rat brain cytosol. The sheets were then washed with PBS for 10 min, fixed, and processed for immunostaining.
Primary antibodies included α-SNAP, monoclonal antibody (mAb) Cl 77.2 (Figures 3, 6, and 7), obtained from Synaptic Systems (Göttingen, Germany), or a rabbit serum raised against recombinant α-SNAP (Hanson et al., 1995); transferrin receptor (mAb H 68.4; Zymed, Berlin, Germany); syntaxin 1, polyclonal rabbit serum R31 (Lang et al., 2001); and monoclonal antibodies HPC-1 (Barnstable et al., 1985) and Cl 78.3 (Chapman et al., 1995). All primary antibodies were diluted 1:200 in PBS-BSA (except Cl 77.2, dilution 1:400 and R31, HPC1, and Cl 78.3 were used for experiments shown in Figure 7, dilution 1:100). As secondary antibodies, Cy3-coupled goat anti-rabbit and Cy5-coupled goat anti-mouse were used (Dianova, Hamburg, Germany). The images acquired in the red and green channel (Figure 5) were aligned by using 0.2-μm Tetraspec microspheres (T-7280; Invitrogen). Quantitation of α-SNAP immunoreactivity is detailed in the Supplemental Materials.
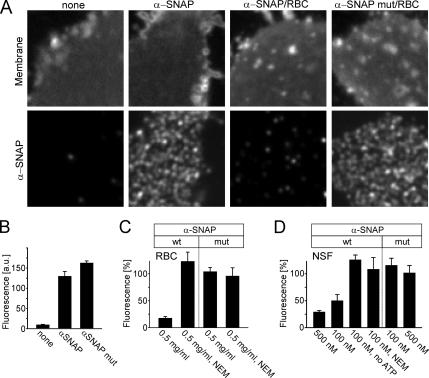
Figure 3.
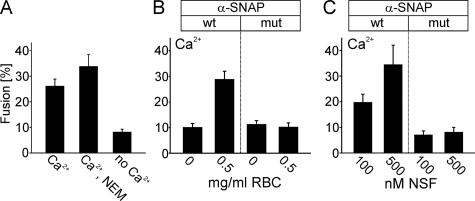
Membrane sheets contain NSF-sensitive binding sites for α-SNAP. (A) Membrane sheets incubated for 5 min with recombinant α-SNAP or α-SNAPL294A were briefly washed, fixed, and immunostained for α-SNAP (bottom). Top, membrane sheets visualized using TMA-DPH (also see Figure 1A). Where indicated, RBC was included in the incubation. (B) Quantification of immunofluorescence intensity in the absence or presence of recombinant α-SNAP or α-SNAPL294A. For better comparison, in the following experiments (C and D; also see Figure 5) values were normalized to the corresponding immunoreactivity stainings in B. (C and D) Binding of α-SNAP (wt) but not of α-SNAPL294A (mut) is prevented by the inclusion of either RBC (C) or purified NSF (D). Addition of NEM (1 mM) or omission of ATP, both known to inactivate NSF, blocked the interference with α-SNAP binding by both rat brain cytosol and purified NSF. The concentrations of RBC (milligrams of protein per milliliter) and recombinant NSF (nanomolar) is given at the bottom of the columns. Values are given as mean ± SEM (n = 3–14 independent experiments, with 35–169 individual membrane sheets analyzed for each experiment).
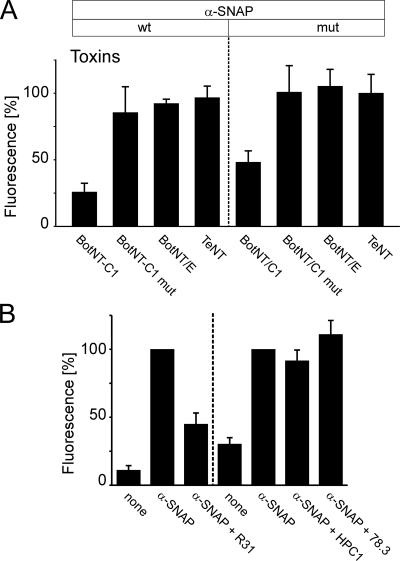
Figure 6.
A-SNAP binds to the SNARE-motif of synatxin 1. (A) A-SNAP binding requires syntaxin 1 but not SNAP-25 or synaptobrevin. Membrane sheets were incubated for 5 min with 2 μM of either the wt- or mutant form of α-SNAP. Where indicated, solutions contained in addition 2 μM purified light chains of either BoNT/C1 cleaving syntaxin 1, BoNT/C1mut (inactive form of BoNT/C1 carrying the mutation E230A), BoNT/E cleaving SNAP-25, or TeNT cleaving synaptobrevin. After brief washing, membrane sheets were processed for immunostaining and analyzed as shown in Figure 3, C and D). Values are given as mean ± SEM (n = 3–4 independent experiments, with 70–120 individual [mean = 106] membrane sheets analyzed for each experiment). (B) Antibodies directed against the SNARE-motif of syntaxin inhibit binding of α-SNAP. Membrane sheets were incubated for 15 min with anti-syntaxin 1 antibodies, washed twice with PBS and followed by 5-min incubation with 2 μM recombinant wild-type α-SNAP. The sheets were then washed, fixed, and immunolabeled for α-SNAP. The antibodies used for preincubation were R31 (polyclonal rabbit antiserum recognizing both the N-terminal domain and the SNARE motif) and HPC1 and Cl 78.3 (independently raised monoclonal antibodies specific for the N-terminal Habc-domain). For the detection of α-SNAP, we used either a monoclonal (Cl 77.2, left) or a polyclonal rabbit antibody (R34, right). In all experiments, fluorescence values were normalized to the immunoreactivity of membrane-bound, recombinant α-SNAP without prior anti-syntaxin 1 antibody treatment. Values are given as mean ± SEM (n = 6–7 independent experiments, with a minimum of 10–144 individual membrane sheets analyzed for each experiment).
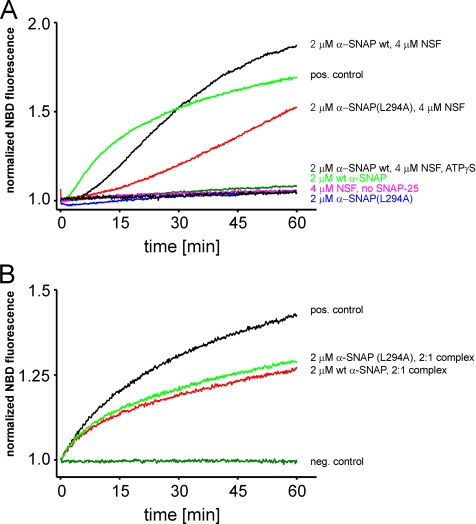
Figure 7.
Inhibition of SNARE-mediated proteoliposome fusion by α-SNAP. Fusion was measured as an increase of NBD fluorescence by using a lipid dequenching assay. Donor liposomes were reconstituted with an N-terminally truncated version of syntaxin 1 (2 μM, residues 183–288) (A) or with a preformed complex of syntaxin 1 (same construct as before) and SNAP-25 (B). Acceptor liposomes contained 10 μM synaptobrevin 2. If syntaxin-containing liposomes were used as donor, the liposomes were combined and preincubated for 10 min at 30°C. Where indicated, solutions contained in addition α-SNAP, α-SNAPL294A, NSF, or ATPγS, and the reaction was started by addition of 10 μM soluble SNAP-25a. (t = 0, reference point for normalization of the signal). In B, donor liposomes contained a preformed syntaxin-SNAP-25 complex, and the reaction was started by mixing donor and acceptor liposomes.
Figure 5.
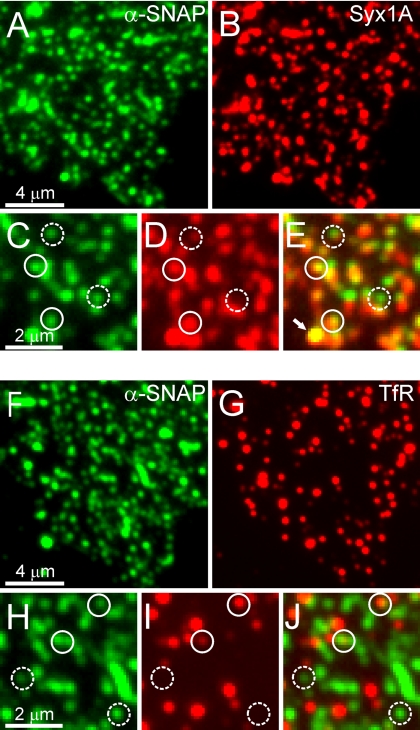
Syntaxin 1 clusters colocalize with α-SNAP binding sites. Membrane sheets were incubated with wild-type α-SNAP as given in Figure 3, and then they were fixed and double labeled for α-SNAP and syntaxin 1 (A–E) or transferrin receptor (TfR) (F–J). (C–E and H–J) Magnified views from A and B and F and G, respectively. Circles were placed on immunoreactive spot in the α-SNAP channel and transferred to identical pixel locations in the corresponding red channels. Immunoreactive spots were rated as colocalized according to Lang et al. (2002). After correction for accidental colocalization, we obtained 58 ± 2% specifically colocalizing spots for α-SNAP/syntaxin 1 and 9 ± 2% for α-SNAP/TfR (n = 10 membrane sheets analyzed per condition, with 50–200 spots analyzed on each membrane sheet; all values mean ± SEM). Solid circles indicate colocalizing, dashed circles noncolocalizing spots. The arrow in E indicates a fluorescent bead visible in all fluorescence channels that was added to the sample and used as a spatial reference for image alignment.
Preparation and Purification of Proteoliposomes
Recombinant synaptobrevin 2, SNAP-25a, and syntaxin 1A (183-288) were expressed and purified as described previously (Schuette et al., 2004). In addition, proteins were further purified by ion-exchange chromatography using an Ákta fast-performance liquid chromatography (FPLC) system (GE Healthcare, Chalfont St. Giles, United Kingdom). For preparation of fluorescently labeled liposomes, lipids (Avanti Lipids, Alabaster, AL) were mixed in chloroform to yield (molar ratios) as follows: phosphatidylcholine (5), phosphatidylethanolamine (1.7), _N_-lissamine-rhodamine-phosphatidylethanolamine (0.15), NBD-phosphatidylethanolamine (0.15), phosphatidylserine (1), phosphatidylinositol (1), and cholesterol (1); for preparation of unlabeled liposomes, only unlabeled phosphatidlethanolamine (2) was used. Lipids were dried under vacuum, and they were resuspended in HB100 (100 mM KCl, 20 mM HEPES, pH7.4, and 1 mM DTT) containing 5% (wt/vol) sodium cholate at a total lipid concentration of 13.5 mM. For preparation of proteoliposomes, lipids and proteins were mixed to yield a molar ratio of 100:1. Liposomes were formed by size exclusion chromatography on a prepacked PC3.2/10 column equilibrated in HB140 (140 mM KCl, 20 mM HEPES, pH 7.4, and 1 mM DTT) by using a SMART FPLC system (GE Healthcare) with a sample-to-column volume ratio of 1:15. Fusion reactions were carried out in HB140 containing 4 mM MgCl2 and 2 mM ATP, with a reaction volume of 30 μl in a microquartz cuvette. The dequenching signal was measured in a FluoroMax II Fluorometer (Horiba Yvon Jobin, Tokyo, Japan) equilibrated to 30°C, by using an excitation wavelength of 460 nm and an emission wavelength of 538 nm.
RESULTS
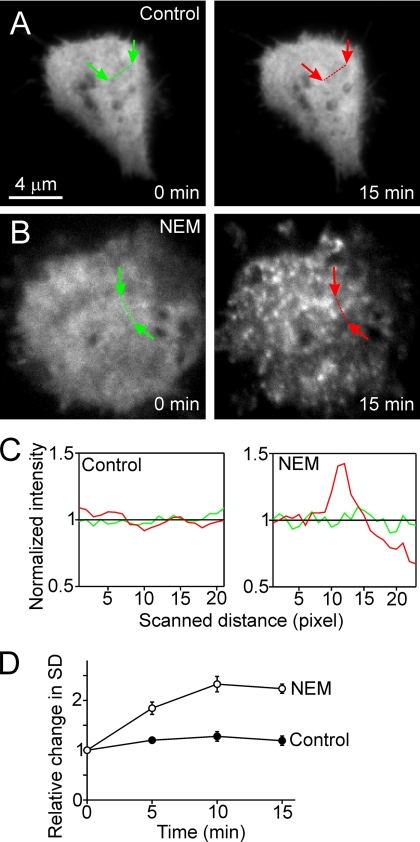
If SNAP proteins are capable of interfering with exocytosis, they need to interact directly or indirectly with the fusion machinery at the plasma membrane. SNAPs are soluble proteins; therefore, we asked whether neuroendocrine PC-12 cells contain a membrane-associated pool of α-SNAP, and if so, whether membrane association is influenced by NSF activity. PC-12 cells were transfected with GFP-labeled α-SNAP, and its distribution was monitored both by epifluorescence and by TIRFM. In untreated live cells, a diffuse staining pattern was observed both by epifluorescence (data not shown) and by TIRF illumination (Figure 1A), documenting that under steady-state conditions, the protein is predominantly localized to the cytosol. However, when cells were treated with 1 mM NEM to inhibit NSF, many fluorescent puncta were noted on the plasma membrane, whereas the overall levels of surface fluorescence remained unchanged (Figure 1B and 1Cd, for quantitation; data not shown).
Figure 1.
Recruitment of α-SNAP to the plasma membrane after NSF inactivation in vivo. (A and B) TIRF images of PC-12 cells expressing GFP-labeled α-SNAP. Cells were imaged for 15 min, taking one image per minute. Images for t = 0 and t = 15 min are shown. (A) Control, homogenous distribution of subplasmalemmal α-SNAP remains unchanged. (B) Treatment with 1 mM NEM causes α-SNAP to concentrate in plasmalemmal spots. Using linescans, the standard deviations of pixel intensities were determined (see Materials and Methods for details; see C for linescans normalized to average intensity) at t = 0 (green linescans) and t = 5, 10, and 15 min (red linescans). (D) Morphological alterations were expressed as relative changes in the SD of pixel intensities.
Because NSF is required for the disassembly of SNARE complexes, a trivial explanation for this finding could be that binding of α-SNAP simply reflects recruitment to _cis_-SNARE complexes. Indeed, unless counteracted by NSF, _cis_-SNARE complexes accumulate in the plasma membrane even if there is no exocytosis (Lang et al., 2002). However, the time course of this accumulation is rather slow. Therefore, we analyzed the time-course of NEM-induced recruitment of α-SNAP, resulting in a half-time of _t_½ ≈ 4 min. This is comparable with the time course of NEM-induced inhibition of exocytosis in bovine chromaffin cells (Xu et al., 1999), but it is several times faster than the accumulation of SNARE complexes (10–15 min; Lang et al., 2002; unpublished observations). Apparently, _cis_-SNARE complexes are not the major target for SNAP binding, raising the question to which receptor α-SNAP is binding, and whether binding of α-SNAP to this receptor is involved in the rapid block of exocytosis upon inactivation of NSF.
To address this question, we have taken advantage of the fact that in neuroendocrine cells Ca2+-dependent exocytosis remains functional for 20–30 min after cell disruption, thereby allowing direct biochemical access to the fusion machinery (Sarafian et al., 1987). We have shown previously that treatment of PC-12 cells with ultrasonic pulses yields inside-out lawns of plasma membrane that retain docked vesicles (Avery et al., 2000). Addition of Ca2+ triggers an exocytotic response that can be directly monitored by fluorescence microscopy when the granules contain a GFP-tagged content marker (Holroyd et al., 2002). For the present study, we have developed this assay further by performing sonication on stage, allowing for monitoring cell-free exocytosis seconds after cell disruption. Figure 2A shows fluorescence images of a single PC12 cell before and immediately after sonication showing that numerous fluorescent secretory granules are attached to the remaining plasma membrane sheet. Addition of Ca2+ resulted in exocytosis of 30–40% of all labeled organelles within 15 min. Exocytosis was visible as an abrupt loss of fluorescence, occasionally preceded by an increase in intensity due to alkalinization of the vesicle interior upon fusion (Figure 2, B and C; also see Holroyd et al., 2002). The extent of exocytosis did not change when the sheets were preincubated in K-Glu-DPTA buffer for up to 10 min before stimulation by Ca2+ (data not shown). Therefore, in all experiments, preincubation time was set to 5 min. Addition of rat brain cytosol did not result in an increase of exocytosis. Independence of cytosol differs from noradrenaline release in permeabilized PC-12 cells, which is reduced upon omission of cytosol (e.g., Hay and Martin, 1992). In contrast, Ca2+-dependent exocytosis required the presence of ATP (Figure 2D), in agreement with previous observations on permeabilized cells (Holz et al., 1989; Bittner and Holz, 1992; Martin et al., 1995).
First, we examined whether binding α-SNAP to the plasma membrane as observed upon NSF inactivation can be reproduced in the cell-free system. When freshly prepared membrane sheets were directly fixed and probed with an antibody against α-SNAP, no specific staining was detectable, regardless of whether the sheets were immediately fixed after preparation (data not shown) or whether they were preincubated for 5 min (Figure 3, A and B), in agreement with the experiments on intact cells described above. However, when the sheets were incubated with recombinant α-SNAP, a strong punctuate staining pattern was observable on the membranes (Figure 3, A and B). The same staining was observed when an α-SNAP mutant (α-SNAPL294A) that binds to SNARE complexes but fails to activate NSF (Barnard et al., 1997) was coincubated with the membranes (Figure 3, A and B). Thus, inverted lawns of plasma membrane contain binding sites for α-SNAP. Next, we investigated whether recruitment is reverted by NSF. On addition of rat brain cytosol (RBC, that contains endogenous NSF) or purified NSF to the binding reaction, only α-SNAPL294A but not wild-type α-SNAP remained bound (Figure 3, A, C, and D). In contrast, both wild-type and mutant α-SNAP remained bound when NSF was inactivated by NEM (Figure 3, C and D). Furthermore, prebound α-SNAP was removed when NSF was added at a later time point (data not shown) documenting that NSF exerts its action by dissociating α-SNAP from its binding sites on the membrane sheets. Together, these data show that α-SNAP accumulates on the plasma membrane if the activity of NSF is impaired. They further demonstrate that binding is reversible when NSF activity is restored.
In the next series of experiments, we investigated whether binding of α-SNAP affects exocytosis. First, we needed to clarify whether exocytosis is dependent on active NSF. Such dependence was observed in most in vitro fusion reactions, including exocytosis from permeabilized PC-12 cells (Banerjee et al., 1996). Apparently, most SNAREs are trapped in inactive _cis_-complexes whose dissociation is rate limiting. However, this does not reflect the physiological steady-state situation where SNAREs are almost quantitatively disassembled (Bethani et al., 2007). Inactive _cis_-complexes only accumulate after cell disruption, with their amount (and thus the need for NSF-activation) being dependent on the time required for membrane preparation (Lang et al., 2002). To inactivate endogenous NSF that may have remained on the sheets after sonication and washing, the sheets were preincubated with 1 mM NEM, a concentration known to completely inhibit NSF in other cell-free fusion reactions (e.g., Rodriguez et al., 1994). As shown in Figure 4A, NEM did not cause any inhibition on Ca2+-dependent exocytosis; thus, any effects on exocytosis elicited by factors added to the assay must be due to influencing steps downstream of SNARE disassembly. We then added α-SNAP to the membrane sheets and measured its influence on exocytosis. As shown in Figure 4B, Ca2+-dependent exocytosis was abolished. The same effect was observed with the α-SNAPL294A mutant (Figure 4B, mut). To test whether the inhibition caused by α-SNAP is reversible by NSF, parallel incubations were carried out in the presence of either cytosol or of purified NSF. As shown in Figure 4, B and C, inhibition by wild-type but not by mutant α-SNAP is reverted in the presence of either cytosol or purified NSF.
Figure 4.
Exocytosis is independent of NSF, but it is inhibited by α-SNAP. All experiments were performed as in Figure 2D, with ATP present in the preincubation and triggering phase and Ca2+ added during the triggering phase. All other additions were present only in the preincubation phase. (A) Preincubation of membrane sheets with 1 mM NEM does not inhibit exocytosis. (B and C) Preincubation with 2 μM α-SNAP (wt) or α-SNAPL294A (mut) inhibits Ca2+-dependent exocytosis. Inhibition by wild-type but not mutant α-SNAP is prevented by the inclusion of either RBC (B) or purified NSF (C). Values are given as mean ± SEM (n = 9–18 membrane sheets for each condition).
The results described so far show 1) inactivation of NSF results in the association of α-SNAP with concentrated spots on the plasma membrane; 2) NSF activity and thus disassembly of _cis_-SNARE complexes is not required for exocytosis of predocked granules; and 3) binding of α-SNAP to the plasma membrane inhibits exocytosis, which is counteracted by active NSF. Apparently, α-SNAP binds to a target protein that is different from _cis_-SNARE complexes and operates downstream of SNARE-disassembly in the fusion pathway. Binding to this target is reverted by NSF, thus resembling NSF-driven dissociation of α-SNAP after disassembly of _cis_-SNARE complexes (Söllner et al., 1993). What could be the identity of this target protein? It was shown earlier that α-SNAP not only binds to fully assembled _cis_-SNARE complexes but also to free syntaxin 1, with the binding site being located in the SNARE motif. Furthermore, NSF dissociates the syntaxin 1/α-SNAP complex in an ATP-dependent manner (Hanson et al., 1995; McMahon and Südhof, 1995). Therefore, we asked whether the inhibitory effects of α-SNAP on exocytosis may be mediated via its binding to free, uncomplexed syntaxin 1.
First, we performed double-labeling experiments to examine whether α-SNAP and syntaxin 1 are colocalized. Comparison of representative images revealed a high degree of overlap (Figure 5, A–E). Quantitative analysis revealed that at least 58% of all α-SNAP–positive puncta colocalize with syntaxin 1. As control, double labeling was performed for the transferrin receptor, a single-pass integral membrane protein that cycles between endosomes and the plasma membrane (Figure 5, F–J). The transferrin receptor also yielded a punctuate staining pattern, although less dense than that of syntaxin 1. The degree of overlap was determined to be only 9% and thus close to the nonspecific background colocalization.
Second, we analyzed binding of α-SNAP in the presence of recombinant light chain of botulinum neurotoxin C1 (BoNT/C1) that selectively cleaves uncomplexed and membrane-bound syntaxin 1(Blasi et al., 1993; Hayashi et al., 1994). Indeed, specific cleavage of syntaxin-1 present on the membrane sheets dramatically reduced the binding of both wild-type and mutant α-SNAP (Figure 6A), suggesting that the contribution of syntaxin 1 to α-SNAP binding is even higher than suggested by the colocalization analysis. No inhibition was observed when a catalytically inactive mutant of the toxin light chain was used. Moreover, treatment with neither tetanus toxin that cleaves synaptobrevin (Schiavo et al., 1992) nor botulinum neurotoxin E that cleaves SNAP-25 (Binz et al., 1994) displayed any negative effect on α-SNAP. Control experiments confirmed that both toxins were highly active, with SNAP-25 immunoreactivity toward an antibody specific for the C-terminal amino acids being reduced to background levels (data not shown).
Finally, we examined whether syntaxin-specific antibodies inhibit α-SNAP binding. Because the binding site of α-SNAP to syntaxin was previously mapped to the SNARE motif (Hanson et al., 1995; McMahon and Südhof, 1995), we used both a polyclonal antibody (R31) that reacts with the SNARE motif and with the N-terminal domain (data not shown), and two monoclonal antibodies (HPC1 and 78.3) known to bind exclusively to the N-terminal domain (Inoue et al., 1992; Chapman et al., 1995). As shown in Figure 6B, incubation with the polyclonal antibody but not with any of the monoclonal antibodies inhibited binding of α-SNAP. Based on these data, we conclude that binding of α-SNAP to the sheets is mediated by the SNARE motif of syntaxin 1.
In conclusion, it seems that binding of α-SNAP to free syntaxin 1 inhibits stimulated exocytosis at a step downstream of NSF-driven SNARE disassembly. The question then arises how exactly the inhibitory action on syntaxin is exerted. It is possible that α-SNAP binding directly competes for SNARE complex formation by means of blocking the SNARE motif. Alternatively, α-SNAP binding may exert its effect further upstream, e.g., by interfering with other (unknown) proteins that are required before _trans_-SNARE complex formation. A third possibility is that α-SNAP exerts its action directly on the _trans_-SNARE complex, e.g., by preventing its Ca2+-dependent activation or by interfering with C-terminal assembly of the complex. To differentiate between these possibilities, we tested the effects of α-SNAP on SNARE-dependent fusion of liposomes (Figure 7). Proteoliposomes containing synaptobrevin fuse spontaneously with liposomes containing syntaxin 1A and SNAP-25 (Weber et al., 1998; Schuette et al., 2004), thus providing a “bare-bones” system for testing the function of SNAREs in membrane fusion and for assessing the impact of potential regulators.
Proteoliposomes containing purified synaptobrevin 2 and syntaxin 1A, respectively, were mixed, and fusion was monitored with a standard dequenching assay. As expected, a robust dequenching signal (Figure 7A, positive [pos.] control) was observed that depended on the presence of SNAP-25 (data not shown; Schuette et al., 2004). Addition of α-SNAP (wild-type and mutant) at twice the syntaxin concentration completely inhibited fusion (Figure 7A). These data show that no other factors are needed for α-SNAP to exert its inhibitory action on SNARE-mediated membrane fusion. Next, we tested whether NSF relieves the block. Recombinant NSF was added simultaneously with α-SNAP and SNAP-25, and ATP was included. In incubations containing wild-type α-SNAP, NSF restored fusion, with a kinetics that was delayed in onset compared with the positive control. This delay likely reflects the time needed for NSF to liberate syntaxin from α-SNAP-syntaxin complexes and for the formation of syntaxin-SNAP-25 acceptor complexes. The higher extent of dequenching observed at the end of the incubation over the positive control is probably due to the continuous action of NSF on _cis_-SNARE complexes that form during fusion, thus increasing the amount of free and fusion-competent SNAREs during the late phase of the reaction. Intriguingly, NSF rescued the block when mutant α-SNAP was used but to a lesser extent compared with the wild type, indicating that the mutant retains residual ability to trigger NSF activity. No fusion was observed when ATP-cleavage by NSF was prevented by using the nonhydrolysable analog adenosine-5′-_O_-(3-thio)triphosphate (ATPγS). Furthermore, NSF on its own did not induce any dequenching (Figure 7A; Brugger et al., 2000).
Finally, we examined whether α-SNAP also inhibits fusion if syntaxin and SNAP-25 are preassembled into a binary complex before reconstitution. This complex is known to form a four-helix bundle in which position of synaptobrevin is occupied by a second syntaxin molecule that needs to be displaced during fusion. Accordingly, fusion activity is reduced in comparison with standard conditions. Intriguingly, this reaction was only slightly inhibited by α-SNAP (Figure 7B). Thus, α-SNAP is unable to inhibit fusion once syntaxin is engaged in partial complexes. We conclude that binding of α-SNAP to the SNARE motif of uncomplexed syntaxin blocks exocytosis by preventing syntaxin from forming SNARE complexes.
DISCUSSION
In the present study, we have shown that α-SNAP potently inhibits exocytosis by binding to the SNARE motif of syntaxin, preventing syntaxin from interacting with its SNARE partners as required for membrane fusion. These findings reveal a second and unexpected site of action of α-SNAP on the SNARE assembly–disassembly cycle, and they provide a molecular explanation for the inhibition of membrane fusion by α-SNAP observed in previous studies (Babcock et al., 2004; Tomes et al., 2005).
To dissect the site of action of α-SNAP on exocytosis we took advantage of a previously established in vitro assay for exocytosis (Avery et al., 2000) that involves the preparation of inside-out lawns of plasma membrane containing docked secretory vesicles. The system was further improved, allowing the triggering of exocytosis with a lag time of less than a minute after cell disruption. Ca2+-dependent exocytosis in this system does not require cytosol, and it does not depend on the disassembly of SNAREs as priming step, setting it apart from all other cell-free assays for membrane fusion, particularly for exocytosis (e.g., Hay and Martin, 1992). Thus, any influence of fusion kinetics exerted by exogenously added factors must be operating downstream of SNARE dissociation, ruling out that the effects of α-SNAP reported here are related in any way to the disassembly of SNARE complexes.
Our data show that α-SNAP specifically binds to membrane-anchored syntaxin 1 that is not complexed with SNAP-25 or engaged in ternary SNARE complexes. Previous studies showed that α-SNAP binds to recombinant syntaxin 1 in vitro (Hanson et al., 1995; McMahon and Südhof, 1995), but it has remained unknown whether this interaction affects the physiological function of syntaxin in intact membranes. Binding was shown to involve a direct interaction with the H3-domain (now referred to as SNARE motif), but neither stoichiometry nor affinity has been determined under equilibrium conditions. In pull-down assays, half-maximal binding of α-SNAP to immobilized syntaxin 1 was observed at a concentration of ∼500 nM, but binding seems to be substoichiometric (Hanson et al., 1995), with the affinity being lower than that observed during binding to assembled SNARE complexes (McMahon and Südhof, 1995), and it was suggested in these studies that α-SNAP-binding activates rather than inhibits the ability of syntaxin to interact with its SNARE partners. NSF disassembled the α-SNAP-syntaxin 1 complex, making it temporarily refractory to rebinding of α-SNAP. These data are in excellent agreement with our current findings. In addition, we observe no effect of BoNT/E and tetanus toxin (TeNT0 on α-SNAP binding, ruling out SNAP-25 and synaptobrevin 2 as putative binding sites. This is in line with previous observations showing that α-SNAP interacts with SNAP-25 only with an at least 10-fold lower affinity and not at all with synaptobrevin (Hanson et al., 1995). Interestingly, our data suggest that α-SNAP does not act on preassembled or partially zippered SNARE complexes that are suggested to reflect the primed state required for the Ca2+ trigger to effect exocytosis. This notion agrees with the observation that in chromaffin cells inhibition of NSF does not block exocytosis of the readily releasable pool (Xu et al., 1999) and that ∼10% of the vesicles in our assay system remain fusion-competent even after addition of α-SNAP.
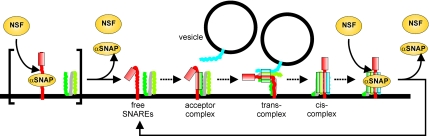
Figure 8 illustrates the sites of action in the SNARE cycle for α-SNAP and NSF. It is evident that the NSF-α-SNAP system needs to be balanced for normal functioning of exocytosis. Regulation of this balance might be exerted at the intracellular concentrations or biological activities of either or both proteins. Although no direct evidence points to such a control for α-SNAP, NSF activity is influenced by at least two mechanisms. Phosphorylation and nitrosylation of NSF have been shown to inhibit its ability to disassemble _cis_-complexes, and they have been correlated with reduced homotypic fusion of intracellular vesicles or decreased exocytosis in vivo (Matsushita et al., 2003; Huynh et al., 2004; Morrell et al., 2005). By blocking NSF activity, both posttranslational modifications shift the NSF-α-SNAP equilibrium toward an increased population of α-SNAP that is bound not only to _cis_-complexes (to which α-SNAP binds with high affinity; see above), but also to free syntaxin. This leads to a decrease in exocytosis as documented in this study. Dephosphorylation or denitrosylation of NSF restores its ability to disassemble both _cis_-complexes and α-SNAP-syntaxin 1 complexes, thereby restoring the proper SNARE cycle.
Figure 8.
Model illustrating the sites of action for α-SNAP and NSF in the SNARE conformational cycle during exocytosis. A-SNAP binds with high affinity to _cis_-SNARE complexes and with lower affinity to free syntaxin. Under normal circumstances, the equilibrium is shifted toward the dissociated part of the cycle, with the steady-state concentration of both _cis_-complexes and α-SNAP/syntaxin complexes being low. However, if the SNAP–NSF system is imbalanced due to overexpression of α-SNAP or down-regulation/inhibition of NSF, free syntaxin rapidly becomes rate limiting, resulting in an inhibition of exocytosis. Please note that free syntaxin is organized in clusters (not shown in the figure), and it is not known whether α-SNAP exhibits any preference for free versus clustered syntaxin. See text for details.
Interactions between α-SNAP and other syntaxins have not yet been investigated; thus, it is presently unknown whether a similar mechanism applies to other intracellular fusion reactions. It is conceivable that binding of α-SNAP to free syntaxin becomes rate limiting for fusion when NSF is impaired or down-regulated. In such situations, α-SNAP-dependent inactivation of free syntaxins may present a quick strategy to shut down membrane fusion before dissociated SNAREs are used up, while rapidly restoring exocytosis when normal NSF activity is restored.
Supplementary Material
[Supplemental Materials]
ACKNOWLEDGMENTS
We thank Dr. Andreas Schönle for help with the setting up of the ImSpector software.
Footnotes
REFERENCES
- Avery J., Ellis D. J., Lang T., Holroyd P., Riedel D., Henderson R. M., Edwardson J. M., Jahn R. A cell-free system for regulated exocytosis in PC12 cells. J. Cell Biol. 2000;148:317–324. doi: 10.1083/jcb.148.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock M., Macleod G. T., Leither J., Pallanck L. Genetic analysis of soluble N-ethylmaleimide-sensitive factor attachment protein function in Drosophila reveals positive and negative secretory roles. J. Neurosci. 2004;24:3964–3973. doi: 10.1523/JNEUROSCI.5259-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Barry V. A., DasGupta B. R., Martin T. F. N-Ethylmaleimide-sensitive factor acts at a prefusion ATP-dependent step in Ca2+-activated exocytosis. J. Biol. Chem. 1996;271:20223–20226. doi: 10.1074/jbc.271.34.20223. [DOI] [PubMed] [Google Scholar]
- Barnard R. J., Morgan A., Burgoyne R. D. Stimulation of NSF ATPase activity by alpha-SNAP is required for SNARE complex disassembly and exocytosis. J. Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnstable C. J., Hofstein R., Akagawa K. A marker of early amacrine cell development in rat retina. Brain Res. 1985;352:286–290. doi: 10.1016/0165-3806(85)90116-6. [DOI] [PubMed] [Google Scholar]
- Bethani I., Lang T., Geumann U., Sieber J. J., Jahn R., Rizzoli S. O. The specificity of SNARE pairing in biological membranes is mediated by both proof-reading and spatial segregation. EMBO J. 2007;26:3981–3992. doi: 10.1038/sj.emboj.7601820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz T., Blasi J., Yamasaki S., Baumeister A., Link E., Südhof T. C., Jahn R., Niemann H. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. J. Biol. Chem. 1994;269:1617–1620. [PubMed] [Google Scholar]
- Bittner M. A., Holz R. W. Kinetic analysis of secretion from permeabilized adrenal chromaffin cells reveals distinct components. J. Biol. Chem. 1992;267:16219–16225. [PubMed] [Google Scholar]
- Blasi J., Chapman E. R., Yamasaki S., Binz T., Niemann H., Jahn R. Botulinum neurotoxin C1 blocks neurotransmitter release by means of cleaving HPC-1/syntaxin. EMBO J. 1993;12:4821–4828. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger B., Nickel W., Weber T., Parlati F., McNew J. A., Rothman J. E., Söllner T. Putative fusogenic activity of NSF is restricted to a lipid mixture whose coalescence is also triggered by other factors. EMBO J. 2000;19:1272–1278. doi: 10.1093/emboj/19.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A. T. Structure and function of SNARE and SNARE-interacting proteins. Q. Rev. Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- Chapman E. R., Hanson P. I., An S., Jahn R. Ca2+ regulates the interaction between synaptotagmin and syntaxin 1. J. Biol. Chem. 1995;270:23667–23671. doi: 10.1074/jbc.270.40.23667. [DOI] [PubMed] [Google Scholar]
- Clary D. O., Griff I. C., Rothman J. E. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- DeBello W. M., O'Connor V., Dresbach T., Whiteheart S. W., Wang S. S., Schweizer F. E., Betz H., Rothman J. E., Augustine G. J. SNAP-mediated protein-protein interactions essential for neurotransmitter release. Nature. 1995;373:626–630. doi: 10.1038/373626a0. [DOI] [PubMed] [Google Scholar]
- Fasshauer D. Structural insights into the SNARE mechanism. Biochim. Biophys. Acta. 2003;1641:87–97. doi: 10.1016/s0167-4889(03)00090-9. [DOI] [PubMed] [Google Scholar]
- Hanley J. G., Khatri L., Hanson P. I., Ziff E. B. NSF ATPase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Otto H., Barton N., Jahn R. The N-ethylmaleimide-sensitive fusion protein and alpha-SNAP induce a conformational change in syntaxin. J. Biol. Chem. 1995;270:16955–16961. doi: 10.1074/jbc.270.28.16955. [DOI] [PubMed] [Google Scholar]
- Hay J. C., Martin T. F. Resolution of regulated secretion into sequential MgATP-dependent and calcium-dependent stages mediated by distinct cytosolic proteins. J. Cell Biol. 1992;119:139–151. doi: 10.1083/jcb.119.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., McMahon H., Yamasaki S., Binz T., Hata Y., Südhof T. C., Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann C., Chow R. H., Neher E., Zucker R. S. Kinetics of the secretory response in bovine chromaffin cells following flash photolysis of caged Ca2+ Biophys. J. 1994;67:2546–2557. doi: 10.1016/S0006-3495(94)80744-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann R., Kachel V., Thoenen H. Relationship between NGF-mediated volume increase and “priming effect” in fast and slow reacting clones of PC12 pheochromocytoma cells. Role of cAMP. Exp. Cell Res. 1983;145:179–190. doi: 10.1016/s0014-4827(83)80019-6. [DOI] [PubMed] [Google Scholar]
- Holroyd P., Lang T., Wenzel D., De Camilli P., Jahn R. Imaging direct, dynamin-dependent recapture of fusing secretory granules on plasma membrane lawns from PC12 cells. Proc. Natl. Acad. Sci. USA. 2002;99:16806–16811. doi: 10.1073/pnas.222677399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz R. W., Bittner M. A., Peppers S. C., Senter R. A., Eberhard D. A. MgATP-independent and MgATP-dependent exocytosis. Evidence that MgATP primes adrenal chromaffin cells to undergo exocytosis. J. Biol. Chem. 1989;264:5412–5419. [PubMed] [Google Scholar]
- Hong W. SNAREs and traffic. Biochim. Biophys. Acta. 2005;1744:120–144. doi: 10.1016/j.bbamcr.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Huynh H., et al. Control of vesicle fusion by a tyrosine phosphatase. Nat. Cell Biol. 2004;6:831–839. doi: 10.1038/ncb1164. [DOI] [PubMed] [Google Scholar]
- Inoue A., Obata K., Akagawa K. Cloning and sequence analysis of cDNA for a neuronal cell membrane antigen, HPC-1. J. Biol. Chem. 1992;267:10613–10619. [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. SNAREs–engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Kibble A. V., Barnard R. J., Burgoyne R. D. Patch-clamp capacitance analysis of the effects of alpha-SNAP on exocytosis in adrenal chromaffin cells. J. Cell Sci. 1996;109:2417–2422. doi: 10.1242/jcs.109.9.2417. [DOI] [PubMed] [Google Scholar]
- Lang T., Bruns D., Wenzel D., Riedel D., Holroyd P., Thiele C., Jahn R. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Margittai M., Holzler H., Jahn R. SNAREs in native plasma membranes are active and readily form core complexes with endogenous and exogenous SNAREs. J. Cell Biol. 2002;158:751–760. doi: 10.1083/jcb.200203088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Wacker I., Steyer J., Kaether C., Wunderlich I., Soldati T., Gerdes H. H., Almers W. Ca2+-triggered peptide secretion in single cells imaged with green fluorescent protein and evanescent-wave microscopy. Neuron. 1997;18:857–863. doi: 10.1016/s0896-6273(00)80325-6. [DOI] [PubMed] [Google Scholar]
- Littleton J. T., Barnard R. J., Titus S. A., Slind J., Chapman E. R., Ganetzky B. SNARE-complex disassembly by NSF follows synaptic-vesicle fusion. Proc. Natl. Acad. Sci. USA. 2001;98:12233–12238. doi: 10.1073/pnas.221450198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. F., Hay J. C., Banerjee A., Barry V. A., Ann K., Yom H. C., Porter B. W., Kowalchyk J. A. Late ATP-dependent and Ca++-activated steps of dense core granule exocytosis. Cold Spring Harb. Symp. Quant. Biol. 1995;60:197–204. doi: 10.1101/sqb.1995.060.01.022. [DOI] [PubMed] [Google Scholar]
- Matsushita K., et al. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. T., Südhof T. C. Synaptic core complex of synaptobrevin, syntaxin, and SNAP25 forms high affinity alpha-SNAP binding site. J. Biol. Chem. 1995;270:2213–2217. doi: 10.1074/jbc.270.5.2213. [DOI] [PubMed] [Google Scholar]
- Morrell C. N., et al. Regulation of platelet granule exocytosis by S-nitrosylation. Proc. Natl. Acad. Sci. USA. 2005;102:3782–3787. doi: 10.1073/pnas.0408310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez L., Stirling C. J., Woodman P. G. Multiple N-ethylmaleimide-sensitive components are required for endosomal vesicle fusion. Mol. Biol. Cell. 1994;5:773–783. doi: 10.1091/mbc.5.7.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafian T., Aunis D., Bader M. F. Loss of proteins from digitonin-permeabilized adrenal chromaffin cells essential for exocytosis. J. Biol. Chem. 1987;262:16671–16676. [PubMed] [Google Scholar]
- Schiavo G., Benfenati F., Poulain B., Rossetto O., Polverino de Laureto P., DasGupta B. R., Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schuette C. G., Hatsuzawa K., Margittai M., Stein A., Riedel D., Kuster P., Konig M., Seidel C., Jahn R. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc. Natl. Acad. Sci. USA. 2004;101:2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T. H. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 2004;16:429–435. doi: 10.1016/j.ceb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Söllner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Sudhof T. C. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Tomes C. N., De Blas G. A., Michaut M. A., Farre E. V., Cherhitin O., Visconti P. E., Mayorga L. S. alpha-SNAP and NSF are required in a priming step during the human sperm acrosome reaction. Mol. Hum. Reprod. 2005;11:43–51. doi: 10.1093/molehr/gah126. [DOI] [PubMed] [Google Scholar]
- Ungermann C., Nichols B. J., Pelham H. R., Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J. Cell Biol. 1998;140:61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ungermann C., Wickner W. The docking of primed vacuoles can be reversibly arrested by excess Sec17p (alpha-SNAP) J. Biol. Chem. 2000;275:22862–22867. doi: 10.1074/jbc.M001447200. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weidman P. J., Melancon P., Block M. R., Rothman J. E. Binding of an N-ethylmaleimide-sensitive fusion protein to Golgi membranes requires both a soluble protein(s) and an integral membrane receptor. J. Cell Biol. 1989;108:1589–1596. doi: 10.1083/jcb.108.5.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart S. W., Brunner M., Wilson D. W., Wiedmann M., Rothman J. E. Soluble N-ethylmaleimide-sensitive fusion attachment proteins (SNAPs) bind to a multi-SNAP receptor complex in Golgi membranes. J. Biol. Chem. 1992;267:12239–12243. [PubMed] [Google Scholar]
- Whiteheart S. W., Matveeva E. A. Multiple binding proteins suggest diverse functions for the N-ethylmaleimide sensitive factor. J. Struct. Biol. 2004;146:32–43. doi: 10.1016/j.jsb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Wilson D. W., Whiteheart S. W., Wiedmann M., Brunner M., Rothman J. E. A multisubunit particle implicated in membrane fusion. J. Cell Biol. 1992;117:531–538. doi: 10.1083/jcb.117.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xu Y., Ellis-Davies G. C., Augustine G. J., Tse F. W. Differential regulation of exocytosis by alpha- and beta-SNAPs. J. Neurosci. 2002;22:53–61. doi: 10.1523/JNEUROSCI.22-01-00053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Ashery U., Burgoyne R. D., Neher E. Early requirement for alpha-SNAP and NSF in the secretory cascade in chromaffin cells. EMBO J. 1999;18:3293–3304. doi: 10.1093/emboj/18.12.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]