Stabilization of Urokinase and Urokinase Receptor mRNAs by HuR Is Linked to Its Cytoplasmic Accumulation Induced by Activated Mitogen-Activated Protein Kinase-Activated Protein Kinase 2 (original) (raw)
Abstract
The mRNAs of urokinase plasminogen activator (uPA) and its receptor, uPAR, contain instability-determining AU-rich elements (AREs) in their 3′ untranslated regions. The cellular proteins binding to these RNA sequences (AREuPA/uPAR) are not known. We show here that the mRNA-stabilizing factor HuR functionally interacts with these sequences. HuR stabilized an AREuPA-containing RNA substrate in vitro and stabilized in HeLa Tet-off cells both endogenous uPA and uPAR mRNAs and a β-globin reporter mRNA containing the AREuPA. RNAi-mediated depletion of HuR in BT-549 and MDA-MB-231 cells significantly reduced the steady-state levels of endogenous uPA and uPAR mRNAs. Furthermore, we show that a constitutively active form of mitogen-activated protein kinase-activated protein kinase 2 (MK2), MK2-EE, has an ARE-mRNA-stabilizing effect that correlates with its ability to enhance the cytoplasmic accumulation of endogenous HuR, but not in cells cotransfected with a dominant negative version of MK2, MK2-K76R. These effects were mimicked by hydrogen peroxide treatment (oxidative stress), which resulted in the phosphorylation of endogenous MK2. In addition, hydrogen peroxide treatment enhanced the cytoplasmic binding of HuR to the AREuPA, which was abrogated in cells transfected with MK2-K76R. These results indicate a role for HuR and MK2 in regulating the expression of uPA and uPAR genes at the posttranscriptional level.
Urokinase plasminogen activator (uPA), when bound to its specific cell surface receptor uPAR, efficiently converts plasminogen to the active serine protease plasmin (54), which initiates the destruction of various extracellular matrix (ECM) proteins. Binding of uPA also enhances the affinity of uPAR to an ECM protein, vitronectin, controlling the dynamic interaction of cells with vitronectin-containing ECM. Thus, with or without involving the proteolytic activity of uPA, the uPA-uPAR system plays an important role in various physiological and pathophysiological processes requiring cell movement, such as wound healing, angiogenesis and tumor metastasis (see references 1, 5, and 30 for reviews). Recent findings suggest the involvement of the uPA-uPAR system in chemotaxis (6, 14) and in the activation of intracellular signaling pathways leading to enhanced cell proliferation, adhesion, and migration (31, 53, 68). Thus, the uPA-uPAR system participates in the regulation of a wide range of cellular activities (see reference 7 for a review).
Steady-state levels of uPA and uPAR mRNAs and proteins in many cancer cells are high due in part to enhanced mRNA stability. In several breast cancer cell lines, such as MDA-MB-231, MDA-MB-436, and BT-549, uPA and uPAR mRNAs are very stable, with half-lives of longer than 10 h (50; H. Tran, unpublished data). This observation is interesting considering that the 3′ untranslated region (UTR) of uPA and uPAR mRNAs contain AU-rich elements (AREs), sequences commonly found in the 3′ UTR of mRNAs with short half-lives.
Regulation of mRNA stability through AREs is a posttranscriptional mechanism that allows cells to fine-tune the expression of important gene products under changing environmental conditions (see references 27 and 56 for reviews). ARE-containing mRNAs encode a wide variety of proteins with diverse functions (3). The most prominent group among them encode early-response gene products, including cytokines, lymphokines, and some proto-oncoproteins (33, 59). AREs are grouped into three classes depending on the absence or presence of the consensus AUUUA pentamer and on how multiple pentameric motifs are arranged within the ARE (11). The AREs of the uPA and uPAR mRNAs belong to the class I ARE, containing one or more nontandem copies of the pentamer motif. In addition, the ARE of the uPA mRNA contains AUUUUA and AUUUUUA motifs common to several cytokine AREs (59). mRNA-destabilizing activity associated with the AREs of uPA and uPAR has been reported (49, 63), but proteins that regulate the stability of these mRNAs through interaction with their AREs have not been identified.
Many ARE-binding proteins (AUBPs) have been identified, but to date HuR is the only AUBP whose functional role in stabilizing ARE-containing mRNAs has been demonstrated by several laboratories (13, 20, 21, 55, 64). Structurally, HuR consists of three RNA recognition motifs (RRMs), with RRM2 and RRM3 connected by a short linker or hinge region (43). HuR binds to the AREs of several mRNAs (9, 25), and in the case of the c-fos ARE, it recognizes a core element of 27 nucleotides that contains AUUUA, AUUUUA, and AUUUUUA motifs, all three of which are required for maximal binding (43). The stabilizing effect of HuR on ARE-containing mRNAs has been confirmed in vivo (13, 20, 55) and in cell-free mRNA decay systems using recombinant HuR and related Hu family proteins (21, 40). Although HuR is localized predominantly in the nucleus, it contains a shuttling sequence termed HNS in the hinge region (19) and is translocated to the cytoplasm under various conditions that positively affect the stability of ARE-containing mRNAs (66, 71). These observations have led to the hypothesis that the mRNA-stabilizing activity of HuR is closely linked to its cytoplasmic localization.
We and others have reported systems in which the stability and/or translation of labile ARE-containing mRNAs is enhanced by a mechanism involving the upregulation and activation of p38 mitogen-activated protein (MAP) kinase or its upstream effectors MKK3 and MKK6 (28, 35, 39, 46, 69). Mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 (MK2), which is phosphorylated and activated by p38, is implicated in cell migration (38) and in the coexport of p38 MAP kinase from the nucleus (17). More importantly, MK2 regulates the stability and/or translation of tumor necrosis factor alpha, cyclooxygenase 2, interleukin-6 (IL-6), IL-8, c-Fos, and granulocyte-macrophage colony-stimulating factor mRNAs through their AREs (37, 39, 51, 69). It is still not known whether HuR can be phosphorylated by one of these kinases or whether its function is under the control of a regulatory pathway involving p38 MAP kinase.
We show here that HuR binds to the AREs of uPA and uPAR mRNAs and that its overexpression stabilizes these mRNAs in the cell. We also propose a link between the activation of MK2 and the cytoplasmic accumulation of HuR, which would subsequently lead to the stabilization of ARE-containing mRNAs. This represents one possible mechanism by which the stress-activated p38 MAP kinase pathway, where MK2 is downstream of p38 MAP kinase, targets ARE-binding proteins to specifically stabilize ARE-containing mRNAs.
MATERIALS AND METHODS
Plasmid DNA constructs.
Plasmid pGEX-2T-HuR was kindly provided by H. Furneaux (43). Glutathione _S_-transferase (GST) was expressed from pGEX-2T (Amersham). To construct a mammalian expression vector for hemagglutinin (HA) epitope-tagged HuR (HA-HuR), a cDNA fragment encompassing amino acids 2 to 326 of HuR was amplified by PCR using pGEX-2T-HuR as a template and inserted into the _Eco_RI-_Xho_I sites of pcDNA3-HA. The construction of pcDNA3-HA has been described previously (34). Plasmids pcDNA3-MK2-WT, pcDNA3-MK2-EE, and pcDNA3-MK2-K76R were kindly provided by M. Gaestel (18, 69). To derive a tetracycline-regulated expression vector for chimeric β-globinΔARE or β-globin-Xho mRNA, β-globin cDNA containing the uPA 3′ UTR without the ARE or β-globin-Xho was excised from pCMV-β-globinΔARE or pCMV-β-globin-Xho (50) with _Hin_dIII-_Sal_I and inserted into the same restriction sites of pPuro-TRE Bluescript (provided by A. Thiele), which harbors a tetracycline-responsive element. pPuro-TRE-β-globin-AREuPA and pPuro-TRE-β-globin-AREuPAmut containing one copy of the ARE from human uPA mRNA and a mutated version of the ARE (both AUUUA motifs mutated to AGGUA), respectively, were prepared as above from the vectors pCMV-β-globin-AREuPA and pCMV-β-globin-AREuPAmut.
Cell culture and DNA/RNA transfection.
MDA-MB-231 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37°C in the presence of 5% CO2. HeLa Tet-off cells (Clontech) were maintained as above and additionally supplemented with 100 ng of G-418 (Invitrogen) per ml. HeLa Tet-off cells were stably transfected with pPuro-TRE-βglobin-Xho, pPuro-TRE-βglobin-AREuPA, pPuro-TRE-βglobin-AREuPAmut, and pPuro-TRE-βglobin-ΔAREuPA by the calcium phosphate precipitation method (Pharmacia) and selected with puromycin (2 μg/ml). Puromycin-resistant clones were pooled, and β-globin mRNA expression was analyzed by Northern blotting. BT-549 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. Transient transfection of plasmid DNA (2 μg per 35-mm dish) using LipofectAMINE 2000 (Invitrogen) was performed as specified by the manufacturer. Transfection of small interfering RNA (siRNA) duplexes was performed as described previously (34). The sequence of siRNA used to target HuR correspond to nucleotides 150 to 168 in human HuR mRNA (where nucleotide 1 is the AUG start codon) and includes a 3′ UU overhang: sense (5′-CUU AUU CGG GAU AAA GUA GUU-3′) and antisense (5′-CUA CUU UAU CCC GAA UAA GUU-3′). The siRNA sequence homologous to luciferase mRNA has been described previously (34).
Measurement of mRNA stability in HeLa Tet-off cells and Northern blotting.
Synthesis of β-globin mRNA in HeLa Tet-off cells was stopped by the addition of 1 μg of doxycycline (tetracycline analogue) per ml. Total RNA was isolated from cells at different times after the addition of doxycycline and subjected to Northern blotting. For analysis of endogenous uPA and uPAR mRNA stability, total RNA was isolated at different times following the addition of 20 μg of 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; Fluka) per ml. A 10-μg portion of total RNA for each time point was resolved on a 1% formaldehyde-agarose gel and transferred to a nylon membrane (Roche). RNA blots were stained with methylene blue to check for equal loading and transfer. Hybridization was performed by the QuikHyb hybridization protocol (Stratagene) using random primed [α-32P]dATP-labeled rabbit β-globin cDNA probe or cDNA probes corresponding to human uPA or uPAR and human glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Hybridization signals were visualized and quantitated with a PhosphorImager (Molecular Dynamics).
Protein extraction, immunoprecipitation, and Western blotting.
Adherent cells were washed with phosphate-buffered saline (PBS) and incubated in lysis buffer (50 mM Tris-HCl [pH 7.4], 120 mM NaCl, 1% NP-40, 1 mM EDTA, 5 mM Na3VO4, 5 mM NaF, 0.5 μg of aprotinin per ml, 1 μg of leupeptin per ml) on ice for 10 min. Cells were collected by scraping and centrifuged at 20,500 × g for 10 min at 4°C. Typically, 10 μg of total protein from the supernatant (whole-cell lysate) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Schleicher & Schuell). Nuclear and cytoplasmic fractions were prepared as described previously (10). MK2 was immunoprecipitated from cell lysates (500 μg) with a rabbit polyclonal antibody (4 μg; Genex Bioscience) coupled to protein A-Sepharose (100 μl; Amersham). The phosphospecific MK2 antibody (Thr 334; Cell Signaling) was used at a dilution of 1:1,000. The primary monoclonal antibodies used were purified anti-HA (12CA5), anti-HuR (3A2; Santa Cruz), anti-cyclin A (from W. Krek), anti-hnRNPC (4F4; from G. Dreyfuss), and anti-β-tubulin (Sigma), all used at a 1:1,000 dilution. We used a horseradish peroxidase-conjugated sheep anti-mouse antibody (Amersham) as a secondary antibody (1:2,000 dilution) or horseradish peroxidase-conjugated protein A (1:5,000). An enhanced chemiluminescence detection method (ECL2; Amersham) was used, and the membrane was exposed to Kodak X-Omat LS Film.
Recombinant GST proteins.
GST fusion proteins were produced in Escherichia coli BL21(DE3) and purified using glutathione-Sepharose beads (Pharmacia) as specified by the manufacturer. Purified proteins were dialyzed against 100 volumes of buffer D (10 mM HEPES-KOH [pH 7.9], 3 mM magnesium acetate, 10% glycerol, 0.1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride 0.5 mM dithiothreitol [DTT]) for 1 h at 4°C and stored at −80°C.
In vitro synthesis of RNA transcripts.
Templates for synthesis of the AREs of uPA or uPAR mRNA were prepared from annealed DNA oligonucleotides containing T7 promoter sequences as described previously (44). The oligonucleotides used were (only sense strands are shown) AREuPA (5′-TAA TAC GAC TCA CTA TAG GGC ACT GAA TAT TTA TAT TTC ACT ATT TTT ATT TAT ATT TTT GTA ATT TTA-3′) and AREuPAR (5′-TAA TAC GAC TCA CTA TAG GGT TAT TAA TTA ATA TTC ATA TTA TTT ATT TTA TAC TTA CAT AAA GAT TTT-3′). Mutant AREs were prepared using oligonucleotides that have a guanosine (G) in place of the thymidine (T) nucleotides underlined above. In vitro transcription using T7 RNA polymerase in the presence of [α-32P]UTP was performed as specified by the manufacturer (Promega). For in vitro RNA decay assays, we prepared RNA transcripts composed of 140 nucleotides (nt) of the rabbit β-globin 3′UTR, with or without the 47-nt AREuPA and with or without a 60-residue poly(A) tail. In vitro transcription of these RNA transcripts was performed in the presence of the mRNA cap analog m7G(5′)ppp(5′)G (Amersham). The detailed procedure has been described previously (22). SP6 RNA polymerase was used as instructed by the manufacturer (Promega). All RNAs were gel purified prior to use.
RNA electrophoretic mobility shift assay (REMSA).
Radiolabeled ARE of uPA or uPAR mRNA (103 cpm) was incubated without or with 1 μg of the indicated antibodies in a 25-μl reaction mixture containing 10 μg of HeLa nuclear extract, 10 mM HEPES-KOH (pH 7.9), 3 mM magnesium acetate, 10% glycerol, 40 ng of poly(A), 0.1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, and 0.5 mM DTT. Reaction mixture were preincubated on ice for 15 min and then incubated for 15 min at 30°C. RNA-protein complexes were resolved on a 6% nondenaturing polyacrylamide gel at 4°C. The gel was dried, and radioactive signals were analyzed using a PhosphorImager.
UV cross-linking and HuR immunoprecipitation assays.
Confluent cells in 100-mm dishes were transfected with or without 10 μg of pcDNA3-MK2-K76R using Lipofectamine 2000 for 24 h. Cells were serum starved for 4 h and, where indicated, treated with 20 μM rottlerin (Calbiochem) for 1 h before the addition of 200 μM hydrogen peroxide (H2O2) for a further 2 h. Nuclear and cytoplasmic fractions were prepared as described previously (10). Total protein (100 μg) from each fraction was incubated with [α-32P]UTP-labeled AREuPA (20 × 103 cpm) in a total reaction volume of 200 μl with buffer components as described above for REMSA. Reaction mixtures were preincubated on ice for 15 min and then incubated for 15 min at 30°C. UV cross-linking was carried out in a Stratalinker (250 mJ/m2) for 5 min on ice. Unbound RNA was digested with RNases A (10 μg) and T1 (10 U) for 15 min at 37°C. Immunoprecipitation was performed by the addition to each reaction mixture of a 50% solution of protein A-Sepharose (40 μl) precoupled with anti-HuR antibodies (800 ng; 3A2). Reaction mixtures were incubated with constant mixing for 1 h at 4°C. Protein A beads were washed once with REMSA buffer and then twice each with TNET buffer (50 mM Tris-HCl [pH 7.8], 140 mM NaCl, 5 mM EDTA, 1% Triton X-100) and TNE buffer (TNET without Triton). Bound proteins were eluted in SDS loading buffer at 95°C for 5 min. Precipitated proteins were resolved by SDS-PAGE, followed by Coomassie staining and autoradiography using a PhosphorImager.
In vitro RNA decay.
HeLa S100 extracts were prepared as described previously (21). A premix containing 50 μg of total HeLa S100 proteins in IVDA buffer (10 mM Tris-HCl [pH 7.5], 100 mM potassium acetate, 2 mM magnesium acetate, 10 mM creatine phosphate, 1 mM ATP, 0.4 mM GTP, 2 mM DTT, 0.1 mM spermine) (67), and, where indicated, 200 ng of GST or GST-HuR recombinant protein was prepared (45 μl total) on ice. Aliquots of 9 μl of the premix were incubated at 37°C with 1 μl of [α-32P]UTP-labeled RNA substrate (103 cpm). Reactions were stopped at different times (0 to 80 min) by the addition of 200 μl of a high-salt buffer (25 mM Tris-HCl [pH 7.6], 0.1% SDS, 400 mM NaCl), 10 μg of calf liver tRNA was used to aid precipitation, and [α-32P]UTP-labeled AREuPAR RNA (103 cpm) was used as an internal control for the subsequent phenol-chloroform extraction and ethanol precipitation of the processed RNA. Precipitated RNA was resuspended in 10 μl of RNA loading buffer (25 mM Tris-HCl [pH 7.6], 8 M urea, 1 mM EDTA) and resolved on a 5% acrylamide gel containing 7 M urea. The gel was dried, and radioactive signals were analyzed using a PhosphorImager.
Immunofluorescence.
Cells were seeded on sterile coverslips in 35-mm dishes (1.5 × 105 cells per dish) and transfected with 2 μg of DNA by using Lipofectamine 2000. After 24 h, the cells were washed with PBS plus Ca2+ and Mg2+ [PBS(+)] and fixed in 1 ml of prewarmed 3% paraformaldehyde in PBS without Ca2+ and Mg2+ [PBS(−)] for 20 min at room temperature. The cells were permeabilized with 0.5% Triton X-100 in PBS(−) for 10 min, blocked with 5% normal goat serum for 20 min, and incubated for 2 h with a monoclonal anti-HuR (2 μg/ml) or anti-hnRNPC (1 μg/ml) antibody diluted in PBS(+) containing 1% goat serum. The cells were washed twice with PBS(+) (10 min per wash), incubated with secondary Alexa488 anti-mouse goat antibody (1:100; Molecular Probes) for 40 min, and washed three times with PBS(+) (10 min per wash). To visualize nuclei, DAPI (4′,6-diamidino-2-phenylindole; 1:5,000) was added during the last wash. The coverslips were mounted on glass slides with Fluoromount (Serva). Fluorescence was visualized with a Zeiss Axioplan 2 fluorescence microscope, and all images were captured at ×600 magnification.
RESULTS
Use of the tetracycline-regulated β-globin reporter system to study AREuPA-mediated mRNA decay in HeLa Tet-off cells.
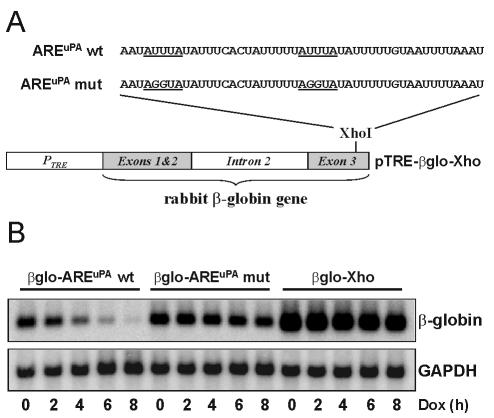
We used a tetracycline-regulated reporter system in HeLa Tet-off cells to study the stability of chimeric β-globin-AREuPA mRNA in vivo. In this system, the addition of doxycycline (a tetracycline derivative) selectively terminates the de novo synthesis of β-globin reporter mRNA, thereby avoiding the use of nonspecific transcription inhibitors known to affect some shuttling RNA binding proteins including HuR (55, 66; H. Tran, unpublished data). We inserted one copy of the AREuPA (wild type or mutant) (Fig. 1A) into the 3′ UTR of a β-globin reporter mRNA, whose expression was under the control of a tetracycline-responsive element (see Materials and Methods). Northern blot analysis of the products of an 8-h doxycycline chase showed that one copy of the AREuPA wild type was sufficient to elicit the rapid degradation of an otherwise stable rabbit β-globin reporter mRNA (Fig. 1B, compare the decay between β-globin-AREuPA wild type [_t_1/2 ≈ 2 h] and β-globin-Xho [_t_1/2 > 20 h]). Introduction of mutations that changed the two AUUUA motifs in the ARE to AGGUA (AREuPA mut) strongly interfered with the destabilizing effect of the AREuPA (Fig. 1B, compare decay between β-globin-AREuPA wild-type and β-globin-AREuPA mutant [_t_1/2 > 12 h]). The results were similar in four independent experiments; we have therefore established a reproducible system to study AREuPA-mediated mRNA decay in vivo.
FIG. 1.
The AREuPA is a functional mRNA-destabilizing element in HeLa Tet-off cells. (A) Schematic representation of tetracycline-regulated pTRE-β-globin constructs with the wild-type (wt) or mutant (mut) AREuPA inserted into the _Xho_I site of pTRE-βglobin-Xho (pTRE-βglo-Xho). (B) Northern blot analysis of the decay of chimeric β-globin mRNAs in HeLa Tet-off cells. Cells stably transfected with the indicated expression vector were subjected to an 8-h doxycycline (Dox) chase. Total RNA was isolated at the indicated times after addition of doxycycline (1 μg/ml), and 10-μg aliquots of RNA were resolved on a formaldehyde-agarose gel. Specific mRNA signals on the same Northern blot were detected using random-primed radiolabeled cDNA probes corresponding to rabbit β-globin or human GAPDH, which was used as a loading control.
HuR binds to the AREs of uPA and uPAR in vitro.
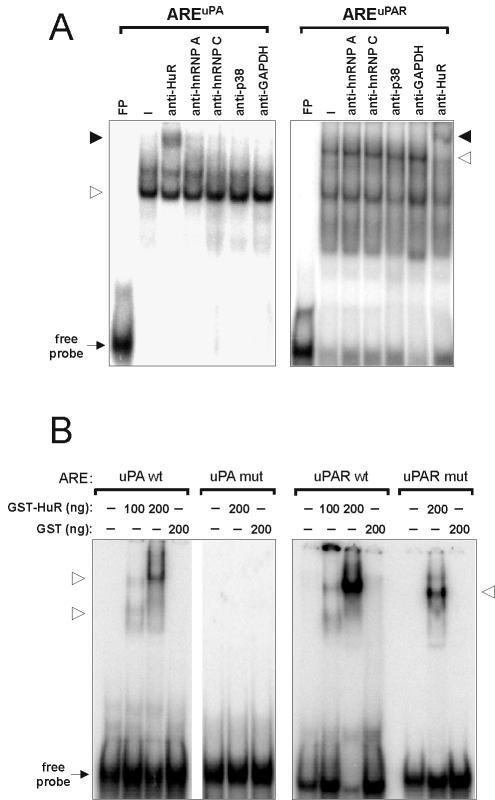
The nuclear protein HuR interacts with AREs of various mRNAs with short half-lives and enhances their stability (9). To examine the possibility that HuR can bind to the AREuPA/uPAR, gel mobility shift assays were performed using HeLa nuclear extracts and radiolabeled AREuPA/uPAR probes. While some of the ARE-protein complexes could be specifically shifted with an anti-HuR monoclonal antibody, monoclonal antibodies directed against the RNA-binding proteins heterogeneous nuclear ribonucleoprotein A (hnRNP A) and hnRNP C or against p38 MAP kinase and GAPDH did not produce any supershift (Fig. 2A). To verify the direct binding of HuR to the AREs, recombinant GST-HuR was expressed in E. coli and purified (data not shown). GST-HuR bound the AREuPA/uPAR in a dose-dependent manner, while GST itself showed no binding (Fig. 2B). When similar probes containing AGGUA stretches instead of the AUUUA pentameric motifs were used, the affinity of binding of GST-HuR to the mutant AREuPAR was much reduced and binding to the mutant AREuPA was even completely abolished (Fig. 2B). These results indicate that the AREs of both uPA and uPAR mRNAs are targets of HuR in vitro.
FIG. 2.
In vitro interaction between HuR and the AREuPA/uPAR. (A) REMSAs using radiolabeled AREuPA/uPAR RNA probes without (FP) or with 10 μg of HeLa nuclear extract in the absence (−) or presence of 1 μg of the indicated monoclonal antibodies. The open arrowhead denotes the specific RNA-protein complex. The solid arrowhead denotes the RNA-protein complex specifically shifted in reaction mixtures containing anti-HuR antibodies. (B) GST (200 ng) or increasing amounts of recombinant GST-HuR fusion proteins were incubated with fixed amounts of radiolabeled AREuPA/uPAR wild-type (wt) or mutant (mut) RNA probes and subjected to EMSA. Open arrowheads denote specific ARE-GST-HuR complexes. REMSA products were resolved on a 6% native polyacrylamide gel.
GST-HuR stabilizes AREuPA-containing RNAs in vitro.
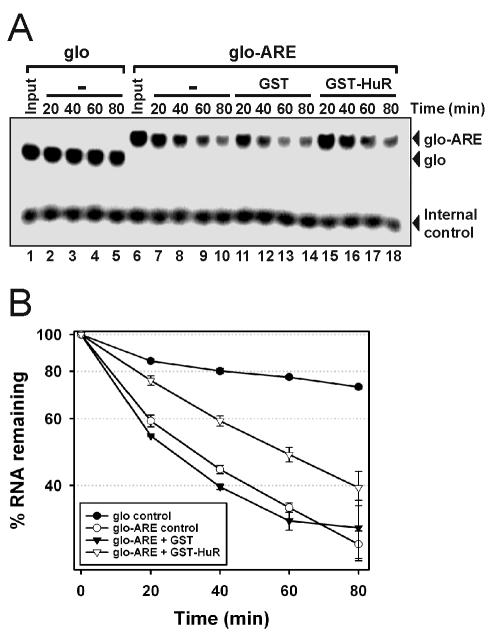
An in vitro, ARE-dependent RNA deadenylation-decay system (17) was used to assess the functional significance of HuR binding to the AREuPA. In this system, decay of the control substrate (glo), a 140-nt β-globin mRNA-derived sequence, was minimal (Fig. 3A, lanes 1 to 5) with an extrapolated _t_1/2 of >3 h (Fig. 3B). However, insertion of the AREuPA (glo-ARE) greatly stimulated the decay of the substrate (Fig. 3A, compare lanes 1 to 5 with lanes 6 to 10) reducing the _t_1/2 to ∼30 min (Fig. 3B). This AREuPA-mediated enhancement of RNA decay was suppressed twofold by the addition of GST-HuR to the decay reaction mixtures (Fig. 3A, lanes 15 to 18; Fig. 3B, _t_1/2 ≈ 60 min). In control reactions, addition of the same amount of GST protein to the decay reaction mixtures did not affect AREuPA-mediated RNA decay (Fig. 3A, lanes 11 to 14; Fig. 3B, _t_1/2 ∼ 30 min). We also used the above RNA substrates containing an additional poly(A) tail of 60 adenylates in decay assays. Interestingly, although RNA decay was suppressed, the inhibitory effect of GST-HuR on RNA deadenylation was not observed (data not shown). These results are consistent with in vivo data showing that HuR slows the decay of the mRNA body without affecting deadenylation kinetics (55) and demonstrate a specific and functional role for HuR in stabilizing AREuPA-containing RNA in vitro.
FIG. 3.
Recombinant HuR proteins stabilize AREuPA-containing RNAs in cell-free RNA decay assays. (A) Radiolabeled β-globin 3′ UTR (140 bp) with (glo-ARE) or without (glo) the AREuPA was added to a reaction mixture containing 50 μg of HeLa S100 extract and 200 ng of GST or GST-HuR (see Materials and Methods). Reaction mixtures were incubated for the indicated times at 37°C before RNA was extracted and resolved on a 7 M urea-5% acrylamide gel. Radiolabeled AREuPAR RNA was added after incubation as an internal control for the recovery of RNA during the extraction procedure. (B) Quantitation of RNA decay signals. The β-globin-to-internal-control ratio at time 0 was arbitrarily set to 100%, and the data shown represent the means and standard errors from two independent experiments.
Overexpression of HuR stabilizes ARE-containing mRNAs in vivo.
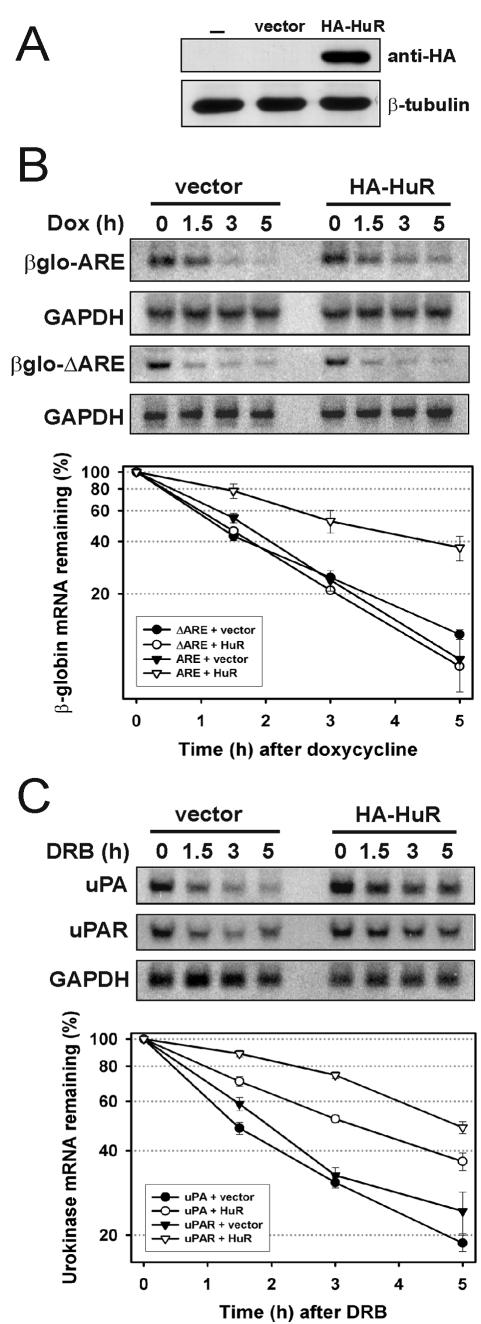
We next asked whether the stabilizing effect of HuR on AREuPA-RNA in vitro reflected its physiological role in the cell. Overexpression of HA-HuR in HeLa Tet-off cells could be achieved with high efficiency (Fig. 4A). This overexpression represents approximately 2.5 times the level of endogenous HuR as estimated from Western blotting (data not shown). The decay rate of β-globin-AREuPA mRNA in these cells was significantly reduced by overexpression of HA-HuR; the _t_1/2 increased from 1.8 to 3.2 h (Fig. 4B). We have previously shown that β-globin-ΔAREuPA mRNA containing the entire 3′ UTR of uPA mRNA except the ARE is also unstable (49). Interestingly, the decay of this mRNA was not affected by the overexpression of HA-HuR (_t_1/2 = 1.2 h in both vector- and HuR-transfected cells), suggesting that the mRNA-stabilizing effect of HuR is dependent on the presence of the AREuPA (Fig. 4B). The HeLa Tet-off cells used in this work express detectable levels of endogenous uPA and uPAR mRNAs, which have a _t_1/2 of 1.5 and 1.9 h, respectively. Similar to the effect on chimeric β-globin-AREuPA mRNA, HuR overexpression increased the stability of endogenous uPA and uPAR mRNAs approximately 2-fold and 2.5-fold (_t_1/2 increased from 1.5 to 3.2 h and from 1.9 to 5 h), respectively (Fig. 4C). The greater stabilizing effect of HuR on uPAR mRNA than on uPA mRNA correlates with the high-affinity interaction between AREuPAR and HuR in vitro, compared with the interaction between AREuPA and HuR (Fig. 2B). HuR overexpression also increased the stability of otherwise unstable uPA mRNA in MCF-10A cells, a cell line derived from normal breast tissue (data not shown). These results indicate that the AREs from uPA and uPAR mRNAs are functional targets of HuR in vivo.
FIG.4.
Overexpression of HuR in HeLa Tet-off cells stabilizes β-globin-AREuPA reporter mRNA and endogenous uPA and uPAR mRNAs. (A) HeLa Tet-off-β-globin-AREuPA cells were transfected without (−) or with 2 μg of pcDNA-HA (vector) or pcDNA-HA-HuR (HA-HuR) for 24 h before being lysed and checked for the expression of HA-tagged HuR by Western blotting using anti-HA and anti-β-tubulin (loading control) antibodies. (B) Total RNA from HeLa Tet-off-β-globin-AREuPA or HeLa Tet-off-β-globin-ΔAREuPA cells transfected as described in panel A were isolated at the times indicated after addition of 1 μg of doxycycline (Dox) per ml and resolved on a 1% formaldehyde-agarose gel. Specific mRNA signals were produced and analyzed as described in Materials and Methods and plotted on a logarithmic scale (lower panel). The β-globin-to-GAPDH ratio at time 0 h was arbitrarily adjusted to 100%, and the graphic data shown represent the means and standard errors from three independent experiments. (C) Total RNA from HeLa Tet-off-β-globin-AREuPA cells transfected as described in panel A were isolated at the times indicated after the addition of 20 μg of DRB per ml and subjected to Northern blot analysis. The same blot was sequentially hybridized with human uPA, uPAR, and GAPDH random primer-labeled cDNA probes. The lower panel shows quantitation and graphic representation of uPA and uPAR mRNA decay. The uPA-to-GAPDH or uPAR-to-GAPDH ratio at time 0 h was arbitrarily adjusted to 100%, and the data shown represent the means and standard errors from two independent experiments.
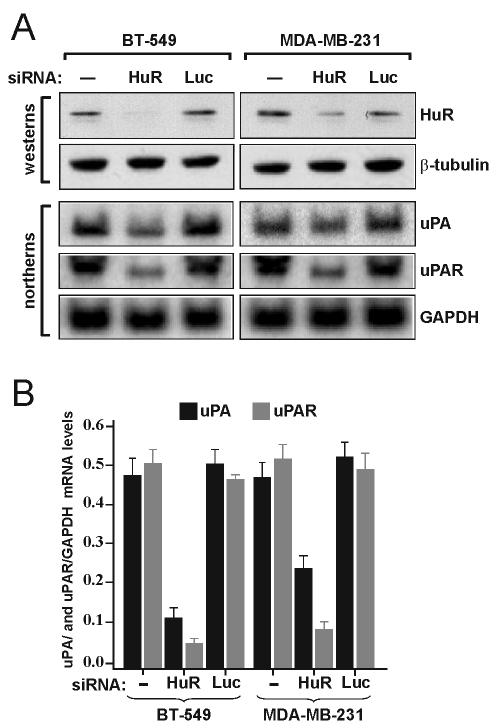
Depletion of HuR by RNA interference reduces steady-state levels of uPA and uPAR mRNAs.
The breast cancer cell lines BT-549 and MDA-MB-231 express significant levels of uPA and uPAR mRNAs, which also exhibit unusually high stability (50; Tran, unpublished data). From the results of overexpression experiments, it was argued that reducing HuR levels in these cells would exhibit effects opposite to those on uPA and uPAR mRNA levels and their stability. To reduce the level of HuR protein, these cells were transfected with an siRNA specific to HuR (siHuR) mRNA. At 48 h after transfection, cells were collected for protein and RNA analysis. HuR protein levels were reduced significantly in BT-549 cells and to a lesser extent in MDA-MB-231 cells (Fig. 5A, western blots). The steady-state levels of uPA and uPAR mRNAs were strongly downregulated in siHuR-transfected cells compared with those in buffer-treated cells or cells transfected with an siRNA specific to luciferase mRNA (Fig. 5A, Northern blots). Quantitation of these results show an approximate fivefold reduction in the levels of uPA and uPAR mRNA in BT-549 cells, as well as a twofold reduction in uPA mRNA levels and a greater than fivefold reduction in uPAR mRNA levels in MDA-MB-231 cells transfected with siRNA against HuR (Fig. 5B). Although mRNA levels were affected, a change in uPA and uPAR mRNA stability as measured by a transcription inhibition chase with DRB was not detected (data not shown; see Discussion).
FIG. 5.
Depletion of HuR by RNA interference reduces steady-state levels of endogenous uPA and uPAR mRNAs in BT-549 and MDA-MB-231 cells. (A) Cells were transfected for 48 h with buffer (−) or with siRNAs against HuR or luciferase (Luc) mRNA. Protein extracts were subjected to Western blotting using anti-HuR and anti-β-tubulin antibodies (westerns). Total RNA extracted from the same cells was subjected to Northern blot analysis. The same blot was sequentially hybridized with human uPA, uPAR, and GAPDH random primer-labeled DNA probes (northerns). (B) Quantitation of the Northern blot mRNA signals described in panel A. The data shown represent the means and standard errors from two independent experiments.
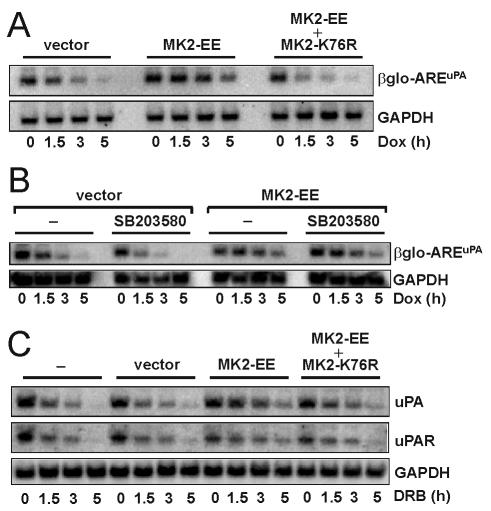
MK2 acts downstream of p38 MAP kinase to stabilize ARE-containing mRNAs.
Previously, we showed the involvement of the p38 MAP kinase pathway in stabilizing uPA and uPAR mRNAs through a mechanism engaging the ARE in MDA-MB-231 breast cancer cells (46). Because MK2 is a downstream target of p38 MAP kinase, its involvement in AREuPA-mediated mRNA stabilization was considered possible. MK2-EE (a constitutively active MK2) or MK2-K76R (a dominant negative MK2) was overexpressed by transient transfection in HeLa Tet-off cells stably expressing β-globin-AREuPA (Fig. 6A). The stability of β-globin-AREuPA mRNA in MK2-EE-transfected cells was increased approximately twofold (_t_1/2 ≈ 3 h) compared to that in vector-transfected cells (_t_1/2 ≈ 1.5 h), but this stabilizing effect was abrogated when the cells were cotransfected with a vector expressing MK2-K76R (_t_1/2 ∼ 1.2 h) (Fig. 6A). A specific inhibitor of p38 MAP kinase, SB203580, which strongly destabilizes ARE-mRNAs in MDA-MB-231 cells (46) and in HeLa Tet-off cells transfected with empty vector (_t_1/2 decreased from 1.5 h [untreated] to 0.5 h [SB203580 treated]) (Fig. 6B), did not inhibit MK2-EE-mediated mRNA stabilization (_t_1/2 ≈ 3 h in MK2-EE-transfected cells, independent of treatment with SB203580). This suggests that MK2-EE acts downstream of p38 MAP kinase. Similarly, overexpression of MK2-EE stabilized endogenous uPA and uPAR mRNAs approximately twofold (Fig. 6C). Coexpression of the dominant negative form of MK2 interfered with the stabilizing effect of MK2-EE on uPA and uPAR mRNAs (Fig. 6C). Taken together, these results indicate that p38 MAP kinase pathway-mediated stabilization of uPA and uPAR mRNA involves MK2 by a mechanism acting through the AREuPA and possibly the AREuPAR.
FIG. 6.
Overexpression of constitutively active MK2 stabilizes the β-globin-AREuPA reporter mRNA, which is not affected by p38 MAP kinase inhibition, and endogenous uPA and uPAR mRNAs. (A) HeLa Tet-off-β-globin-AREuPA cells were transfected with 2 μg of pcDNA3 (vector) or pcDNA3-MK2-EE (MK2-EE) or cotransfected with 2 μg each of pcDNA3-MK2-EE and pcDNA3-MK2-K76R (MK2-K76R) for 24 h and then treated with 1 μg of doxycycline (Dox) per ml for the times indicated, and total RNA was prepared. Northern blot analysis was performed to detect specific β-globin (βglo) and GAPDH mRNA signals. The data shown are representative of three independent experiments. (B) HeLa Tet-off-β-globin-AREuPA cells were transfected with 2 μg of pcDNA3 (vector) or pcDNA3-MK2-EE (MK2-EE) for 24 h, treated with 10 μM of SB203580 for 1 h or left untreated (−), and then treated with 1 μg of doxycycline per ml for the times indicated, and total RNA was prepared. Northern blots were sequentially probed to detect β-globin and GAPDH mRNA signals. The data shown are representative of two independent experiments. (C) HeLa Tet-off-β-globin-AREuPA cells were transfected with the vectors described in panel A or left untreated (−), and total RNA was isolated at the times indicated after the addition of 20 μg of DRB per ml and subjected to Northern blot analysis. The same blot was sequentially hybridized with human uPA, uPAR, and GAPDH random primer-labeled DNA probes. The data shown are representative of two independent experiments.
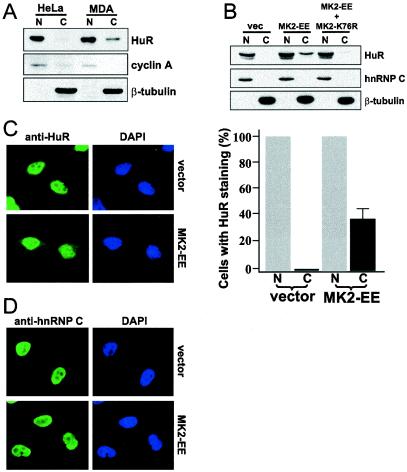
Overexpression of constitutively active MK2 induces cytoplasmic accumulation of HuR.
It has been observed that the predominantly nuclear HuR translocates to the cytoplasm under various conditions that stabilize ARE-containing mRNAs (66). We have also observed that globin-AREuPA reporter mRNA is unstable in HeLa cells, in contrast to its high stability in MDA-MB-231 cells (46). In addition, the steady-state levels and stabilities of uPA and uPAR mRNAs are high in MDA-MB-231 cells and can be downregulated by the inhibition of p38 MAP kinase (46). To see whether there is a positive correlation between cytoplasmic localization of HuR and the stability of ARE-containing mRNAs, we examined the subcellular localization of HuR in HeLa and MDA-MB-231 cells. As shown in the Western blot in Fig. 7A, HuR could be detected in the cytoplasmic fractions of MDA-MB-231 cells but not of HeLa cells, although the protein was present in the nuclear fractions of both cell lines to a similar extent. This could suggest that a positive correlation exists between the cytoplasmic localization of HuR and the stabilities of uPA, uPAR, and β-globin reporter mRNAs in these cells. In this experiment, the quality of cell fractionation was confirmed by staining the same blot with antibodies against cyclin A (nuclear) and β-tubulin (cytoplasmic) (Fig. 7A). Next we asked whether the MK2-EE-dependent stabilization of uPA, uPAR, and β-globin-AREuPA reporter mRNAs shown in Fig. 6 could be associated with changes in the cytoplasmic translocation of HuR. HeLa cells were transfected with an expression vector encoding constitutively active MK2 (MK2-EE) or empty vector, and the subcellular distribution of HuR was analyzed (Fig. 7B). HuR was not detectable in the cytoplasm of empty vector-transfected cells. When cells were transfected with MK2-EE, the level of HuR in the cytoplasm increased dramatically, whereas cotransfection with dominant negative MK2-K76R abrogated such a change. The same membrane was probed with antibodies against hnRNP C (nuclear) or β-tubulin (cytoplasmic) to confirm the quality of cell fractionation and to argue against a possible cross-contamination between nuclear and cytoplasmic fractions during the extraction procedure. These results were further confirmed by immunofluorescence studies. In cells transfected with the empty vector, HuR was visible only in the nucleus (Fig. 7C, anti-HuR/vector). In MK2-EE-overexpressing cells, although HuR was still localized predominantly in the nucleus, it was now also detectable in the cytoplasm of 35% of the cells (Fig. 7C, anti-HuR/MK2-EE and graph). This number may reflect the efficiency of DNA transfection. MK2-EE overexpression did not affect the distribution of the nuclear protein hnRNP C (Fig. 7D, anti-hnRNP C/vector and anti-hnRNP C/MK2-EE). These results demonstrate a clear correlation between MK2 activation and the redistribution of HuR to the cytoplasm.
FIG. 7.
Overexpression of constitutively active MK2 induces cytoplasmic accumulation of HuR. (A) Subcellular localization of HuR in HeLa and MDA-MB-231 cells. Nuclear (N) or cytoplasmic (C) extracts were subjected to Western blotting, and the same blot was sequentially stained with antibodies against HuR, cyclin A, and β-tubulin. (B) Subcellular fractionation of HeLa Tet-off cells transfected as described in the legend to Fig. 6A. Western blotting was performed as described in panel A, except that antibodies against HuR, hnRNP C, and β-tubulin were used. vec, vector. (C) Indirect immunofluorescence in HeLa Tet-off-β-globin-AREuPA cells grown on coverslips and transfected with 2 μg of pcDNA3 (vector) or pcDNA3-MK2-EE (MK2-EE) for 24 h. The cells were fixed, permeabilized, and stained using antibodies against HuR or hnRNP C and Alexa488-conjugated anti-mouse goat secondary antibodies. They were also treated with DAPI to visualize nuclei. Quantitation of HuR signals from the cytoplasm or nucleus of transfected cells is shown on the right. Fifteen random fields were chosen through the microscope, and fluorescent signals were assessed in 150 cells for each transfection condition. Data shown represent the means from two independent experiments; the error bar indicates the standard error of the mean. (D) All cells transfected as in panel C and stained with hnRNP C antibodies show only nuclear signals.
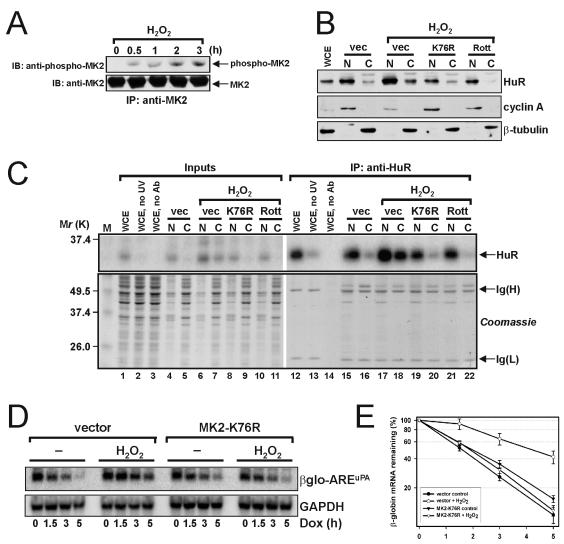
Increased binding of cytoplasmic HuR to the AREuPA and stabilization of β-globin-AREuPA mRNA by oxidative stress require MK2.
To demonstrate a physiological role for MK2 in the elevation of cytoplasmic HuR, we subjected cells to oxidative stress using hydrogen peroxide (H2O2), which is known to redistribute a significant fraction of cellular HuR to the cytoplasm (66). To test whether H2O2 can activate MK2, total MK2 was immunoprecipitated from HeLa Tet-off cells that had been serum starved for 4 h and then treated with H2O2 for 0.5, 1, 2, and 3 h. Using a phosphospecific MK2 antibody, we observed that MK2 was phosphorylated after 0.5 h and that levels of MK2 phosphorylation increased linearly up to 3 h (Fig. 8A). In subsequent experiments, we used cells treated with H2O2 for 2 h or left untreated. The cells were fractionated into nuclear and cytoplasmic fractions and analyzed by Western blotting. Whereas cytoplasmic HuR was barely detected in control (transfected with empty vector) untreated cells, H2O2-stimulated cells displayed enhanced cytoplasmic accumulation of HuR (Fig. 8B). Importantly, this effect was not observed when H2O2 was added to cells expressing dominant-negative MK2-K76R (K76R) or cells pretreated with rottlerin, an effective and specific chemical inhibitor of both MK2 and PRAK (12). We asked if the elevated levels of cytoplasmic HuR in H2O2-treated cells corresponded to increased binding of HuR to the AREuPA. Nuclear and cytoplasmic lysates prepared from cells treated as above were incubated with radiolabeled AREuPA, UV cross-linked, digested with RNases A and T1, and immunoprecipitated with monoclonal antibodies against HuR. Cross-linked RNA-protein complexes were resolved by SDS-PAGE and analyzed by autoradiography. As expected, we found increased binding of HuR (PhosphorImager signal corresponding to a ca. 36-kDa band) to the AREuPA by using cytoplasmic lysates from H2O2-treated cells but not lysates from untreated cells or cells preincubated with rottlerin or transfected with MK2-K76R (Fig. 8C, upper panel). Coomassie blue staining of the SDS-polyacrylamide gel showed equivalent loading both for the 5% inputs and the immunoprecipitates (Fig. 8C, Coomassie). Finally, the stability of β-globin-AREuPA mRNA in H2O2-treated cells was examined. The half-life of β-globin-AREuPA mRNA in untreated cells was ca. 1.8 h (Fig. 8D and E, vector, −). In contrast, the half-life of β-globin-AREuPA mRNA in H2O2-treated cells increased twofold to ca. 4 h (Fig. 8D and E, vector, H2O2). However, H2O2 treatment in cells expressing MK2-K76R did not affect the half-life of β-globin-AREuPA mRNA (Fig. 8D and E, K76R, H2O2). These results suggest that enhanced cytoplasmic accumulation of HuR and stabilization of AREuPA-containing mRNAs in response to oxidative stress require the activity of MK2.
FIG. 8.
Increased binding of cytoplasmic HuR to the AREuPA and stabilization of β-globin-AREuPA mRNA by oxidative stress requires MK2. (A) MK2 was immunoprecipitated from whole-cell lysates of HeLa Tet-off cells treated with 200 μM H2O2 for the times indicated. The cells were serum starved for 4 h before addition of H2O2. Western blotting was performed with a polyclonal antibody recognizing the phosphorylated Thr 334 of MK2. IP, immunoprecipitation; IB, immunoblotting. (B) HeLa Tet-off cells were transfected with pcDNA3 (vec) or pcDNA3-MK2-K76R (K76R) for 24 h, serum starved for 4 h, treated with 20 μM rottlerin (Rott) for 1 h or left untreated, and then treated with 200 μM H2O2 for 2 h. Preparation of nuclear (N) and cytoplasmic (C) lysates followed immediately after the treatments. WCE, whole-cell extract. Western blotting was performed with antibodies against HuR, cyclin A, and β-tubulin. (C) Cell lysates prepared from cells treated as described in panel B were incubated with radiolabeled AREuPA RNA, UV cross-linked (except for those in lanes 2 and 13), treated with RNases A and T1, and immunoprecipitated with anti-HuR antibodies (except those in lanes 3 and 14). Cross-linked RNA-protein complexes were resolved by SDS-PAGE and analyzed by autoradiography (upper panel). The most intense signal corresponded to a band migrating at ca. 36 kDa, indicated by an arrow and labeled HuR. Before autoradiographic exposure, the gel was stained with Coomassie brilliant blue to assess the equivalence of inputs and anti-HuR immunoprecipitates (lower panel). Data shown in panels A to C are representative of two independent experiments. Ig., immunoglobulin. (D) HeLa Tet-off-β-globin-AREuPA cells were transfected with 2 μg of pcDNA3 (vector) or 2 μg of pcDNA3-MK2-K76R (MK2-K76R) for 24 h. The cells were left untreated (−) or treated with 200 μM H2O2 for 2 h before the addition of 1 μg of doxycycline (Dox) per ml for the times indicated, and total RNA was prepared. Northern blot analysis was performed to detect specific β-globin and GAPDH mRNA signals. (E) Quantitation of the Northern blot mRNA signals described in panel D. Data shown represent the means and standard errors from three independent experiments.
DISCUSSION
In the present study, we have identified two proteins, HuR and MK2, involved in the regulation of uPA and uPAR mRNA stability. We found that HuR bound directly to the AREs of uPA and uPAR mRNAs and selectively stabilized RNA containing these AREs in vitro and in vivo. The degree of ARE mRNA stabilization by overexpression of HuR in our experiments (2- to 2.5-fold) is in good agreement with previous results (13, 20, 55) from experiments with different AREs and mammalian cell lines. This suggests a common mechanism by which HuR binds and stabilizes ARE-containing mRNAs. Our results are also consistent with the results of the previous report that described a role for MK2 as the main downstream substrate and effector of p38 MAP kinase in stabilizing uPA mRNA by a mechanism targeting the AREuPA (28). Importantly, we demonstrated here a novel link between the activation of MK2 and the increased cytoplasmic accumulation of HuR.
Contrary to previous reports which showed that depletion of HuR levels led to a decrease in p21, cyclin A, and cyclin B1 mRNA stability (64, 66), we did not observe such effects on uPA and uPAR mRNA stability. However, we did observe a decrease in steady-state levels of uPA and uPAR mRNAs after HuR depletion (Fig. 5). A decrease in the steady-state levels of p21, cyclin A, and cyclin B1 mRNAs was also observed in HuR-depleted cells (64, 66). The differences between our observation and that of others with respect to the effect of HuR depletion on mRNA destabilization may lie in the method used for HuR depletion and/or the cell lines used, i.e., generation of human colorectal carcinoma (RKO) cell lines constitutively expressing antisense HuR (64, 66) versus transfection of siRNAs targeting HuR in breast cancer cell lines (Fig. 5). A simpler explanation for our results is that HuR affects the processing or export of uPA and uPAR pre-mRNAs; therefore, its depletion caused the observed reduction in steady-state levels of uPA and uPAR mRNAs (Fig. 5) or, indeed, p21 and cyclin A/B1 mRNAs (64, 66). Furthermore, we cannot rule out the possibility that the remaining HuR in our siHuR-treated cells (ca. 20 to 30%) is sufficient to exert normal regulation on the stabilities of the reduced levels of uPA and uPAR mRNAs in these cells. Further experiments are needed to solve this issue.
Stabilization of ARE-containing mRNA by various stimuli that activate stress-signaling pathways has been demonstrated (2, 26, 40, 42, 45) and, on at least one occasion, was suggested to be linked to the enhanced cytoplasmic localization of HuR (66). We have provided evidence to support this idea by using the constitutively active, stress-inducible kinase MK2. We showed that HuR indeed accumulates in the cytoplasm, in conjunction with an increase in the stability of ARE-containing mRNAs, when cells transiently overexpress constitutively active MK2 (Fig. 6 and 7) or when cells are subjected to oxidative stress, which is known to activate p38 MAP kinase (47) and to cause the phosphorylation and likely activation of MK2 (Fig. 8). Similarly, conditions of hypoxic stress are known to stabilize ARE mRNAs (40) and to activate MK2 (32). How does activation of MK2 induce the cytoplasmic accumulation of HuR? Direct cellular targets of MK2 include hsp27 (39) and hnRNP A0 (57). Both these proteins have been implicated in MK2-regulated ARE mRNA stabilization (39, 57). If and how they may be involved in increasing the cytoplasmic concentration of HuR is at present unclear. However, it is unlikely that HuR is a direct cellular substrate of MK2 because (i) extensive analysis of macrophage AUBPs revealed only two major phosphorylated substrates of MK2, hnRNP A0 and RBM7 (57), and (ii) phosphorylated forms of HuR have not been found despite some effort in looking for them (8).
The mechanisms that regulate the localization or function of HuR are not fully understood, but recent studies have provided some important clues. It was shown that HuR associates with four proteins in vivo, three of which (SETα, SETβ, and pp32) are inhibitors of protein phosphatase 2A (8). These four proteins (including APRIL) interact with RRM3 and the hinge region of HuR, domains that are important for the ability of HuR to shuttle between the nucleus and cytoplasm (19). Interestingly, APRIL and pp32 are known phosphoproteins (61, 62). This is consistent with accumulating evidence that the nuclear-cytoplasmic localization of HuR is modulated by signal transduction pathways. For example, AMP-activated protein kinase (AMPK), a metabolic stress sensor, was shown to reduce the cytoplasmic levels of HuR (65). However, stimulation of glucose transport by AMP-activated protein kinase is mediated through its activation of p38 MAP kinase (70). These two independent observations are conflicting if, according to our model, the p38 MAP kinase pathway is involved in the positive regulation of cytoplasmic HuR. However, they raise the possibility that activation of p38 MAP kinase is not solely responsible for the translocation of HuR to the cytoplasm. Presumably, the activation of MK2 may be sufficient, based on speculation that it can target some ARE-binding proteins (36). Our results partly support this hypothesis because inhibition of p38 MAP kinase did not interfere with MK2-EE-induced ARE-mRNA stabilization (Fig. 6B). This also suggests that the specificity of p38 MAP kinase-mediated AREuPA-mRNA stabilization reported earlier (46) is carried out by its downstream effector, MK2. Another possibility is that the extracellular cues that lead to AMPK-mediated activation of p38 MAP kinase do not impinge on the function of these kinases in the regulation of cytoplasmic HuR. Recently, it was reported that HuR is posttranslationally modified by methylation on arginine residues residing within the hinge region (41). Interestingly, HuR methylation is increased in lipopolysaccharide-stimulated macrophages leading to HuR-mediated stabilization of tumor necrosis factor alpha mRNA (41). It is not yet clear whether this modification can modulate the interaction of HuR with its ligands, thus affecting its nucleocytoplasmic shuttling. Whatever the mechanism, this result points to the possibility that methylation and not phosphorylation of HuR may be the key regulatory modification important for its cellular localization (41). In this context, the kinase activity of MK2 may regulate the activity of the protein-arginine methyltransferase CARM1, which methylates HuR (41).
Two HuR ligands, APRIL and pp32, participate in the nuclear export of HuR mediated by the mammalian export receptor CRM1 specifically during heat shock (23). One major response of cells to heat shock is the activation of stress-signaling pathways, the most prominent of which is the p38 MAP kinase pathway (see reference 15 for a review). However, on heat shock, HuR dissociates from cytoplasmic poly(A)+ RNA and associates with nuclear poly(A)+ RNA, and a fraction of it accumulates in discrete cytoplasmic foci (24). It was postulated that increased association of HuR to nuclear poly(A)+ RNA was sufficient to produce the observed stabilizing effect on ARE-mRNAs during this specific stress condition (24). Our findings show that the distribution of HuR in the cytoplasm on transfection of constitutively active MK2 appears to be uniform (Fig. 7C). Clearly, the effect of activated MK2 on the cellular location of HuR after heat shock, if any, is occurring through an alternative mechanism. One model for HuR function suggests that HuR associates with ARE mRNAs in the nucleus and during nuclear export to protect these mRNAs from nuclear decay. This protection continues in the cytoplasm, and once the mRNAs have been translated, HuR is recycled back to the nucleus to carry out another round of ARE-binding protection (33). This model, which would presumably apply under normal cellular conditions or under a variety of different stimuli, but not heat shock, is more in line with our findings.
NF90 is a double-stranded RNA-binding protein that was recently described as a novel AUBP (60), and we have observed an RNA-dependent interaction between NF90, also known as NFAR1 (58), and HuR (unpublished data). The export of NF90 from the nucleus of activated T cells was shown to be required for the stabilization of ARE-containing IL-2 mRNA (60). Furthermore, ARE mRNA stabilization during amino acid starvation correlated with an increase in the cytoplasmic concentration of HuR (71). Notably, heat shock and other cellular stresses can cause phosphorylation-dependent nuclear coexport of MK2 and p38 MAP kinase, suggesting their role in the phosphorylation of cytoplasmic substrates (4, 16, 17). The data we have presented here indicate that the kinase activity of MK2 is important for the nuclear export of HuR. Just how MK2 can influence this event and whether it is coexported with HuR (directly or indirectly) or can modulate the function of HuR in the cytoplasm will be the focus of future studies. Taken together, these results support the idea that stabilization of labile ARE mRNAs in the cell requires the nuclear export of several proteins, including HuR and possibly MK2.
Stabilization of some ARE-containing mRNAs has been found previously in certain tumors (29, 48). Some of our observations have led us to propose that the altered distribution of HuR, and possibly other proteins, contributes to (or is important for) neoplastic transformation. HuR is abundant in the cytoplasm of a highly metastatic breast cancer cell line, MDA-MB-231 (Fig. 7A), and we have shown that constitutively active p38 MAP kinase levels are high in these cells, which coincidentally express an extremely stable uPA mRNA (46). This observation provides a plausible scenario in which the activated p38 MAP kinase pathway leads to elevated levels of cytoplasmic HuR, which carries out its role in sustaining the stability of uPA, uPAR, and other ARE-containing mRNAs in MDA-MB-231 cells. We suggest that this model is a phenomenon relevant to many other metastatic cancer cells. In light of the present finding that MK2 and HuR act downstream of p38 MAP kinase in the regulation of uPA and uPAR mRNA decay, we postulate that inhibition or expression of dominant negative forms of MK2 and HuR in MDA-MB-231 cells would affect mRNA decay and cell invasiveness in a similar manner to p38 MAP kinase inhibition (46). Indeed, our preliminary, unpublished observations using rottlerin (12), which effectively inhibits both MK2 and PRAK, a p38-regulated/activated protein kinase (52), caused a significant decrease in the stability of uPA mRNA in MDA-MB-231 cells, similar to the decreased stability exerted by inhibition of p38 MAP kinase using SB203580 (46). A distinct role for MK2 in cell migration was recently described (38). Together with the data presented here and knowledge of the established roles for the uPA-uPAR system in cell adhesion and migration, we suggest an exciting link between MK2 and the uPA-uPAR system in the regulation of this essential cellular process.
In summary, the AREs of uPA and uPAR mRNAs have been identified as new targets of the mRNA-stabilizing protein HuR. We also showed that MK2 acts downstream of p38 MAP kinase to specify stress-related, ARE mRNA-stabilizing signals, and we have linked activation of MK2 to the cytoplasmic accumulation of HuR. It remains to be seen mechanistically how stress-induced signals can cause the increased shuttling of HuR (or other proteins) from the nucleus to cytoplasm to affect the stability of labile ARE-containing mRNAs. This knowledge is required to understand how changes in mRNA stability could perpetuate or maintain certain pathological conditions.
Acknowledgments
We are grateful to A. Thiele, G. Dreyfuss, H. Furneaux, W. Krek, and M. Gaestel for the gifts of antibodies and plasmids. We thank P. King for reading the manuscript, S. Thiry for providing technical assistance, and A. Faisal for engaging in stimulating discussions.
REFERENCES
- 1.Andreasen, P. A., R. Egelund, and H. H. Petersen. 2000. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell. Mol. Life Sci. 57**:**25-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, G. K., M. A. Harding, J. P. Calvet, and E. D. Adamson. 1987. The heat shock response in HeLa cells is accompanied by elevated expression of the c-fos proto-oncogene. Mol. Cell. Biol. 7**:**3452-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakheet, T., B. R. Williams, and K. S. Khabar. 2003. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 31**:**421-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Levy, R., S. Hooper, R. Wilson, H. F. Paterson, and C. J. Marshall. 1998. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 8**:**1049-1057. [DOI] [PubMed] [Google Scholar]
- 5.Besser, D., P. Verde, Y. Nagamine, and F. Blasi. 1996. Signal transduction and the u-PA/u-PAR system. Fibrinolysis 10**:**215-237. [Google Scholar]
- 6.Blasi, F. 2001. u-PA and cell migration: urokinase receptor as a ligand for a chemotactic G-protein-coupled receptor. Haemostasis 31(Suppl. 1)**:**59. [PubMed] [Google Scholar]
- 7.Blasi, F., and P. Carmeliet. 2002. uPAR: a versatile signalling orchestrator. Nat. Rev. Mol. Cell. Biol. 3**:**932-943. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151**:**1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58**:**266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caruccio, L., and R. Banerjee. 1999. An efficient method for simultaneous isolation of biologically active transcription factors and DNA. J. Immunol. Methods 230**:**1-10. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20**:**465-470. [DOI] [PubMed] [Google Scholar]
- 12.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351**:**95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, J. L., R. Wait, K. R. Mahtani, G. Sully, A. R. Clark, and J. Saklatvala. 2001. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 21**:**721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degryse, B., S. Orlando, M. Resnati, S. A. Rabbani, and F. Blasi. 2001. Urokinase/urokinase receptor and vitronectin/alpha(v)beta(3) integrin induce chemotaxis and cytoskeleton reorganization through different signaling pathways. Oncogene 20**:**2032-2043. [DOI] [PubMed] [Google Scholar]
- 15.Dorion, S., and J. Landry. 2002. Activation of the mitogen-activated protein kinase pathways by heat shock. Cell Stress Chaperones 7**:**200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engel, K., A. Ahlers, M. A. Brach, F. Herrmann, and M. Gaestel. 1995. MAPKAP kinase 2 is activated by heat shock and TNF-alpha: in vivo phosphorylation of small heat shock protein results from stimulation of the MAP kinase cascade. J. Cell. Biochem. 57**:**321-330. [DOI] [PubMed] [Google Scholar]
- 17.Engel, K., A. Kotlyarov, and M. Gaestel. 1998. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 17**:**3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel, K., H. Schultz, F. Martin, A. Kotlyarov, K. Plath, M. Hahn, U. Heinemann, and M. Gaestel. 1995. Constitutive activation of mitogen-activated protein kinase-activated protein kinase 2 by mutation of phosphorylation sites and an A-helix motif. J. Biol. Chem. 270**:**27213-27221. [DOI] [PubMed] [Google Scholar]
- 19.Fan, X. C., and J. A. Steitz. 1998. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 95**:**15293-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17**:**3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ford, L. P., J. Watson, J. D. Keene, and J. Wilusz. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13**:**188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fritz, D. T., L. P. Ford, and J. Wilusz. 2000. An in vitro assay to study regulated mRNA stability. Sci. STKE 2000**:**PL1. [DOI] [PubMed] [Google Scholar]
- 23.Gallouzi, I. E., C. M. Brennan, and J. A. Steitz. 2001. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 7**:**1348-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 97**:**3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg-Cohen, I., H. Furneauxb, and A. P. Levy. 2002. A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J. Biol. Chem. 277**:**13635-13640. [DOI] [PubMed] [Google Scholar]
- 26.Gorospe, M., X. Wang, and N. J. Holbrook. 1998. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol. Cell. Biol. 18**:**1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 265**:**11-23. [DOI] [PubMed] [Google Scholar]
- 28.Han, Q., J. Leng, D. Bian, C. Mahanivong, K. A. Carpenter, Z. K. Pan, J. Han, and S. Huang. 2002. Rac1-MKK3-p38-MAPKAKPK2 pathway promotes urokinase plasminogen activator mRNA stability in invasive breast cancer cells. J. Biol. Chem. 107**:**48379-48385. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch, H. H., A. P. Nair, V. Backenstoss, and C. Moroni. 1995. Interleukin-3 mRNA stabilization by a trans-acting mechanism in autocrine tumors lacking interleukin-3 gene rearrangements. J. Biol. Chem. 270**:**20629-20635. [DOI] [PubMed] [Google Scholar]
- 30.Irigoyen, J. P., P. Munoz-Canoves, L. Montero, M. Koziczak, and Y. Nagamine. 1999. The plasminogen activator system: biology and regulation. Cell. Mol. Life Sci. 56**:**104-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jo, M., K. S. Thomas, A. V. Somlyo, A. P. Somlyo, and S. L. Gonias. 2002. Cooperativity between the Ras-ERK and Rho-Rho kinase pathways in urokinase-type plasminogen activator-stimulated cell migration. J. Biol. Chem. 277**:**12479-12485. [DOI] [PubMed] [Google Scholar]
- 32.Kayyali, U. S., C. M. Pennella, C. Trujillo, O. Villa, M. Gaestel, and P. M. Hassoun. 2002. Cytoskeletal changes in hypoxic pulmonary endothelial cells are dependent on MAPK-activated protein kinase MK2. J. Biol. Chem. 277**:**42596-42602. [DOI] [PubMed] [Google Scholar]
- 33.Keene, J. D. 1999. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA 96**:**5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kisielow, M., S. Kleiner, M. Nagasawa, A. Faisal, and Y. Nagamine. 2002. Isoform-specific knockdown and expression of adaptor protein ShcA using small interfering RNA. Biochem. J. 363**:**1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kontoyiannis, D., A. Kotlyarov, E. Carballo, L. Alexopoulou, P. J. Blackshear, M. Gaestel, R. Davis, R. Flavell, and G. Kollias. 2001. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 20**:**3760-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotlyarov, A., and M. Gaestel. 2001. Is MK2 (mitogen-activated protein kinase-activated protein kinase 2) the key for understanding post-transcriptional regulation of gene expression? Biochem. Soc. Trans. 30**:**959-963. [DOI] [PubMed] [Google Scholar]
- 37.Kotlyarov, A., A. Neininger, C. Schubert, R. Eckert, C. Birchmeier, H. D. Volk, and M. Gaestel. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1**:**94-97. [DOI] [PubMed] [Google Scholar]
- 38.Kotlyarov, A., Y. Yannoni, S. Fritz, K. Laass, J. B. Telliez, D. Pitman, L. L. Lin, and M. Gaestel. 2002. Distinct cellular functions of MK2. Mol. Cell. Biol. 22**:**4827-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasa, M., K. R. Mahtani, A. Finch, G. Brewer, J. Saklatvala, and A. R. Clark. 2000. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol. 20**:**4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy, N. S., S. Chung, H. Furneaux, and A. P. Levy. 1998. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273**:**6417-6423. [DOI] [PubMed] [Google Scholar]
- 41.Li, H., S. Park, B. Kilburn, M. A. Jelinek, A. Henschen-Edman, D. W. Aswad, M. R. Stallcup, and I. A. Laird-Offringa. 2002. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. J. Biol. Chem. 277**:**44623-44630. [DOI] [PubMed] [Google Scholar]
- 42.Lindstein, T., C. H. June, J. A. Ledbetter, G. Stella, and C. B. Thompson. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244**:**339-343. [DOI] [PubMed] [Google Scholar]
- 43.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271**:**8144-8151. [DOI] [PubMed] [Google Scholar]
- 44.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15**:**8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ming, X. F., M. Kaiser, and C. Moroni. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17**:**6039-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montero, L., and Y. Nagamine. 1999. Regulation by p38 mitogen-activated protein kinase of adenylate- and uridylate-rich element-mediated urokinase-type plasminogen activator (uPA) messenger RNA stability and uPA-dependent in vitro cell invasion. Cancer Res. 59**:**5286-5293. [PubMed] [Google Scholar]
- 47.Naderi, J., M. Hung, and S. Pandey. 2003. Oxidative stress-induced apoptosis in dividing fibroblasts involves activation of p38 MAP kinase and over-expression of Bax: resistance of quiescent cells to oxidative stress. Apoptosis 8**:**91-100. [DOI] [PubMed] [Google Scholar]
- 48.Nair, A. P., S. Hahn, R. Banholzer, H. H. Hirsch, and C. Moroni. 1994. Cyclosporin A inhibits growth of autocrine tumour cell lines by destabilizing interleukin-3 mRNA. Nature 369**:**239-242. [DOI] [PubMed] [Google Scholar]
- 49.Nanbu, R., P. A. Menoud, and Y. Nagamine. 1994. Multiple instability-regulating sites in the 3′ untranslated region of the urokinase-type plasminogen activator mRNA. Mol. Cell. Biol. 14**:**4920-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nanbu, R., L. Montero, D. D'Orazio, and Y. Nagamine. 1997. Enhanced stability of urokinase-type plasminogen activator mRNA in metastatic breast cancer MDA-MB-231 cells and LLC-PK1 cells down-regulated for protein kinase C—correlation with cytoplasmic heterogeneous nuclear ribonucleoprotein C. Eur. J. Biochem. 247**:**169-174. [DOI] [PubMed] [Google Scholar]
- 51.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H. D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277**:**3065-3068. [DOI] [PubMed] [Google Scholar]
- 52.New, L., Y. Jiang, M. Zhao, K. Liu, W. Zhu, L. J. Flood, Y. Kato, G. C. Parry, and J. Han. 1998. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 17**:**3372-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen, D. H., D. J. Webb, A. D. Catling, Q. Song, A. Dhakephalkar, M. J. Weber, K. S. Ravichandran, and S. L. Gonias. 2000. Urokinase-type plasminogen activator stimulates the Ras/Extracellular signal-regulated kinase (ERK) signaling pathway and MCF-7 cell migration by a mechanism that requires focal adhesion kinase, Src, and Shc. Rapid dissociation of GRB2/Sps-Shc complex is associated with the transient phosphorylation of ERK in urokinase-treated cells. J. Biol. Chem. 275**:**19382-19388. [DOI] [PubMed] [Google Scholar]
- 54.Parry, M. A., X. C. Zhang, and I. Bode. 2000. Molecular mechanisms of plasminogen activation: bacterial cofactors provide clues. Trends Biochem. Sci. 25**:**53-59. [DOI] [PubMed] [Google Scholar]
- 55.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17**:**3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ross, J. 1996. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 12**:**171-175. [DOI] [PubMed] [Google Scholar]
- 57.Rousseau, S., N. Morrice, M. Peggie, D. G. Campbell, M. Gaestel, and P. Cohen. 2002. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 21**:**6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders, L. R., D. J. Perkins, S. Balachandran, R. Michaels, R. Ford, A. Mayeda, and G. N. Barber. 2001. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 276**:**32300-32312. [DOI] [PubMed] [Google Scholar]
- 59.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46**:**659-667. [DOI] [PubMed] [Google Scholar]
- 60.Shim, J., H. Lim, J. R. Yates, and M. Karin. 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 10**:**1331-1344. [DOI] [PubMed] [Google Scholar]
- 61.Ulitzur, N., C. Rancano, and S. R. Pfeffer. 1997. Biochemical characterization of mapmodulin, a protein that binds microtubule-associated proteins. J. Biol. Chem. 272**:**30577-30582. [DOI] [PubMed] [Google Scholar]
- 62.Walensky, L. D., D. S. Coffey, T. H. Chen, T. C. Wu, and G. R. Pasternack. 1993. A novel M(r) 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal. Cancer Res. 53**:**4720-4726. [PubMed] [Google Scholar]
- 63.Wang, G. J., M. Collinge, F. Blasi, R. Pardi, and J. R. Bender. 1998. Posttranscriptional regulation of urokinase plasminogen activator receptor messenger RNA levels by leukocyte integrin engagement. Proc. Natl. Acad. Sci. USA 95**:**6296-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, W., M. C. Caldwell, S. Lin, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19**:**2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, W., J. Fan, X. Yang, S. Furer-Galban, I. Lopez de Silanes, C. von Kobbe, J. Guo, S. N. Georas, F. Foufelle, D. G. Hardie, D. Carling, and M. Gorospe. 2002. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell. Biol. 22**:**3425-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20**:**760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, Z., N. Day, P. Trifillis, and M. Kiledjian. 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 19**:**4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Webb, D. J., D. H. Nguyen, and S. L. Gonias. 2000. Extracellular signal-regulated kinase functions in the urokinase receptor-dependent pathway by which neutralization of low density lipoprotein receptor-related protein promotes fibrosarcoma cell migration and matrigel invasion. J. Cell Sci. 113**:**123-134. [DOI] [PubMed] [Google Scholar]
- 69.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18**:**4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xi, X., J. Han, and J. Z. Zhang. 2001. Stimulation of glucose transport by AMP-activated protein kinase via activation of p38 mitogen-activated protein kinase. J. Biol. Chem 276**:**41029-41034. [DOI] [PubMed] [Google Scholar]
- 71.Yaman, I., J. Fernandez, B. Sarkar, R. J. Schneider, M. D. Snider, L. E. Nagy, and M. Hatzoglou. 2002. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J. Biol. Chem. 277**:**41539-41546. [DOI] [PMC free article] [PubMed] [Google Scholar]