Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins (original) (raw)
Abstract
The nature of chaperone action in the eukaryotic cytosol that assists newly translated cytosolic proteins to reach the native state has remained poorly defined. Actin, tubulin, and Gα transducin are assisted by the cytosolic chaperonin, CCT, but many other proteins, for example, ornithine transcarbamoylase (OTC), a cytosolic homotrimeric enzyme of yeast, do not require CCT action. Here, we observe that yeast cytosolic OTC is assisted to its native state by the SSA class of yeast cytosolic Hsp70 proteins. In vitro, refolding of OTC diluted from denaturant was assisted by crude yeast cytosol and ATP and found to be directed by SSA1/2. In vivo, when OTC was induced in a temperature-sensitive SSA-deficient strain, it exhibited reduced specific activity, and nonnative subunits were detected in the soluble fraction. These findings indicate that, in vivo, the Hsp70 system assists in folding at least some newly translated cytosolic enzymes, most likely functioning in a posttranslational manner.
An important question in the study of cellular-protein folding concerns the nature of chaperone action in assisting newly translated eukaryotic cytosolic proteins to the native state. Studies of the bacterial system have established an essential role at all temperatures of the double-ring chaperonin, GroEL, in cooperation with its cochaperonin, GroES, in assisting a large number of proteins to reach native form through the actions of binding and folding in a large central cavity (1–4). However, it has been unclear whether there is similar assistance of folding of newly made proteins in the eukaryotic cytosol (5). Although the cytosolic chaperonin CCT (also known as TCP1 complex or TRiC) is present in the cytosol and has been shown to be essential in Saccharomyces cerevisiae, it seems to assist only a limited set of substrates, including actin, tubulin, and Gα-transducin (6, 7). Likewise, the Hsp90 class of chaperone is essential but seems to act on only a limited range of proteins, including steroid receptor molecules and signal-transducing kinases (8–10). Thus it remains unclear whether other newly translated eukaryotic cytosolic proteins require chaperone assistance to reach the native state.
A variety of experiments have supported the idea that chaperones are involved in the folding of newly translated eukaryotic cytosolic proteins. Early studies in HeLa cells, for example, identified interaction of a broad collective of newly translated polypeptides with Hsc70 by coimmunoprecipitation (11). Subsequent studies with both intact yeast (12) and derived translation lysates (13) showed interaction of nascent chains with the SSB class of yeast Hsp70 proteins. A functional requirement for Hsp70 action was indicated in studies translating the peroxisomal protein, firefly luciferase, in reticulocyte lysate—in an Hsp70-immunodepleted lysate, the newly translated protein failed to reach native form (14). Other studies suggested an involvement of the essential Hsp90 chaperone in folding cytosolic proteins distinct from recognized substrates such as steroid receptors and signal-transducing kinases. For example, purified Hsp90 could trap nonnative conformations of β-galactosidase after dilution from denaturant; these complexes slowly produced the native state when Hsp70, Hdj1, and ATP were added (15). In other studies, renaturation of firefly luciferase was enhanced by Hsp90 (16–18). However, in vivo studies that expressed these same proteins in a conditional lethal Hsp90 yeast mutant at nonpermissive temperatures did not detect any effect on the acquisition of normal specific activity (10).
To assess further the role of cytosolic chaperones in assisting the folding of newly translated eukaryotic cytosolic proteins, we selected a yeast cytosolic enzyme, ornithine transcarbamoylase (OTC), as reporter, and examined its folding both in vitro and in vivo in yeast lysate and intact cells.
METHODS
Proteins.
Yeast OTC with a COOH-terminal myc tag was generated by the addition of 10 codons, encoding EQKLISEEDL, to the COOH terminus of the yeast OTC (ARG3) gene. OTC and OTC-myc proteins were produced in Escherichia coli by expression of pET14a derivatives carrying the respective coding regions in BL21 cells. For purification, a soluble fraction from sonicated isopropyl β-d-thiogalactoside-induced cells (centrifuged at 15,000 × g for 15 min) was subjected to two steps of HiTrapQ chromatography (Pharmacia), first at pH 6.0 in 50 mM potassium phosphate, then at pH 8.5 in 50 mM Tris⋅HCl, followed by gel filtration on S300 (Pharmacia). For renaturation studies, purified OTC or OTC-myc (homotrimer) was denatured at 23°C for 2 h in 6 M guanidine⋅HCl and 10 mM DTT. For assay of ATP-dependent refolding, the denatured enzyme (100 μM in monomer) was diluted 100-fold into cytosol or chromatographic fractions of cytosol that had been first dialyzed for 4 h at 4°C against 50 mM Hepes, pH 7.4/100 mM KCl/5 mM MgCl2/1 mM DTT, then split into equal portions, each 100 μl in volume, one of which was supplemented with ATP to 5 mM. The pairs of mixtures were incubated at 30°C for 1 h, then 2–5 μl were assayed for OTC enzymatic activity according to Kalousek et al. (ref. 19; protocol available on request).
SSA1/2 proteins were purified from the pep4-deficient strain, JHRY20–2C (20), according to the method of Levy et al. (21), employing anion-exchange chromatography followed by ATP-agarose chromatography.
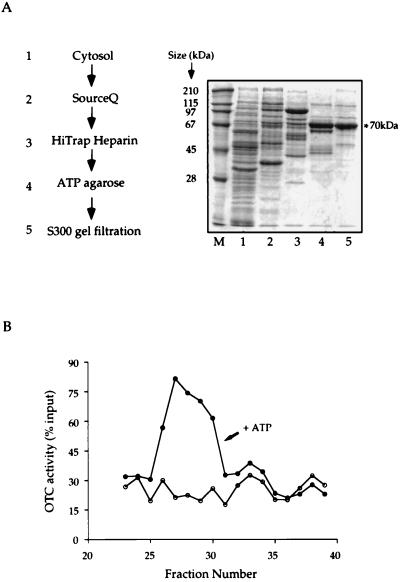
Steps of enrichment for OTC renaturing activity were carried out with the pep4-deficient strain, JHRY20–2C, which was grown to late logarithmic phase in yeast extract/peptone medium containing 2% dextrose. Spheroplasts were prepared by Zymolyase treatment (22), and lysates were prepared by Dounce homogenization in 20 mM Hepes, pH 7.4/50 mM KCl/1 mM EDTA/1 mM DTT/10% glycerol/2 mM phenylmethylsulfonyl fluoride. The lysate was cleared by 15,000 × g centrifugation for 20 min at 4°C. Notably, no mitochondrial Hsp60 was detected in the cleared lysate on immunoblotting with antiyeast Hsp60 antibodies. Subsequent chromatographic steps were carried out as indicated in Fig. 2. A gradient of KCl concentration was used to elute the SourceQ and HiTrap heparin columns. For ATP-agarose chromatography, elution was carried out with 10 mM ATP.
Figure 2.
Fractionation of yeast cytosolic extract to enrich the OTC-myc renaturing activity. (A) Steps of fractionation and SDS/PAGE analysis of fractions with peak renaturing activity from the various steps. (B) Typical profile of renaturing activity of fractions eluted from Q-Sepharose Fast Flow chromatography. Crude yeast cytosol was prepared and subjected to the sequence of fractionation steps shown (see Methods for details). A fraction from the peak of renaturing activity from each of the various steps was displayed in SDS/PAGE. In B the peak of ATP-dependent renaturing activity from Q-Sepharose FastFlow fractionation is shown, expressed as recovery of input denatured OTC in the assay of each fraction. The peak elutes at fractions 26–30 and corresponds to elution at 280–320 mM NaCl.
Peptide sequencing was carried out on tryptic peptides prepared from SDS/PAGE bands according to the method of Stone and Williams (23). The peptides identified were RLIGRNFNDPEVQGDMK, matching the sequence of SSA2, and NFTPEQISSMVLGKMK, matching the sequences of both SSA1 and SSA2.
Strains.
For preparation of yeast cytosolic extract, the strain JHRY20–2C was employed (pep4_∷_URA3 his4–519 ura3–52 leu2–3,112 trp1; ref. 20). SSA1WT (WY12) and ssa1ts (WY13) strains (produced in E.A.C.’s lab; ref. 24) carried respectively a single intact SSA gene, SSA1, or a temperature-sensitive (ts) version in the chromosomal locus, ssa1–45, in the background _MAT_α _his3–11,15 leu2–3,112 ura3–52 trp1-Δ_1 lys2 ssa2_∷_LEU2 ssa3_∷_TRP1 ssa4_∷_LYS2 pep4_∷_HIS3. The Hsp90 wild-type and mutant strains, p82a and 1–101a, kindly supplied by Susan Lindquist, are W303 derivatives (MATa leu2–3,112, trp1–1 ura3–1 ade2–1 his3–11,15 hsp82_∷_leu2 hsc82_∷_leu2) bearing CEN-TRP plasmids encoding either wild-type HSP82 or a ts version with a G170D codon change (25). The ts CCT strains, with conditional substitutions in α- or β-subunits, have been described (26) and were all studied in the background of YPH500 (_MAT_α _ura3–52 lys2–801 ade2–101 trp1-_Δ_63 his3-Δ_200 leu2_Δ_1; ref. 27). The strains ts in _CCT_ζ, alleles 25 and 47, were produced by plasmid shuffling (28) a hydroxylamine-mutagenized _CCT_ζ CEN-His3 plasmid with an unmutagenized (URA) plasmid, in the chromosomal _CCT_ζ-disrupted YPH500 derivative that contains a LEU2 marker replacing the _CCT_ζ coding sequence and carries a Ycp50 plasmid with _CCT_ζ. The strain deleted of the SSB1 and SSB2 genes, JN208, and an isogenic wild-type strain, JN54, (from E.A.C.’s lab) were _MAT_α _his3–11,15 leu2–3,112 lys2 trp1-Δ_1 ura3–52 (12). An HSP104 disruptant was produced by transformation of YPH500 with a linear DNA containing the HSP104 gene replaced between the _Eco_RI and _Kpn_I sites with URA3.
Cell Studies.
For induction of OTC-myc in yeast, wild-type or chaperone-deficient transformants (see Strains), containing the plasmid, pGALOTC-myc, bearing the OTC-myc coding segment adjoined with a GAL1 operon promoter in the plasmid, pCGS109 (29), were grown to logarithmic phase in yeast extract/peptone medium containing 2% ethanol/3% glycerol/2% raffinose and then, for induction, were transferred to yeast extract/peptone medium containing 2% galactose that had been preequilibrated to the temperature of induction. After 2 h, cells were collected, washed with 10 mM NaN3, and disrupted by vortexing (three times for 45 sec at 4°C) with an equal volume of acid-washed glass beads (≈50 μl/OD600 of cells) in a buffer containing 20 mM Hepes (pH 7.4), 50 mM KCl, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride. The lysate was centrifuged at 15,000 × g for 15 min, and the supernatant was either dialyzed, as above, in preparation for OTC enzymatic assay, or directly fractionated in SDS/10% PAGE or in a nondenaturing gel. Gels were analyzed by immunoblotting with anti-myc 9E10 monoclonal antibodies (Santa Cruz Biotechnology) as described (26). Known amounts of purified OTC-myc protein were applied in parallel with cell extract and dilutions thereof (particularly of wild-type extract) to provide standards for quantitative analysis. A linear range of densitometric signal was observed between 1 and 10 ng of OTC-myc. In the case of the SSA1WT and ssa1ts strains, OTC enzymatic activity was also measured in anti-myc immunoprecipitates of cell extract to measure directly the amount of activity from the induced OTC-myc protein as distinct from any significant activity contributed by the endogenous ARG3 gene product. This result was particularly important for specific-activity determination in the ssa1ts strain at 37°C, where only a low level of activity was detected. OTC enzyme assay was carried out by addition of ornithine and carbamoyl phosphate substrates to the protein A Sepharose beads washed in enzyme assay buffer. After 5 min at 30°C, the beads were removed, and the citrulline produced was measured colorimetrically according to the method described by Kalousek et al. (19). Results are expressed as nanomoles of citrulline produced per minute (milliunits). The amounts of enzymatic activity measured in the immunoprecipitates proved to be the same as those measured directly on the soluble fraction, supporting the idea that endogenous ARG3 does not contribute significant levels of activity in these studies.
RESULTS AND DISCUSSION
Cytosolic Yeast OTC as a Reporter of Chaperone Activity.
Yeast cytosolic OTC, a product of the ARG3 gene, like the well-characterized homologous mammalian mitochondrial urea-cycle enzyme, is a homotrimer of 36-kDa subunits (30). The mammalian OTC subunit, after import into mitochondria, requires the action of the matrix-localized chaperonin, Hsp60, to reach native form (31). Analogously, when the mature subunit of the mammalian enzyme is expressed in E. coli, the bacterial chaperonin, GroEL, is required to produce active enzyme (2). The yeast OTC subunit bears 38% identity (48% similarity) to the mammalian subunit and, like the mitochondrial protein, largely misfolds after dilution from 6 M guanidine⋅HCl (Fig. 1A, bar 2). In contrast to the folding of its mammalian homologue by Hsp60, however, the yeast OTC subunit does not seem to be recognized or assisted in folding by the homologously localized CCT. In particular, when yeast OTC subunits were expressed at nonpermissive temperature in ts lethal CCT-deficient yeast strains, they reached active form with the same kinetics as in wild-type cells (data not shown). Correspondingly, yeast OTC subunits, either translated in reticulocyte lysate, which contains CCT, or diluted from denaturant into a mixture with purified CCT, failed to become associated with the chaperonin (not shown).
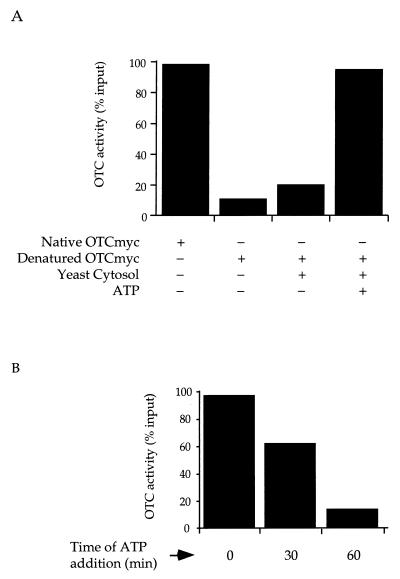
Figure 1.
(A) Crude yeast cytosol mediates ATP-dependent renaturation of OTC or a COOH-terminal myc-tagged derivative. (B) Extent of renaturation is reduced when ATP is added at later times. OTC or OTC-myc was diluted 100-fold from 6 M guanidine⋅HCl into buffer or yeast cytosolic extract prepared as described in Methods, with or without ATP. OTC enzymatic activity was assayed after 1 h. Activity was expressed as a percentage of the activity of input material that had not been subjected to denaturation.
ATP-Dependent Renaturation of Yeast OTC by Crude Yeast Cytosolic Extract.
To aid in the immunological identification of yeast OTC, a variant was produced that contained a COOH-terminal myc epitope, OTC-myc. After OTC-myc was overexpressed in E. coli and purified, it had the same physical characteristics and activity as wild-type yeast OTC (not shown). To determine whether there is an activity in yeast cytosol that assists OTC folding, purified OTC-myc was denatured in 6 M guanidine⋅HCl and diluted into a crude postmitochondrial yeast cytosolic extract. When unsupplemented extract was used, only ≈20% activity was recovered, as compared with ≈10% recovered when diluted into buffer (Fig. 1A, bar 3). However, when the extract was supplemented with 5 mM ATP at the time of dilution, nearly full recovery of OTC activity was achieved (Fig. 1A, bar 4). These results (identical for wild-type OTC) are consistent with the presence of an ATP-dependent chaperone(s) in the yeast cytosol that supports the renaturation of nonnative OTC subunits. Interestingly, when the addition of ATP to the extract containing denatured OTC was delayed, recovery was reduced progressively (Fig. 1B). Because OTC becomes incapable of reaching native form if ATP is withheld from the mixture initially, it seems that, in the absence of nucleotide, the chaperone component(s) does not compete successfully against misfolding. This behavior differs from that of chaperonins, such as Hsp60 and GroEL, which can, in the absence of nucleotide, both capture nonnative mammalian OTC and stably bind it in a productive state for a protracted period.
ATP-Dependent Renaturation Is Mediated by SSA Proteins, a Family of Yeast Cytosolic Hsp70 Proteins.
To identify the component(s) involved in OTC renaturation in the cytosolic extract, steps of chromatographic separation were carried out as outlined in Fig. 2A. A peak of renaturing activity was detected for each of these steps. The peak of a typical first step, as determined by SourceQ chromatography, is shown in Fig. 2B, which illustrates an ATP-dependent renaturation activity eluting between 280 and 320 mM NaCl. Despite excellent recovery of activity at this step, subsequent fractionation steps produced only ≈10- to 20-fold more enrichment of renaturing activity in relation to total protein concentration, with a large loss of total activity. Neither reordering the fractionation steps, nor use of alternative separation procedures (e.g., sucrose gradient fractionation), nor supplementation with nucleotide during fractionation steps could overcome these losses. Although the activity loss could reflect separation of a component that cooperates with the ATP-dependent one, recombining fractions after chromatography was also unsuccessful in restoring activity. Despite the modest enrichment of activity, there was, nevertheless, a progressive enrichment of a 70-kDa protein species associated with the active fractions.
Tryptic digestion of the 70-kDa protein species and amino acid sequencing of peptides identified peptides matching the SSA2 protein, a constitutively expressed member of the abundant cytosolic Hsp70 chaperone family, consisting of four related proteins (SSA1–4). Although members of this family are functionally redundant, at least one must be active (at relatively high concentration) for viability of yeast (32). To date, the SSA family has been implicated in posttranslational maintenance of precursor proteins in conformations competent for import into the endoplasmic reticulum and mitochondria (24, 33, 34). This action in the cytosol contrasts with the cotranslational interaction of the other class of cytosolic yeast Hsp70 proteins, the SSB class, with nascent chains exiting from the ribosome (12, 13).
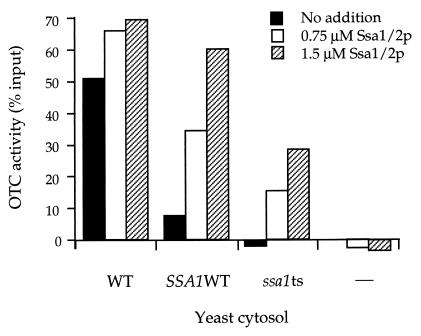
To determine whether the SSA proteins were, in fact, responsible for the OTC-renaturing activity observed in the yeast cytosolic fraction, extracts were prepared from SSA-deficient yeast strains, and their renaturing activity was compared with that of wild-type yeast cytosolic extract (Fig. 3). When an extract from a strain containing only a single intact SSA gene, SSA1, was tested, it exhibited only ≈20% of the level of activity of the wild-type extract (Fig. 3, _SSA1_WT, black bar). (SSA1 is normally present at only 30–40% of the level of SSA2 under nonstress conditions.) Even more strikingly, when an extract from a strain bearing a single SSA with a ts mutational defect (_ssa1_ts) was examined, no significant renaturing activity was detected (Fig. 3, _ssa1_ts, black bar). These results suggested that the absolute level of functional SSA protein in the cytosolic extract determined the level of renaturation observed. To test this conclusion further, purified SSA protein (a mixture of SSA1 and SSA2) obtained from a wild-type strain was added to the respective extracts, and the renaturing activity was measured. In each case, an increment of additional renaturation was observed, with the most significant effects observed on the SSA1 and _ssa1_ts extracts (Fig. 3, SSA1 WT, open and hatched bars, and _ssa1_ts, open and hatched bars). Thus the levels of functional SSA protein present in the cytosolic extract, whether determined by endogenous levels present in the cells from which extract is prepared or adjusted by exogenous supplementation, directly correlated with the extent to which OTC was renatured. These results support the idea that SSA is the ATP-dependent component that mediates the renaturation of OTC in the yeast cytosolic extract.
Figure 3.
Extent of OTC-myc renaturation by yeast cytosolic extract is related to the level of SSA (Hsp70) protein, influenced either in vivo, by the SSA genotype of the strain from which extract is prepared, or in vitro, by addition of purified SSA1/2 protein. Assay for renaturation of OTC-myc was carried out as in Fig. 1 for extracts prepared from three different yeast strains: wild-type strain (WT), with all four SSA genes intact; _SSA1_WT strain, with only SSA1 intact and SSA2–4 disrupted; or _ssa1_ts strain, with a ssa1–45 ts version of SSA1 and SSA2–4 disrupted. A control was also carried out with buffer only (–). The extracts were tested for renaturing activity unsupplemented (black bars), supplemented with the mixture of purified SSA1 and SSA2 proteins obtained from wild-type yeast grown at 30°C to levels of either 0.75 μM (open bars), or 1.5 μM (hatched bars). Activity is expressed by subtracting activity measured in the absence of ATP from that in its presence and expressing this difference as a percentage of the total input enzymatic activity. Note that provision of SSA1 and SSA2 proteins to a buffer extract is insufficient to promote OTC-myc renaturation (right-hand bars), consistent with requirement for other components that are present in cytosolic extract.
Production of Native OTC in Intact Yeast Is Assisted by SSA Proteins.
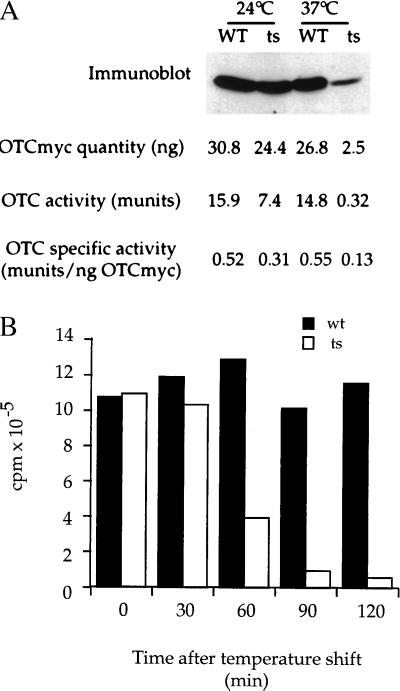
To assess whether SSA proteins play a role in OTC biogenesis in vivo, we programmed galactose-inducible expression of OTC-myc in yeast bearing either SSA1 as the single intact SSA (referred to as SSA/WT in Fig. 3) or only a ts ssa1 allele (_ssa1_ts). The two strains were grown in glucose medium at 24°C and were then shifted to galactose-containing medium at 37°C. After 90 min, cells were disrupted, and a postmitochondrial supernatant was prepared. One portion of this extract was analyzed directly for the amount of induced OTC-myc protein by solubilization, SDS/PAGE, and immunoblotting with anti-myc antibody, whereas the other portion was assayed for the amount of OTC enzymatic activity (Fig. 4A and see Methods). From extracts of identical amounts of SSA1 and _ssa1_ts mutant cells, the amounts of enzymatic activity were compared with the amounts of OTC-myc protein detected. These data provided a measurement of the specific activity of the induced OTC, indicating how efficiently the newly made protein had folded/assembled to its native active form. At 24°C, there was a 2-fold reduction in the level of enzyme activity in _ssa1_ts as compared with SSA1 cells, even though nearly the same amounts of OTC-myc protein were produced (Fig. 4A, left-hand lanes), indicating that the specific activity of the protein in the mutant strain was reduced to ≈60%. Thus, at 24°C, only ≈60% of the newly translated OTC-myc subunits reached enzymatically active form in _ssa1_ts.
Figure 4.
Specific activity of newly translated OTC-myc is reduced in SSA-deficient cells as compared with wild-type (SSA1+ SSA2–4-deleted) cells at both permissive (24°C) and nonpermissive (37°C) growth temperatures. (A) Cells grown at 24°C were placed in galactose-containing medium at either 24°C or 37°C. After 90 min, equal amounts of cells were harvested and extracted as described in Methods (note that ts SSA cells arrest growth but not protein synthesis immediately after shift to 37°C). Extracts were analyzed both for amount of induced OTC-myc protein by immunoblotting and for amount of induced OTC enzymatic activity by assay as described in Methods. The immunoblot signal for OTC-myc was scanned densitometrically to compare the relative amounts of OTC-myc in wild-type and mutant cells against standards applied to the same gels (see Methods), and specific activity was calculated by dividing the amount of OTC enzymatic activity by the relative amount of OTC-myc protein. (B) To measure total translation, cells were shifted to 37°C (t0) and pulse-radiolabeled with [35S]methionine for 5 min at each time point. Radiolabeled proteins were quantitated by trichloroacetic acid precipitation and scintillation counting. Wild-type cells (wt), solid bars; ts SSA-deficient cells (ts), open bars. Note that OTC enzymatic activity is reduced by ≈50% in ts mutant cells at 24°C as compared with wild-type cells, whereas synthesis of OTC-myc protein is reduced by only ≈20%. At 37°C, both translation and activity are greatly reduced relative to wild-type cells, with activity affected to a greater extent (45-fold vs. 10-fold effect on OTC-myc synthesis). Reduced translation of OTC-myc at 37°C in mutant cells reflects a general defect of translation (B). To establish that the measurements of reduced levels of OTC-myc protein in the ts mutant cells at 37°C determined by densitometric gel scanning were accurate, reduced amounts of wild-type cell extract were loaded side-by-side with mutant cell extract, so as to produce identical signals (not shown). The level of OTC-myc protein produced with a 90-min induction at 37°C in mutant cells is ≈10% that of wild-type cells. To measure the amounts of OTC-myc protein produced, known amounts of purified OTC-myc were immunoblotted from the same gels (see Methods).
At 37°C, SSA-deficiency affected both the synthesis of OTC subunits and their activity. The amount of OTC-myc protein in _ssa1_ts was reduced to ≈10% that in SSA1, reflecting a general defect of translation observed in pulse-labeling studies (Fig. 4B), but OTC enzymatic activity was reduced to an even greater degree in _ssa1_ts, measuring only ≈2% of that in SSA1. These relative amounts of OTC activity and protein indicate that the specific activity of OTC in the mutant strain at 37°C was only ≈25% that in SSA1 (Fig. 4A, right-hand lanes). Assuming that virtually all of the newly made OTC-myc subunits in SSA1 cells reach the native homotrimeric state, it would thus seem that only ≈25% of newly translated OTC-myc becomes active homotrimer in the SSA-deficient strain at nonpermissive temperature, with the remainder in an inactive, presumably misfolded state.
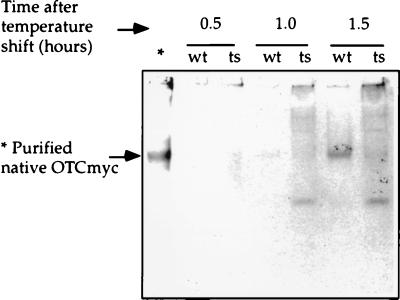
To assess the fate of nonnative OTC-myc subunits in _ssa1_ts cells, we first addressed whether they became lodged in large, insoluble aggregates. Extracts were subjected to a high-speed centrifugation step (340,000 × g for 10 min), and supernatant and pellet fractions were examined by immunoblotting. No significant amount of OTC-myc was found in the insoluble fraction (not shown). Because the nonnative subunits apparently remained soluble, we sought to observe them in nondenaturing gel electrophoresis (Fig. 5). When SSA1 extract was applied, followed by immunoblotting, virtually all of the OTC-myc was found at a single position that corresponded to that of purified homotrimer. When mutant extracts were examined at various times after temperature shift, however, only a small portion of the OTC-myc migrated to this position, and the majority was detected at other positions in the gel. In particular, 1.5 h after temperature shift (the time used in Fig. 4A), ≈80% of the subunits migrated to other positions. This behavior is in agreement with the specific activity measurements, which showed that ≈80% of induced OTC-myc in mutant cells had failed to reach active form at this time after shift. The nonnative protein seemed to occupy several different conformational states. A significant portion was detected in the gel slot, as compared with little or no signal at this position in SSA1, suggesting that some subunits had, in fact, aggregated. Another portion was found as a distinct species migrating faster than homotrimer. These molecules might be a population of unassembled OTC monomers, or alternatively, might comprise a population of OTC-myc bound to a cellular component. Whatever the exact fate of such species, we conclude that a substantial portion of the newly translated OTC produced in the SSA-deficient cells failed to be incorporated into native, active homotrimer.
Figure 5.
Nondenaturing gel analysis of soluble fractions from wild-type (wt) and SSA-deficient (ts) cells after galactose induction of OTC-myc at 37°C. At each time point, equal amounts of wild-type and mutant cells were harvested, and lysates were prepared. After ultracentrifugation at 340,000 × g for 10 min, supernatants were fractionated in an 8% polyacrylamide gel, pH 8.8, at 4°C and immunoblotting was then carried out with anti-myc monoclonal antibody 9E10. ∗ Indicates control lane displaying OTC-myc purified from E. coli.
Studies carried out here with both yeast cytosolic extracts and intact cells indicate that the newly translated subunits of OTC, a yeast cytosolic enzyme, are assisted in reaching their native homotrimeric state by the SSA class of yeast cytosolic Hsp70 proteins. Because OTC is a housekeeping enzyme of the yeast cytosol, its assistance by this class of chaperone suggests that SSA proteins may play a general role in assisting newly translated cytosolic proteins to reach the native state. Further studies examining other substrate proteins in vivo will be needed to establish such a role. Notably, the lack of insolubility of nonnative OTC subunits in SSA-deficient cells indicates that it is unlikely that other substrates could be identified readily by their selective insolubility in the mutant cells. Indeed, when _ssa1_ts cells were subjected to pulse–chase and extracts were separated into soluble and insoluble fractions, we did not observe any change in the amount or identity of insoluble proteins after two-dimensional gel electrophoresis of the urea-solubilized precipitates (S.K., unpublished results).
SSA Action in Polypeptide-Chain Folding vs. Oligomeric Assembly.
The action of SSA proteins in facilitating OTC biogenesis likely lies at the level of assisting OTC monomers to reach an assembly-competent conformation, because SSA proteins function to maintain newly made mitochondrial and endoplasmic-reticulum precursor proteins in translocation-competent states apparently by binding and conformational adjustment of monomeric species (24, 33, 34). Likewise, the action of Hsp70 proteins at the “trans” side of membranes in driving membrane translocation seems to involve interaction with monomeric species (35–39). Perhaps even more directly indicative is the fact that purified SSA1 protein, when in the presence of its cooperating partner protein YDJ1, was able to assist the refolding of monomeric firefly luciferase diluted from denaturant (21). Nevertheless, a role in assisting already folded OTC monomers to assemble into native homotrimer cannot be ruled out, particularly in light of the observation of a discrete species that may be unassembled monomers in the nondenaturing gel analysis of SSA-deficient mutant extract (Fig. 5).
SSA Assistance Is Likely to Be Posttranslational.
The action of SSA proteins in assisting biogenesis of cytosolic OTC seems to be exerted posttranslationally. In the in vitro studies with cytosolic extract, full-length OTC subunits were the target of assistance by SSA. More significantly, however, both in a yeast-cell-free translation system and in vivo, SSA proteins do not seem to form cotranslational interactions with translating polypeptides; such interactions are mediated, rather, by the class of Hsp70 proteins known as SSB proteins (12, 13). In the studies referred to below, we observed that mutational alteration of this class of proteins did not interfere with OTC biogenesis. Also consistent with posttranslational action of SSA proteins are the observations concerning their action in maintaining precursor-protein translocation competence, which also seems to be mediated post-translationally on full-length polypeptide chains (24, 33, 34).
Cooperation with SSA by Other Components.
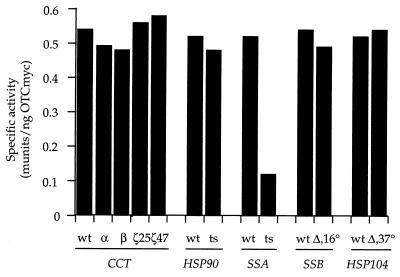
Having observed a role for SSA proteins in assisting biogenesis of OTC in the yeast cytosol, it seems appropriate to ask whether the normal cooperating partner of SSA proteins, the yeast DnaJ homologue YDJ1 (24, 40), also participates. In preliminary immunodepletion experiments in vitro that examined ATP-dependent OTC renaturation by a yeast cytosolic extract that had been immunodepleted of either SSA proteins or of YDJ1, it seems that YDJ1 is also required. Although the cooperating chaperones SSA and YDJ1 seem to be involved in folding OTC in the context of a cytosolic extract, notably, the two purified chaperones were unable to renature OTC in vitro (data not shown). This result raises questions as to whether other components participate or whether a characteristic assembly of SSA and YDJ1 must be formed (41). Parenthetically, we note that in the in vivo experiments, because translation was affected, another involved component may have become limiting at nonpermissive temperature. We tested whether other major cytosolic chaperone systems are involved in vivo by placing the galactose-inducible yeast OTC-myc gene into yeast strains bearing ts lethal versions of Hsp90 or of subunits of CCT or into strains with disrupted SSB or Hsp104 genes, the latter two of which are cold-sensitive (12) or thermointolerant (42), respectively. For each strain, OTC-myc was induced after shift to the nonpermissive temperature (except for Hsp104 where 37°C was employed), and specific activity of the newly made OTC-myc was measured as it had been in the SSA-deficient strain (Fig. 6). This survey showed that OTC biogenesis was affected only in the SSA-deficient strain.
Figure 6.
Specific activity of OTC-myc induced in a variety of chaperone-deficient mutant strains under nonpermissive growth conditions. OTC specific activity is expressed as nanomoles of citrulline produced per minute (munits) per nanogram of OTC-myc protein. Wild-type or mutant strains for the indicated cytosolic chaperones were placed at the temperature nonpermissive for growth of the mutant for 2 h (excepting Hsp104, where 37°C does not significantly affect growth) in media containing galactose to induce OTC-myc. Extracts were prepared from equal amounts of wild-type and mutant cells, as in Fig. 4 and were analyzed as in Fig. 4 both for amount of induced OTC-myc protein by immunoblotting and for amount of induced OTC enzymatic activity. For each strain, the specific activity of the induced OTC-myc protein is displayed, measured as the amount of OTC enzymatic activity per nanogram of OTC-myc protein.
The mechanism by which an Hsp70 system alone could assist folding to native form is interesting to consider. Whereas folding at chaperonin assemblies occurs in an encapsulated central cavity, this mechanism does not seem to be followed by the Hsp70 class of chaperones. Rather, it seems most likely that SSA/YDJ1 bind to local, most likely, hydrophobic, segments of nonnative cytosolic OTC and that productive folding and assembly occur after ATP-directed release from the chaperones. We conclude that, at least for this cytosolic enzyme, the major chaperone assistance in folding/assembly of its newly translated subunits is mediated posttranslationally by the SSA system. Further studies should resolve whether such action is also required by other proteins of the eukaryotic cytosol.
Acknowledgments
We thank S. Lindquist for Hsp90 yeast strains, I. Mellman and D. Toft for antibodies, and Wayne Fenton, George Farr, Bob Schumacher, and other members of the Horwich lab for valuable discussions. This work was supported by grants from the National Institutes of Health and by the Howard Hughes Medical Institute.
ABBREVIATIONS
OTC
ornithine transcarbamoylase
ts
temperature-sensitive
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- Fayet O, Ziegelhoffer T, Georgopoulos C. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horwich A L, Low K B, Fenton W A, Hirshfield I N, Furtak K. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- 3.Ewalt K L, Hendrick J P, Houry W A, Hartl F-U. Cell. 1997;90:491–500. doi: 10.1016/s0092-8674(00)80509-7. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Johnson J L, Craig E A. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 6.Willison K, Horwich A L. In: The Chaperonins. Ellis R J, editor. New York: Academic; 1996. pp. 107–136. [Google Scholar]
- 7.Farr G W, Scharl E C, Schumacher R J, Sondek S, Horwich A L. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 8.Rutherford S L, Zuker C S. Cell. 1994;79:1129–1132. doi: 10.1016/0092-8674(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 9.Bohen S P, Kralli A, Yamamoto K R. Science. 1995;268:1303–1304. doi: 10.1126/science.7761850. [DOI] [PubMed] [Google Scholar]
- 10.Nathan D F, Vos M H, Lindquist S. Proc Natl Acad Sci USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckmann R P, Mizzen L, Welch W. Science. 1990;248:850–856. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- 12.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 13.Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke B, Lopez-Buesa P, Walter W, Weidmann M, Craig E. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frydman J, Nimmesgern E, Ohtsuka K, Hartl F U. Nature (London) 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 15.Freeman B C, Morimoto R I. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher R J, Hurst R, Sullivan W P, McMahon N J, Toft D O, Matts R L. J Biol Chem. 1994;269:9493–9499. [PubMed] [Google Scholar]
- 17.Thulasiraman V, Matts R L. Biochemistry. 1996;35:13443–13450. doi: 10.1021/bi9615396. [DOI] [PubMed] [Google Scholar]
- 18.Yonehara M, Minami Y, Kawata Y, Nagai J, Yahara I. J Biol Chem. 1996;271:2641–2645. doi: 10.1074/jbc.271.5.2641. [DOI] [PubMed] [Google Scholar]
- 19.Kalousek F, Francois B, Rosenberg L E. J Biol Chem. 1978;253:3939–3944. [PubMed] [Google Scholar]
- 20.Rothman J H, Hunter C P, Valls C A, Stevens T H. Proc Natl Acad Sci USA. 1987;83:3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy E J, McCarty J, Bukau B, Chirico W. FEBS Lett. 1995;368:435–440. doi: 10.1016/0014-5793(95)00704-d. [DOI] [PubMed] [Google Scholar]
- 22.Sherman F, Wakem J. Methods Enzymol. 1991;194:38–56. doi: 10.1016/0076-6879(91)94006-x. [DOI] [PubMed] [Google Scholar]
- 23.Stone K L, Williams K R. In: A Practical Guide to Proteins and Peptide Purification for Microsequencing. 2nd Ed. Matsudaira P T, editor. New York: Academic; 1993. pp. 43–69. [Google Scholar]
- 24.Becker J, Walter W, Yan W, Craig E A. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathan D F, Lindquist S. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miklos D, Caplan S, Mertens D, Hynes G, Pitluk Z, Kashi Y, Harrison-Lavoie K, Stevenson S, Brown C, Barrell B, et al. Proc Natl Acad Sci USA. 1994;91:2743–2747. doi: 10.1073/pnas.91.7.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski R S, Boeke J S. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 29.Johnston M, Davis R W. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penninckx M, Simon J P, Wiame J-M. Eur J Biochem. 1974;49:429–442. doi: 10.1111/j.1432-1033.1974.tb03848.x. [DOI] [PubMed] [Google Scholar]
- 31.Cheng M Y, Hartl F-U, Martin J, Pollock R A, Kalousek F, Neupert W, Hallberg E M, Hallberg R L, Horwich A L. Nature (London) 1989;337:620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- 32.Werner-Washburne M, Stone D E, Craig E A. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshaies R J, Koch B D, Werner-Washburne M, Craig E A, Schekman R. Nature (London) 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- 34.Chirico W J, Waters M G, Blobel G. Nature (London) 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- 35.Kang P-J, Ostermann J, Schilling J, Neupert W, Craig E A, Pfanner N. Nature (London) 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 36.Vogel J P, Misra L M, Rose M D. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders S, Whitfield K, Vogel J, Rose M, Schekman R. Cell. 1992;69:353–366. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- 38.Matlack K E S, Plath K, Misselwitz B, Rapoport T A. Science. 1997;277:938–941. doi: 10.1126/science.277.5328.938. [DOI] [PubMed] [Google Scholar]
- 39.Matouschek A, Azem A, Ratliff K, Glick B S, Schmid K, Schatz G. EMBO J. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caplan A J, Cyr D M, Douglas M G. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 41.Motohashi K, Taguchi H, Ishii N, Yoshida M. J Biol Chem. 1994;269:27074–27079. [PubMed] [Google Scholar]
- 42.Sanchez Y, Lindquist S L. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]