Role of hybrid tRNA-binding states in ribosomal translocation (original) (raw)
Abstract
During translation, tRNAs must move rapidly to their adjacent sites in the ribosome while maintaining precise pairing with mRNA. This movement (translocation) occurs in a stepwise manner with hybrid-state intermediates, but it is unclear how these hybrid states relate to kinetically defined events of translocation. Here we analyze mutations at position 2394 of 23S rRNA in a pre-steady-state kinetic analysis of translocation. These mutations target the 50S E site and are predicted to inhibit P/E state formation. Each mutation decreases growth rate, the maximal rate of translocation (_k_trans), and the apparent affinity of EF-G for the pretranslocation complex (i.e., increases _K_1/2). The magnitude of these defects follows the trend A > G > U. Because the C2394A mutation did not decrease the rate of single-turnover GTP hydrolysis, the >20-fold increase in _K_1/2 conferred by C2394A can be attributed to neither the initial binding of EF-G nor the subsequent GTP hydrolysis step. We propose that C2394A inhibits a later step, P/E state formation, to confer its effects on translocation. Replacement of the peptidyl group with an aminoacyl group, which is predicted to inhibit A/P state formation, decreases _k_trans without increasing _K_1/2. These data suggest that movements of tRNA into the P/E and A/P sites are separable events. This mutational study allows tRNA movements with respect to both subunits to be integrated into a kinetic model for translocation.
Keywords: elongation factor G, translation, exit site, GTPase
Movement of tRNAs through the ribosome is believed to occur in a stepwise manner (1). Chemical protection experiments showed that, after peptidyl transfer, the acceptor ends of the tRNAs can move spontaneously with respect to the 50S subunit to form the hybrid state. In the hybrid state, the deacylated tRNA occupies the 30S P site and 50S E site (P/E site) while the peptidyl-tRNA occupies the 30S A site and 50S P site (A/P site). It was proposed that hybrid state formation precedes codon–anticodon movement within the 30S subunit, and only the latter event requires catalysis by elongation factor G (EF-G). Support for the hybrid-state model came from Förster resonance energy transfer (FRET) experiments in which an ≈20-Å movement of the 5′ end of the newly deacylated tRNA toward ribosomal protein L1 was inferred after peptide bond formation, whereas the position of the peptidyl group changed little (2). Because L1 lies near the 50S E site, these data are consistent with movement of tRNA into the P/E site. Further evidence came from single-molecule FRET studies that monitored the distance between probes attached to the elbows of tRNAs in the A and P sites. Fluctuations between two distinct configurations were observed that were structurally consistent with the classical and hybrid states (3). More recently, a third state of the pretranslocation (PRE) complex was observed by single-molecule FRET, consistent with one tRNA bound to the P/E site and the other bound to the A/A site (4). Experiments that monitored mRNA in ribosomal complexes showed that movement of tRNA from P/P to P/E destabilizes the codon–anticodon helix, providing additional evidence for the hybrid-state model (5). These data suggest that rotation of tRNA about the pivot-point anticodon necessary for P/E binding distorts and thereby weakens the codon–anticodon helix. Finally, the P/E-bound tRNA has been visualized directly by cryo-EM (6–8).
Several models have been proposed to describe translocation in kinetic terms. Early experiments showed that single-turnover translocation can be catalyzed by EF-G in the presence of nonhydrolyzable GTP analogs (reviewed in ref. 9). These data indicated that GTP hydrolysis is not strictly required for translocation, and it was proposed that hydrolysis is necessary for dissociation of EF-G from the post-translocation (POST) complex instead. However, more recent pre-steady-state studies showed that GTP hydrolysis precedes and accelerates codon–anticodon movement, suggesting that the energy of hydrolysis is used to drive translocation (10, 11). Experiments in which the rates of tRNA movement and inorganic phosphate (Pi) release were monitored suggested that GTP hydrolysis is followed by a conformational rearrangement, termed unlocking, that limits both codon–anticodon movement and Pi release (12). Deletion of domains 4 and 5 of EF-G slowed codon–anticodon movement and Pi release identically, supporting the existence of a rate-limiting rearrangement that precedes both events. Effects of antibiotics and ribosomal mutations indicated that, although both events are limited by the unlocking step, codon–anticodon movement and Pi release are independent of each other and probably occur in random order (12–14). Finally, ribosomal rearrangements must occur to “relock” the tRNAs in their new sites, followed by release of EF-G·GDP from the POST complex, although the kinetics of these events have yet to be fully characterized.
A number of studies suggest an important role for hybrid-state formation in the mechanism of EF-G-dependent translocation (11, 15–17). However, it remains unclear how the P/P-to-P/E and A/A-to-A/P transitions relate to the kinetically defined events of translocation. Here, we address this question by analyzing mutations in rRNA and substitutions in the translocated tRNA that inhibit movement of tRNA into the P/E and A/P sites, respectively. Based on our findings, we present a kinetic model for EF-G-dependent translocation that incorporates movement of tRNA within both subunits of the ribosome.
Results
Mutations of C2394 Confer Moderate Defects in Cell Growth.
To investigate the role of the P/E hybrid state in translocation, we mutagenized C2394, a key nucleotide of the 50S E site that interacts with the 3′-terminal adenosine (A76) of tRNA (1, 18–21). Plasmids containing rrnB without or with mutation C2394A, C2394G, or C2394U were moved into an Escherichia coli strain lacking all chromosomal rrn operons (Δ7 prrn), replacing the resident plasmid containing rrnC. Although C2394 is highly conserved, ribosomes with any base substitution at this position supported cell growth with only moderate defects in growth rate (control = 1.7 ± 0.08, C2394A = 1.1 ± 0.06, C2394G = 1.2 ± 0.05, and C2394U = 1.4 ± 0.14 doublings per hour). Similar data regarding mutation C2394G have been reported previously (22).
Mutations of C2394 Confer Defects in EF-G-Catalyzed Translocation.
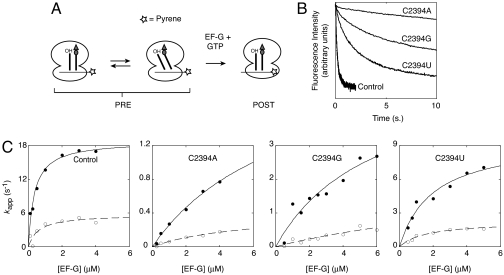
To study the effects of the E-site mutations on translocation, we used a single-turnover assay that monitors the rate of codon–anticodon movement in the ribosome by fluorescence stopped-flow (23). This assay employs an mRNA labeled at its 3′ end with pyrene, a fluorophore sensitive to the local environment. In the pretranslocation (PRE) complex, the pyrene is highly fluorescent, but when the mRNA translocates by three nucleotides toward or into the mRNA channel, the fluorescence is quenched (Fig. 1A).
Fig. 1.
Effects of 50S E-site mutations on EF-G-dependent translocation. (A) Schematic of the stopped-flow assay used to monitor codon–anticodon movement in the ribosome. Pretranslocation (PRE) complexes were formed by incubating ribosomes with pyrene-labeled message m433 and tRNATyr2 to bind the P site, and then adding Ac-Val-tRNAVal to bind the A site. Experiments were performed by rapidly mixing PRE complex (0.25 μM) with a mixture of EF-G and GTP in a stopped-flow spectrometer. Translocation of mRNA results in quenching of fluorescence as the pyrene moves toward or into the mRNA channel of the ribosome. (B) Examples of fluorescence traces for ribosomes without and with mutations at position 2394 of 23S rRNA (as indicated). The data were fit to a double-exponential function from which apparent rate constants for the fast and slow process were obtained. (C) Apparent rate constants (_k_app) for the fast (●) and slow (○) processes were plotted versus EF-G concentration for control and mutant ribosomes (as indicated). Resultant plots were fit to the equation _k_app = ([EF-G]·_k_trans)/([EF-G] + _K_1/2) to yield _k_trans, the maximal rate of translocation, and _K_1/2, the concentration of EF-G at which half-maximal rate was observed (see Table 1).
Ribosomes were purified from each Δ7 prrn strain. There were no obvious differences between control and mutant 70S ribosomes based on sucrose gradient sedimentation and SDS/PAGE analyses (data not shown). Moreover, the efficiency of tRNA binding to the P and A sites and the extent of translocation were comparable among the ribosome preparations [supporting information (SI) Fig. S1].
Control and mutant ribosomes were used to assemble PRE complexes containing pyrene-labeled mRNA (m433), tRNATyr2 in the P site, and _N_-acetyl-Val-tRNAVal (Ac-Val-tRNAVal) in the A site. These complexes were rapidly mixed with EF-G in the presence of GTP, and decreased pyrene fluorescence was observed at different rates depending on the mutation (Fig. 1B). In each case, the decrease in fluorescence intensity over time did not fit a single-exponential function, but instead fit a double-exponential function (Figs. S2–S5). This phenomenon was also seen when ribosomes isolated from E. coli MRE600 were used (data not shown) and has been observed previously by others (ref. 13 and S. Joseph, personal communication). Presumably, the two phases reflect two populations of ribosomes undergoing translocation at different rates, although the basis for this putative population heterogeneity remains unclear. The fraction amplitude corresponding to the fast (_A_1 ≈ 0.5) and slow (_A_2 ≈ 0.5) processes remained constant regardless of whether the ribosomes harbored an E-site mutation. Thus, this putative PRE complex heterogeneity seems unrelated to the ability of the ribosomes to adopt the hybrid-state conformation. In fact, the fast and slow processes were similarly affected by the mutations (see below), suggesting that both processes involve movement of tRNA through the P/E site.
Mutations of C2394 Decrease _k_trans and Increase _K_1/2.
Apparent rates were measured at several concentrations of EF-G to derive _k_trans, the maximal rate of translocation, and _K_1/2, the concentration of EF-G at which half-maximal rate was observed (Fig. 1C and Table 1). In control ribosomes, when Ac-Val-tRNAVal was translocated to the P site, _k_trans for the fast process was found to be 19 s−1, with a _K_1/2 of 0.36 μM, consistent with previously reported values (11, 12). Mutation C2394A increased _K_1/2 by 23-fold and decreased _k_trans by 8-fold; C2394G increased _K_1/2 by 13-fold and decreased _k_trans by 4-fold; and C2394U increased _K_1/2 by 4-fold and decreased _k_trans by 2-fold. Kinetic parameters for the slow process were also derived, and these parameters were similarly influenced by substitution of C2394 (Table 1). The degree to which these E-site mutations affected each kinetic parameter followed the trend A > G > U, the same trend that was observed when growth rate was measured (see above).
Table 1.
Kinetic parameters for EF-G-dependent translocation in control and mutant ribosomes
| Ribosomes | tRNA translocated to the P site | Fast process | Slow process | ||
|---|---|---|---|---|---|
| _k_trans, s−1 | _K_1/2, μM | _k_trans, s−1 | _K_1/2, μM | ||
| Control | Ac-Val-tRNAVal | 19 | 0.36 | 5.8 | 0.61 |
| C2394A | Ac-Val-tRNAVal | 2.4 | 8.1 | 0.42 | 5.8 |
| C2394G | Ac-Val-tRNAVal | 4.7 | 4.8 | ND | ND |
| C2394U | Ac-Val-tRNAVal | 8.0 | 1.3 | 2.1 | 1.2 |
| Control | Val-tRNAVal | 4.3 | 0.48 | 1.1 | 0.28 |
| Control | Ac-Phe-tRNAPhe | 39 | 1.1 | NA | NA |
| Control | Phe-tRNAPhe | 0.21 | 0.14 | 0.05 | 1.4 |
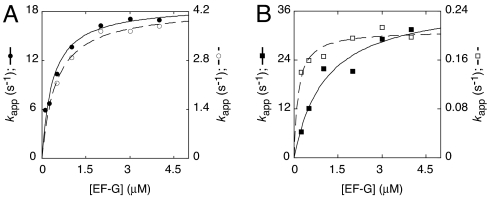
Replacing the _N_-Acetylaminoacyl Group with an Aminoacyl Group Decreases _k_trans but Does Not Increase _K_1/2.
Chemical protection and single-molecule FRET experiments provided evidence that aminoacyl-tRNA is defective in A/P state formation (1, 3, 4). The aminoacyl group decreases the apparent rate and extent of translocation, suggesting an important role for the A/P state in translocation (5, 24, 25). Mutation G2553U, predicted to destabilize tRNA in the A/A site, suppressed the inhibitory effect of the aminoacyl group, further suggesting that this group inhibits the A/A-to-A/P transition (5). To investigate the role of the A/P state in translocation, we analyzed the kinetics of mRNA movement when aminoacyl-tRNA was translocated to the P site (Fig. 2). When Val-tRNAVal was translocated, _k_trans was reduced by 4-fold compared with the control (Ac-Val-tRNAVal), whereas _K_1/2 of EF-G for the PRE complex was unaffected (Fig. 2A, Table 1, Fig. S6). We repeated the experiment in a different context, comparing translocation of Ac-Phe-tRNAPhe to that of Phe-tRNAPhe. Replacing Ac-Phe with Phe decreased _k_trans by 190-fold, whereas no increase in _K_1/2 was observed (Fig. 2B, Table 1, Figs. S7 and S8). If anything, the Phe substitution seemed to enhance the apparent affinity of EF-G for the PRE complex. However, analysis of the slow process did not indicate a decrease in _K_1/2 (Table 1), suggesting that the _K_1/2 value for the fast process may be an underestimate. Nevertheless, these data are similar to the Val-tRNAVal case, except that the rate decrease conferred by the Phe substitution was much larger.
Fig. 2.
Substitutions predicted to inhibit A/P state formation decrease _k_trans but do not increase _K_1/2. PRE complexes were formed by incubating control ribosomes with pyrene-labeled mRNA and tRNATyr2 to bind the P site, followed by addition of Ac-Val-tRNAVal, Val-tRNAVal, Ac-Phe-tRNAPhe, or Phe-tRNAPhe to bind the A site. Apparent rates of translocation were measured at several concentrations of EF-G to obtain _K_1/2 and _k_trans for each A-site tRNA species translocated: Ac-Val-tRNAVal (A, ●), Val-tRNAVal (A, ○), Ac-Phe-tRNAPhe (B, ■), and Phe-tRNAPhe (B, □). Plots include only the apparent rate constants for the fast process.
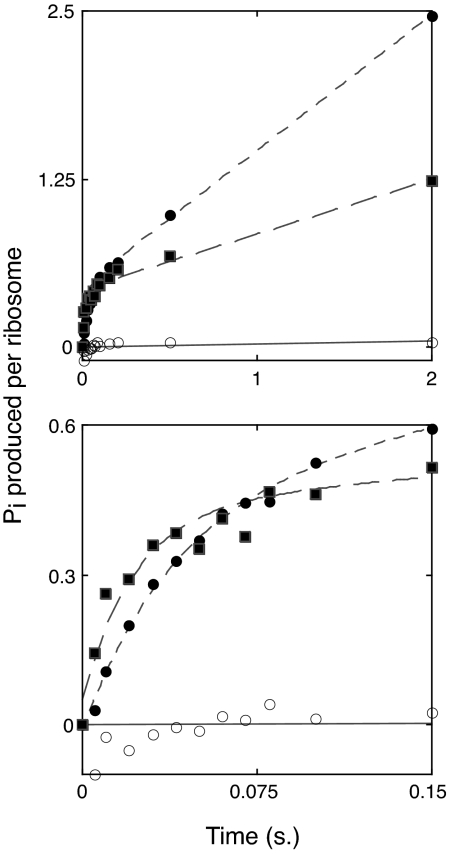
Mutation C2394A Does Not Decrease the Rate of EF-G-Dependent GTP Hydrolysis.
Next, we tested whether mutation C2394A affected the GTP hydrolysis step of EF-G-dependent translocation (Fig. 3). To measure the rate of single-turnover GTP hydrolysis, we used chemical quench flow as described in ref. 26. PRE complexes were assembled with control or C2394A ribosomes and then mixed with EF-G·[γ-32P]GTP for various periods of time before quenching with 0.6 M perchloric acid. The amount of GTP hydrolyzed at each time point was then determined by extraction and quantification of 32Pi. As shown in Fig. 3, under the conditions used (25 μM GTP, 1.5 μM EF-G, 1 μM PRE complex), a burst phase corresponding to single-turnover GTP hydrolysis was observed, followed by a linear phase corresponding to multiple-turnover hydrolysis. The apparent single-turnover (burst) rate was similar in control (25 s−1) and C2394A (44 s−1) ribosomes. The burst amplitude was also similar, although somewhat (15%) lower in the mutant ribosomes. Multiple-turnover GTP hydrolysis was noticeably inhibited by C2394A, which may result from substantially slower translocation in these ribosomes (Fig. 1C). The fact that C2394A did not decrease the rate of single-turnover GTP hydrolysis suggests that the mutation affects neither the initial binding of EF-G nor the GTP hydrolysis step. Rather, C2394A inhibition of overall translocation must be due to inhibition of a step following GTP hydrolysis, presumably that of P/E state formation. Our findings are consistent with an earlier study showing that the acylation state of the P-site tRNA (which strongly influences the P/P-to-P/E transition) does not affect the rate of single-turnover GTP hydrolysis (10).
Fig. 3.
Mutation C2394A does not affect the apparent rate of GTP hydrolysis. PRE complexes containing tRNATyr in the P site and Ac-Val-tRNAVal in the A site were rapidly mixed with EF-G·[γ-32P]GTP for indicated amounts of time before quenching with 0.6 M perchloric acid and quantifying the amount of 32Pi produced. For complexes without (●) or with (■) mutation C2394A, the reaction exhibited burst kinetics with exponential and linear phases corresponding to single- and multiple-turnover GTP hydrolysis, respectively. Burst rates (control = 25 s−1; mutant = 44 s−1) and amplitudes (control = 0.46; mutant = 0.39) deduced from curve-fitting were similar for the two complexes. Substantially less GTP hydrolysis was evident in the absence of ribosomes (○). Upper and Lower represent the same experiment; Lower shows a narrower time window.
Discussion
Here, we study the contribution of a key E-site nucleotide, C2394, to EF-G-dependent translocation. Each substitution for C2394 decreases _k_trans and increases _K_1/2, and the magnitude of the defects follows the trend A > G > U. The same trend is observed when growth rate in the Δ7 prrn background is measured, suggesting that translocation limits growth rate in those strains expressing mutant ribosomes. The same trend (A > G > U > WT) is also observed when the stability of mRNA in complexes containing P-site deacylated tRNA is assessed in an mRNA repositioning assay (S.E.W., K. G. McGarry, and K.F., unpublished data). In this assay, mRNA repositioning correlates with the ability of tRNA to bind the P/E site (5, 27). Thus, effects of these E-site mutations on growth, translocation, and P/E-binding all correlate. The fact that purine substitutions at position 2394 are more deleterious can be rationalized in structural terms. In the 50S E site, the 3′-terminal adenosine of tRNA (A76) intercalates between 23S rRNA nucleotides G2421 and A2422 and forms hydrogen bonds with C2394 (19, 21). Replacement of C2394 with either purine should not only disrupt H-bonding to A76 but also sterically inhibit stacking of A76 between G2421 and A2422.
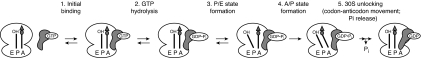
In contrast to the effects of substitutions for C2394 on both _k_trans and _K_1/2, substituting Val-tRNAVal for Ac-Val-tRNAVal in the PRE complex decreases _k_trans with little effect on _K_1/2 (Table 1). The model presented in Fig. 4 to describe translocation provides a qualitative explanation of these results. In the model, EF-G·GTP binding and rapid, reversible GTP hydrolysis (steps 1 and 2; refs. 10 and 11) are followed first by movement of deacylated tRNA to give the P/E state (step 3) and then by movement of peptidyl-tRNA to give the A/P state (step 4), which is probably equivalent to the INT complex described earlier (11). The A/P state is then converted to the POST complex by codon–anticodon movement, which coincides with 30S unlocking and Pi release (step 5; ref. 12) and is the rate-determining step for the process. Steps 4 and 5 may be conformationally linked: thus, EF-G-dependent translocation is accelerated by either mutation of the 50S A site or lengthening of the peptidyl group (17), and each of these changes promotes the A/A-to-A/P transition (4).
Fig. 4.
Model for EF-G-dependent translocation that incorporates tRNA movements with respect to both subunits. See text for details.
Substitution at C2394 should have the greatest effect on the rate constant for step 3, whereas substitution of Val-tRNAVal for Ac-Val-tRNAVal should principally affect the rate constant for step 4. Kinetic modeling based on the model presented in Fig. 4 shows that, under certain assumptions and in accord with the results presented in Table 1, reductions in the value of _k_3 can lead to larger effects on _K_1/2 than on _k_trans, whereas reductions in _k_4 can be confined largely to _k_trans (SI Appendix).
Independent formation of the P/E and A/P states is consistent with results of recent kinetic and single-molecule FRET experiments (4, 11). In the work of Pan et al. (11), which examined translocation of a PRE complex containing tRNAfMet in the P site and fMetPhe-tRNAPhe in the A site, the P/E state could be demonstrated only in the presence of viomycin, leading to the conclusion that, in the absence of antibiotic, _k_4≫_k_3. The large reduction in _k_4 that should result from substituting Val-tRNAVal for Ac-Val-tRNAVal might also lead to some accumulation of the P/E state.
According to our model, EF-G·GTP can dissociate from the PRE complex but EF-G·GDP cannot (Fig. 4). We have considered an alternative model in which EF-G·GDP can dissociate from the PRE complex, allowing multiple GTP molecules to be hydrolyzed per translocation event (SI Appendix). One prediction of this alternative model is that the apparent rates of GTP hydrolysis and translocation would be identical, which is clearly not the case (Figs. 1 and 3; refs. 10 and 11). Thus, the idea of a futile cycle of GTP hydrolysis in either control or mutant ribosomes is incompatible with the data.
Binding of EF-G to ribosomal complexes can be stabilized in the presence of nonhydrolyzable GTP analogs or certain antibiotics such as fusidic acid. A number of groups have analyzed structural differences between ribosomal complexes with EF-G trapped in the GTP form [e.g., 5-guanylyl imidodiphosphate (GDPNP)] versus the GDP form in an effort to understand the role of GTP hydrolysis in the mechanism of EF-G-dependent translocation (6, 27–31). A particularly intriguing observation from these studies was that bound EF-G promotes an intersubunit rotation (termed ratchet subunit rotation or RSR) in complexes containing P-site deacylated tRNA. This RSR correlates with P/E-bound tRNA and movement of the L1 stalk toward the 50S E site. In a complementary study, it was shown that deacylation of P-site tRNA enhances EF-G binding, suggesting that the RSR conformation (with P/E-tRNA) stabilizes EF-G (32). On the basis of these studies, it was recently proposed that initial binding of EF-G·GTP promotes hybrid-state formation, which is followed by GTP hydrolysis (8). Here the timing of hybrid-state formation versus GTP hydrolysis is largely based on results obtained with EF-G·GDPNP, which is taken as a model for EF-G·GTP. However, recent kinetic studies (11), along with our present results, indicate that EF-G catalyzes GTP hydrolysis before hybrid state formation. This apparent disagreement is readily resolved if it is assumed that, during translocation, ribosome-bound EF-G·GDPNP can be a model for bound EF-G·GDP·Pi, as suggested elsewhere (11), with hybrid state formation preceding Pi release. Support for this assumption is provided by the recent structural characterization of an archaeal initiation factor aIF2·GDP·Pi complex, in which the position of the switch 2 region is similar to that seen in the aIF2·GDPNP complex (33).
Integral to our model is that step 2, GTP hydrolysis, is reversible. This is consistent with studies showing that, within the active sites of ATPases and GTPases, including eukaryotic initiation factor eIF2 (34), the free energy of NTP hydrolysis approaches zero, in sharp contrast to the thermodynamics in solution (35–39). Lorsch and colleagues (34) have provided compelling evidence for an internal equilibrium between GTP and GDP·Pi within the nucleotide-binding pocket of eIF2 with an estimated equilibrium constant of 0.5. When eIF2·GTP·Met-tRNA interacts with a model preinitiation complex, hydrolysis of GTP occurs rapidly regardless of whether a start codon is present or absent. However, in the latter case, the amplitude of GTP hydrolysis is reduced and the subsequent step of Pi release is blocked. The authors propose that before start codon recognition an internal equilibrium between GTP and GDP·Pi is established. Subsequent recognition of the start codon allows Pi release to occur, making hydrolysis of GTP irreversible. The mechanism of EF-G-dependent translocation is similar in that Pi release occurs much more slowly than GTP hydrolysis (12, 26) and is limited by a conformational rearrangement (12, 40). Further experiments will be required to determine the internal equilibrium position of GTP and GDP·Pi within ribosome-bound EF-G.
Materials and Methods
Strains and Plasmids.
Strains expressing homogeneous populations of ribosomes harboring each substitution of C2394 were constructed as described in ref. 41. Plasmid replacement was confirmed by purification of plasmid DNA from each strain and sequencing the relevant region of the 23S rRNA gene.
Biochemical Reagents.
Tight-couple ribosomes, acylated tRNAs, mRNAs, and EF-G were prepared as described in ref. 25. Messages m432 (5′-AAGGAAAUAAUGUUUACUUUGUU-3′) and m433 (5′-AAGGAAAUAACAUUUACGUAGCU-3′) contained a 2′-amino-pyrene modification on the 3′-terminal uridine and were purchased from Dharmacon.
Translocation Experiments.
To form PRE complexes, control or mutant ribosomes (1.5 μM) were first incubated with mRNA (m432 or m433; 1.25 μM) and tRNATyr2 (1.5 μM) in buffer 1 [50 mM Tris (pH 7.6), 100 mM NH4Cl, 15 mM MgCl2] for 10 min at 37°C to fill the P site. Then, Ac-Val-tRNAVal, Ac-Phe-tRNAPhe, Val-tRNAVal, or Phe-tRNAPhe (1.5 μM) was added (as indicated) and the reactions were incubated for 10 more minutes to bind the A site. PRE complexes were then diluted 5-fold (to 0.25 μM) with buffer 1 containing GTP (1 mM) just before performing an experiment. To form EF-G·GTP, EF-G (at various concentrations as indicated) was incubated with GTP (1 mM) in buffer 2 [50 mM Tris (pH 7.6), 30 mM NH4Cl, 70 mM KCl, 5 mM MgCl2, 6 mM 2-mercaptoethanol] for 5 min at 37°C. To measure the rate of mRNA movement, equal volumes of PRE complex (0.25 μM) and EF-G·GTP (as indicated), were rapidly mixed at 37°C in an SX18-MV stopped-flow spectrometer (Applied Photophysics) essentially as described in ref. 23. Data were fit to a single- or double-exponential curve by using the SX18-MV software. Apparent rates were then plotted versus EF-G concentration and fit to the equation _k_app = ([EF-G]·_k_trans)/([EF-G] + _K_1/2).
GTPase Experiments.
The rate of single-turnover GTP hydrolysis was determined essentially as described in ref. 26. To form PRE complexes, control or mutant ribosomes (2 μM) were first incubated with message m404 (4 μM; see ref. 42) and tRNATyr2 (3 μM) in buffer 3 [50 mM Tris (pH 7.6), 30 mM NH4Cl, 70 mM KCl, 7 mM MgCl2, 1 mM DTT] for 10 min at 37°C to fill the P site. Then, Ac-Val-tRNAVal (2.5 μM) was added to bind the A site and the complexes were placed on ice. To form EF-G·GTP, EF-G (3 μM) was incubated with [γ-32P]GTP (Perkin–Elmer; 50 μM, ≈1,000 dpm/pmol) in buffer 3 for 2 min at 37°C and then placed on ice. Equal volumes of PRE complex (1 μM final) and EF-G·GTP (1.5 μM final) were then rapidly mixed at 25°C in a quench-flow apparatus (Kintek) for indicated times before quenching with 0.6 M HClO4 containing 1.8 mM KH2PO4, as described (26). Data were fit to the equation y = A_·exp(−_k_app1·_t) + k_app2·_t + C; where A is the burst amplitude, _k_app1 is the apparent burst rate, t is time, _k_app2 is the apparent turnover rate, and C is a constant.
Supplementary Material
Supporting Information
Acknowledgments.
We thank C. Squires and S. Quan for E. coli strain SQZ10; M. Ibba, J. Tomsic, and Z. Suo for useful discussions; H. Roy and J. Tomsic for expert technical advice; V. Gopalan for use of his Kintek quench flow apparatus; and M. Ibba and D. Qin for comments on the manuscript. This work was supported by National Institutes of Health Grants GM072528 (to K.F.) and GM071014 (to B.S.C.) and a Herta Camerer Gross Fellowship from Ohio State University (to S.E.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 2.Odom OW, Picking WD, Hardesty B. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry. 1990;29:10734–10744. doi: 10.1021/bi00500a004. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon–anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal RK, et al. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor DJ, et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spirin AS. Ribosomal translocation: Facts and models. Prog Nucleic Acid Res Mol Biol. 1985;32:75–114. doi: 10.1016/s0079-6603(08)60346-3. [DOI] [PubMed] [Google Scholar]
- 10.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 11.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 13.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 14.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lill R, Robertson JM, Wintermeyer W. Binding of the 3′ terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg JS, Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc Natl Acad Sci USA. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bocchetta M, Xiong L, Shah S, Mankin AS. Interactions between 23S rRNA and tRNA in the ribosomal E site. RNA. 2001;7:54–63. doi: 10.1017/s1355838201001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 21.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 22.Sergiev PV, et al. Function of the ribosomal E-site: A mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 24.Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 25.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 26.Pan D, Kirillov S, Zhang CM, Hou YM, Cooperman BS. Rapid ribosomal translocation depends on the conserved 18–55 base pair in P-site transfer RNA. Nat Struct Mol Biol. 2006;13:354–359. doi: 10.1038/nsmb1074. [DOI] [PubMed] [Google Scholar]
- 27.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 29.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 30.Wilson KS, Nechifor R. Interactions of translational factor EF-G with the bacterial ribosome before and after mRNA translocation. J Mol Biol. 2004;337:15–30. doi: 10.1016/j.jmb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 32.Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 33.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc Natl Acad Sci USA. 2007;104:18445–18450. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 35.White HD, Taylor EW. Energetics and mechanism of actomyosin adenosine triphosphatase. Biochemistry. 1976;15:5818–5826. doi: 10.1021/bi00671a020. [DOI] [PubMed] [Google Scholar]
- 36.Bowater R, Zimmerman RW, Webb MR. Kinetics of ATP and inorganic phosphate release during hydrolysis of ATP by rabbit skeletal actomyosin subfragment 1. Oxygen exchange between water and ATP or phosphate. J Biol Chem. 1990;265:171–176. [PubMed] [Google Scholar]
- 37.Allin C, Ahmadian MR, Wittinghofer A, Gerwert K. Monitoring the GAP catalyzed H-Ras GTPase reaction at atomic resolution in real time. Proc Natl Acad Sci USA. 2001;98:7754–7759. doi: 10.1073/pnas.131549798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips RA, Hunter JL, Eccleston JF, Webb MR. The mechanism of Ras GTPase activation by neurofibromin. Biochemistry. 2003;42:3956–3965. doi: 10.1021/bi027316z. [DOI] [PubMed] [Google Scholar]
- 39.Chakrabarti PP, Suveyzdis Y, Wittinghofer A, Gerwert K. Fourier transform infrared spectroscopy on the Rap. RapGAP reaction, GTPase activation without an arginine finger. J Biol Chem. 2004;279:46226–46233. doi: 10.1074/jbc.M405603200. [DOI] [PubMed] [Google Scholar]
- 40.Seo HS, et al. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 41.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13:2348–2355. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker SE, Fredrick K. Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J Mol Biol. 2006;360:599–609. doi: 10.1016/j.jmb.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information