The NY-1 Hantavirus Gn Cytoplasmic Tail Coprecipitates TRAF3 and Inhibits Cellular Interferon Responses by Disrupting TBK1-TRAF3 Complex Formation (original) (raw)
Abstract
Pathogenic hantaviruses replicate within human endothelial cells and cause two diseases, hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. In order to replicate in endothelial cells pathogenic hantaviruses inhibit the early induction of beta interferon (IFN-β). Expression of the cytoplasmic tail of the pathogenic NY-1 hantavirus Gn protein is sufficient to inhibit RIG-I- and TBK1-directed IFN responses. The formation of TBK1-TRAF3 complexes directs IRF-3 phosphorylation, and both IRF-3 and NF-κB activation are required for transcription from the IFN-β promoter. Here we report that the NY-1 virus (NY-1V) Gn tail inhibits both TBK1-directed NF-κB activation and TBK1-directed transcription from promoters containing IFN-stimulated response elements. The NY-1V Gn tail coprecipitated TRAF3 from cellular lysates, and analysis of TRAF3 deletion mutants demonstrated that the TRAF3 N terminus is sufficient for interacting with the NY-1V Gn tail. In contrast, the Gn tail of the nonpathogenic hantavirus Prospect Hill virus (PHV) failed to coprecipitate TRAF3 or inhibit NF-κB or IFN-β transcriptional responses. Further, expression of the NY-1V Gn tail blocked TBK1 coprecipitation of TRAF3 and infection by NY-1V, but not PHV, blocked the formation of TBK1-TRAF3 complexes. These findings indicate that the NY-1V Gn cytoplasmic tail forms a complex with TRAF3 which disrupts the formation of TBK1-TRAF3 complexes and downstream signaling responses required for IFN-β transcription.
Hantaviruses predominantly replicate within vascular endothelial cells and cause two human diseases, hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome (6, 32, 53). Viruses causing hemorrhagic fever with renal syndrome include Hantaan, Seoul, and Puumala viruses, while NY-1 virus (NY-1V), Sin Nombre virus, and Andes virus (ANDV) are among hantaviruses that cause hantavirus pulmonary syndrome. In contrast, Prospect Hill virus (PHV) and Tula virus are hantaviruses which are not associated with any human disease (44). Hantaviruses are negative-stranded RNA viruses, which contain three genome segments and encode four viral proteins (43, 44). The 142-residue-long cytoplasmic tail of the NY-1V Gn protein (also called G1) has recently been shown to regulate cellular interferon (IFN) responses (1). NY-1V and Hantaan virus (HTNV) inhibit the induction of IFN-stimulated genes (ISGs) at early times after infection while PHV fails to regulate early ISG responses (14, 26). Consistent with this finding, the Gn tail of PHV does not inhibit IFN transcription (1). These findings demonstrate a prominent difference between pathogenic and nonpathogenic hantaviruses and focus attention on cellular IFN pathways and signaling proteins that are regulated by pathogenic hantaviruses.
Viral induction of type I IFN (alpha/beta IFN [IFN-α/β]) is a vital component of innate cellular immune responses that limit viral replication (42, 46). Viruses are detected by intracellular and extracellular sensors, which direct signaling pathway activation and result in IFN-β transcriptional responses (20, 30, 37, 51). Intracellularly, viral components are detected by RIG-I and MDA5, which direct signaling responses through mitochondrial proteins IPS-1/MAVS/Cardiff/VISA and in turn bind TRAF3 (23, 41, 52). TRAF3 connects upstream sensory responses to downstream effector functions by forming a complex with TBK1 and directing TBK1 phosphorylation of the transcription factor IRF-3 (10, 33). Phosphorylated IRF-3 dimerizes, translocates to the nucleus, and directs transcription by binding to the IFN-β promoter (18, 19, 35, 36, 48). Transcription from the IFN-β promoter requires the activation of both IRF-3 and NF-κB, and following IFN induction and secretion, IFN directs autocrine and paracrine transcriptional responses through the activation of IFN receptor-directed signaling pathways (19, 48).
Viral replication requires the successful negotiation and regulation of innate cellular responses, and many viruses block signaling pathways that direct IFN transcription in order to bypass cellular regulatory mechanisms (51). Hantaviruses are grown in Vero E6 cells which are themselves deficient in the induction of type I IFN (8). However, following infection of IFN-competent human endothelial cells it is reported that pathogenic, but not nonpathogenic, hantaviruses regulate the early induction of ISGs (14, 26). The Gn cytoplasmic tail of NY-1V, but not PHV, blocks RIG-I- and TBK1-directed IFN-β transcriptional responses but is unable to inhibit transcription directed by a constitutively active IRF-3 protein (1). These findings suggested that the NY-1V Gn tail regulates IFN signaling responses at the level of the TBK1 complex, although specific cellular targets of Gn tail regulation have not been defined.
Components of TBK1 complexes are compelling targets for IFN regulation since TBK1 directs the activation of both transcription factors required for IFN transcription (38). TBK1 activates NF-κB through interactions with TRAF2, and TBK1-TRAF3 complexes link upstream viral sensors to IRF-3-directed transcriptional responses. In fact, TRAF3 appears to be indispensable for IFN-β transcription since TRAF3-knockout cells fail to induce type I IFN responses directed by virtually all upstream pathway activators (33).
In this report we demonstrate that the expressed NY-1V Gn tail coprecipitates the N-terminal domain of TRAF3 and disrupts the formation of TRAF3-TBK1 complexes. Infection of human endothelial cells by pathogenic NY-1V, but not nonpathogenic PHV, also blocked TBK1-TRAF3 complex formation. Thus, the disruption of TBK1-TRAF3 complexes by the recombinant expressed Gn tail is also observed within hantavirus-infected cells. The NY-1V Gn tail also blocked TBK1- and TRAF2-directed NF-κB activation, suggesting that the Gn tail is capable of regulating both NF-κB- and IRF-3-directed transcriptional responses. These findings suggest a mechanism whereby pathogenic hantaviruses inhibit the induction of IFN-β by preventing TBK1-TRAF3 complex formation and demonstrate that TRAF3 is a cellular target for pathogenic hantavirus regulation of IFN responses.
MATERIALS AND METHODS
Cells and virus.
Biosafety level 3 facilities were used for cultivating hantaviruses. HEK 293 cells and Vero E6 cells were grown in Dulbecco's modified Eagle's medium (DMEM), 10% fetal calf serum, l-glutamine, penicillin, and streptomycin (Gibco). NY-1V and PHV were cultivated in Vero E6 cells, and their titers were determined as previously described (39). Viral stocks contained 5 × 105 focus-forming units per ml.
Transfection and hantavirus infection.
Vero E6 cells were transfected with TBK1 and TRAF3 expression vectors using Lipofectamine HD (Invitrogen) in six-well plates. Virus was adsorbed to cells 48 h posttransfection at a multiplicity of infection of 1 for 1 h at 37°C. Cells were washed and maintained in DMEM with 2% fetal calf serum. Two days postinfection cells were lysed in 200 μl of coimmunoprecipitation lysis buffer (20 mM HEPES, pH 7.45, 75 mM KCl, 1.5 mM MgCl, 0.5 mM EDTA, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 μM orthovanadate, 5 μM sodium pyrophosphate, 1 μM NaF) per well of a six-well plate (16). Lysates were clarified by centrifugation (30 min at 13,000 × g), and TBK1 was immunoprecipitated with an anti-myc monoclonal antibody. Protein A/G Plus agarose beads (Santa Cruz Biotechnology) were added, and immunoprecipitated proteins were analyzed as indicated.
Antibodies.
Monoclonal anti-GAL4 antibody (GAL4 [RK5C1], sc-510) was purchased from Santa Cruz Biotechnology, and monoclonal anti-FLAG M2 (F3165) antibody was purchased from Sigma-Aldrich. Anti-myc monoclonal antibody (R950-25) was purchased from Invitrogen. Antinucleocapsid rabbit polyclonal serum directed at the NY-1V nucleocapsid protein was used to detect N protein from both NY-1V and PHV as previously described (12, 13). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit immunoglobulin G (IgG) antibodies were purchased from Amersham Biosciences.
Protein expression plasmids.
NY-1V Gn cytoplasmic tail (pBIND-NY1G1cyto) and PHV Gn cytoplasmic tail (pBIND-PHVG1cyto) were expressed as C-terminal fusions to upstream GAL4 tags as previously described (45). pRK-FLAG-TRAF2 and pRK-FLAG-TRAF3 expression vectors have been previously described (40). Plasmids expressing TRAF3 truncations (pRK-T3 N415) were generated by inserting stop codons at amino acid position 415 in TRAF3 using a QuikChange site-directed mutagenesis kit (Stratagene). The pc-TBK1 expression vector has been previously described (38).
Transfections.
Transfections were performed in HEK 293 cells in six-well plates (Corning, Inc.) using Lipofectamine 2000 transfection reagent (Invitrogen) as previously described (45). In coimmunoprecipitation experiments, cells were transfected with 1 μg of each plasmid as indicated for 16 h and washed with phosphate-buffered saline, and samples were analyzed 48 h posttransfection. For luciferase assays, cells were transfected using calcium phosphate as previously described (1).
Western blot analysis.
Gn cytoplasmic tail, TRAF3, and TBK1 expression was evaluated following cotransfection of HEK 293 cells treated with MG132 (50 μM) or the MG132 diluent, dimethyl sulfoxide, by Western blotting as previously described (45). Cells were lysed in 2× Laemmli sample buffer and analyzed by Western blotting 48 h posttransfection using anti-GAL4 (Gn tail) (1:2,000) or anti-myc (TBK1) (1:1,000). Blots were washed with Tris-buffered saline (100 mM NaCl, pH 7.4, 0.1% Tween 20) and incubated with horseradish peroxidase-conjugated anti-mouse IgG (1:3,000). Western blots were developed by fluorography in the presence of ECL reagent (Amersham), and blotting assays were performed three times with similar results.
Immunoprecipitation assays.
HEK 293 cells were transfected with 1 μg each of plasmids expressing Gn tail, TRAF3, or TBK1. Two days posttransfection, cells were washed with phosphate-buffered saline and lysed in coimmunoprecipitation lysis buffer (16). The NY-1V Gn tail is proteasomally degraded and stabilized by the presence of MG132. MG132 (50 μM) was added 6 h prior to cell lysis unless otherwise indicated. Lysates were clarified by centrifugation, and Gn tail or TBK1 proteins were immunoprecipitated as above with anti-GAL4 or anti-myc monoclonal antibodies, respectively, and Protein A/G Plus agarose beads. Coprecipitated proteins were analyzed by Western blotting as indicated. The inability to immunoprecipitate the full-length Gn protein and assay it by Western blotting precluded similar analysis with the full-length Gn protein.
Luciferase assays.
HEK 293 cells were cotransfected with a κB-Luc reporter or the pISRE-luc reporter (Clontech Laboratories) along with a constitutively active Renilla luciferase expression vector (pRL) (Promega) as previously described (1). In addition cells were transfected with TBK1 or TRAF2 expression vectors and NY-1V or PHV Gn tail or N-protein expression vectors as indicated (1). Cells were transfected with a constant amount of total DNA through the addition of empty pBIND vector. Cells were lysed 48 h posttransfection in passive lysis buffer (Promega) and analyzed using a Turner Designs TD 20/20 luminometer. Firefly luciferase activity was normalized to Renilla luciferase levels to standardize cell and transfection differences between samples. Luciferase assays were performed at least three times with similar results.
RESULTS
The NY-1V Gn cytoplasmic tail inhibits IFN-β and NF-κB transcriptional responses.
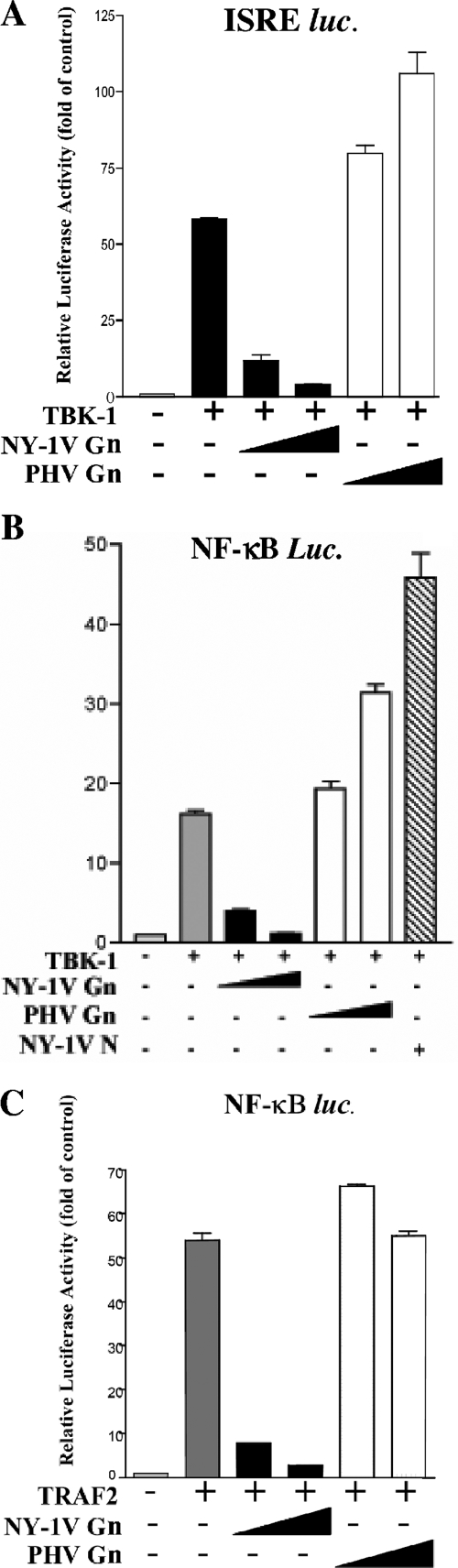
We have previously shown that the NY-1V Gn cytoplasmic tail inhibits RIG-I- and TBK1-directed IFN-β transcriptional responses while the Gn tail from the nonpathogenic hantavirus PHV failed to regulate transcription (1). IFN-β induction requires IRF-3 phosphorylation as well as NF-κB activation, and here we tested whether expression of the NY-1V Gn tail specifically inhibited NF-κB-directed transcriptional responses. Cells transfected with a plasmid expressing the NY-1V Gn cytoplasmic tail inhibited transcription from both ISRE and NF-κB luciferase reporters (Fig. 1A and B). In contrast, cells transfected with increasing amounts of the plasmids expressing the PHV Gn cytoplasmic tail or the NY-1V N protein failed to regulate ISRE or NF-κB transcriptional responses and instead slightly enhanced transcriptional responses (Fig. 1A and B).
FIG. 1.
NY-1V Gn cytoplasmic tail inhibits TBK1- and TRAF2-directed NF-κB activation. HEK 293 cells were transfected with 250 ng ISRE promoter (A) or NF-κB luciferase reporters (B and C) with or without TBK1 (500 ng) or TRAF2 (500 ng) expression vector. Where indicated, cells were transfected with increasing amounts of NY-1V or PHV Gn cytoplasmic tail expression vectors (1 to 2 μg), NY-1V nucleocapsid protein expression vector (2 μg), or control vector in order to transfect cells with a constant amount of DNA. Luciferase reporter activity was assayed 48 h posttransfection, was normalized to Renilla luciferase levels, and is reported as the increase over controls lacking TBK1 or TRAF2 activation.
TRAF2 reportedly forms a complex with TBK1 and activates NF-κB (38). In order to determine whether the NY-1V Gn tail blocks TRAF2-directed responses, we cotransfected cells with the NY-1V or PHV Gn tail and TRAF2 expression plasmids and assessed NF-κB transcriptional responses using a luciferase reporter. TRAF2 overexpression activated κB luciferase reporter gene expression over 50-fold, and coexpression of the PHV Gn tail had no effect on NF-κB activation (Fig. 1C). In contrast, coexpression of the NY-1V Gn tail resulted in the dose-dependent inhibition of TBK1 (Fig. 1B)- and TRAF2 (Fig. 1C)-directed NF-κB activation.
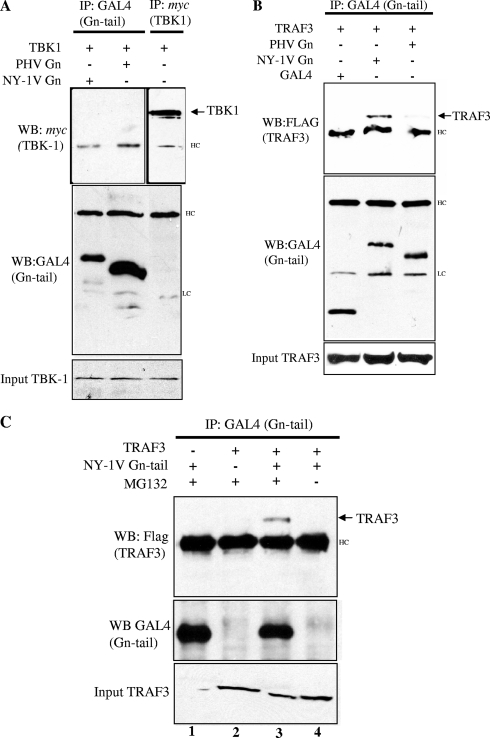
The NY-1V Gn cytoplasmic tail coprecipitates TRAF3.
TRAF3 forms a signaling complex with TBK1, which is required for IFN-β induction in response to viral infection. The NY-1V Gn tail blocks TBK1-directed IFN transcriptional responses upstream of IRF-3 at the level of TBK1 and TRAF3 (1). To determine if the NY-1V Gn tail directly binds TBK1 or TRAF3 proteins, we coexpressed the NY-1V or PHV Gn tail in the presence of TBK1 or TRAF3 and assayed for coimmunoprecipitation of TBK1 or TRAF3 with the expressed Gn tail protein. Figure 2A indicates that TBK1 and Gn tails of NY-1V and PHV were well expressed and present in cell lysates; however, neither the NY-1V nor the PHV Gn tail was coprecipitated by TBK1 (Fig. 2A). In contrast, an identical experiment performed with TRAF3 resulted in the coprecipitation of TRAF3 by the NY-1V, but not PHV, Gn tail (Fig. 2B). The NY-1V Gn tail is proteasomally degraded in the absence of the proteasome inhibitor MG132 (45), and Fig. 2C demonstrates that coprecipitation of TRAF3 by the NY-1V Gn tail was dependent on proteasome inhibition, which permits Gn tail accumulation. Although we have not been able to demonstrate Gn tail coprecipitation of endogenous TRAF3, interactions between endogenous TRAF3 and TBK1 have also not been reported (33). These findings indicate that the NY-1V Gn tail interacts with TRAF3-containing complexes and that the pathogenic NY-1V Gn tail and the nonpathogenic hantavirus PHV Gn tail differ in their ability to interact with TRAF3.
FIG. 2.
The NY-1V Gn cytoplasmic tail binds TRAF3. (A) HEK 293 cells were transfected with TBK1, NY-1V Gn tail, or PHV Gn tail expression plasmids as indicated. Cells were treated with MG132 (50 μM) for 6 h prior to cell lysis. Cells were lysed 2 days posttransfection. GAL4- or _myc_-tagged proteins were immunoprecipitated (IP) with anti-GAL4 or anti-myc antibodies. Immunocomplexes were isolated on protein A/G agarose beads, and immunoprecipitated proteins were detected by anti-GAL4 (Gn tail) or anti-myc (TBK1) Western blotting (WB). (B) HEK 293 cells were transfected with TRAF3, NY-1V Gn tail, PHV Gn tail, or GAL4 expression plasmids as indicated. Transfected cells were treated with MG132 (50 μM) for 6 h prior to cell lysis. FLAG-tagged TRAF3 protein expression was analyzed by anti-FLAG Western blotting and is shown in the bottom panel. GAL4-tagged Gn tail proteins were immunoprecipitated, and immunocomplexes were isolated as described for panel A. Immunoprecipitated Gn tail proteins were analyzed by anti-GAL4 Western blotting (center panel), and coimmunoprecipitated FLAG-TRAF3 protein was analyzed by anti-FLAG Western blotting (top panel). (C) HEK 293 cells were transfected with NY-1V Gn tail expression vector alone (lane 1) or TRAF3 expression vector alone (lane 2) or cotransfected with both NY-1V Gn tail and TRAF3 expression vectors (lanes 3 and 4). The coprecipitation of TRAF3 by the NY-1V Gn tail was performed in the presence (lane 3) or absence (lane 4) of MG132. HC, IgG heavy chain; LC, IgG light chain.
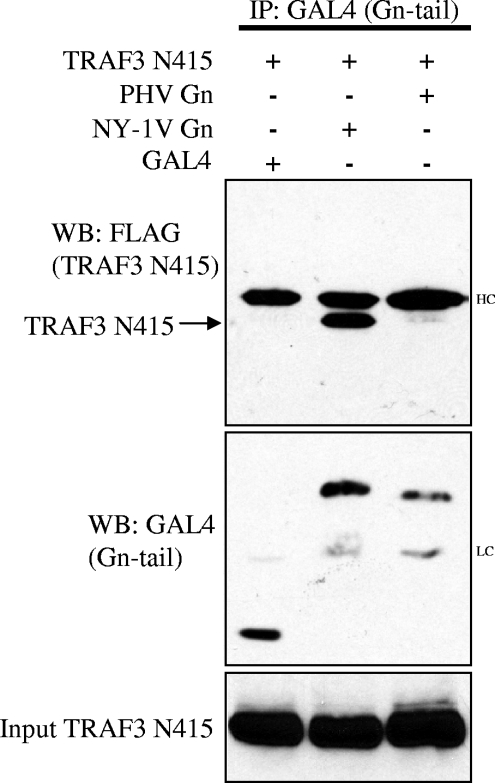
The NY-1V Gn tail interacts with the N-terminal domain of TRAF3.
TRAF3 contains a unique C-terminal domain that mediates binding to IPS-1 and links upstream signals to downstream TBK1-directed IFN induction (41). To determine if the NY-1V Gn tail interacts with N- or C-terminal TRAF domains, we evaluated the ability of the NY-1V Gn tail to bind a C-terminally truncated TRAF3 protein (TRAF3-N415). Figure 3 demonstrates that the NY-1V, but not PHV, Gn tail coprecipitated the N-terminal domain of TRAF3. This finding indicates that the N-terminal domain of TRAF3 is sufficient for interaction with the NY-1V Gn tail and is discrete from TRAF3 domains that mediate binding to IPS-1. NY-1V Gn tail interactions with the TRAF3 N terminus provide a potential mechanism for the pathway-specific regulation of IFN-β transcriptional responses by pathogenic hantaviruses.
FIG. 3.
The NY-1V Gn cytoplasmic tail binds the N-terminal domain of TRAF3. HEK 293 cells were cotransfected with GAL4, NY-1V Gn, or PHV Gn expression plasmids along with FLAG-TRAF3 (N415). Transfected cells were treated with MG132 (50 μM) for 6 h prior to cell lysis and analysis. Two days posttransfection and following anti-GAL4 antibody immunoprecipitation (IP), NY-1V and PHV Gn tail expression was analyzed by anti-GAL4 Western blotting (center panel). Immunocomplexes were isolated on protein A/G agarose beads, and coimmunoprecipitated TRAF3 (N415) protein was analyzed by anti-FLAG Western blotting (WB) (top panel). TRAF3 (N415) expression was determined by anti-FLAG Western blotting (bottom panel). IgG heavy chain (HC) and light chain (LC) are indicated.
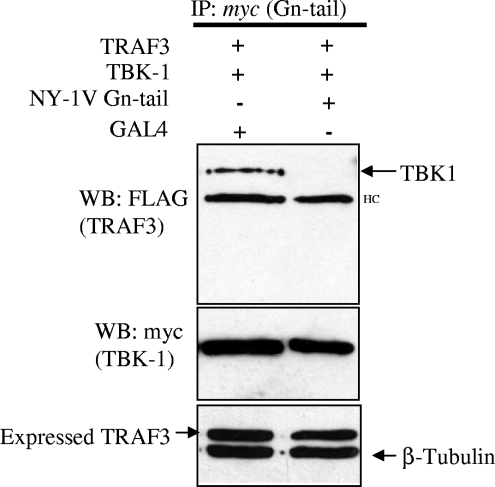
TBK1 binding to TRAF3 is inhibited by the NY-1V Gn cytoplasmic tail.
TBK1 and TRAF3 form a signaling complex that directs downstream IFN-β transcriptional responses (16, 33). Since the NY-1V Gn tail interacts with complexes containing TRAF3 and disrupts TBK1-directed signaling responses, we reasoned that the NY-1V Gn tail might interfere with TBK1-TRAF3 interactions. To assess this, we evaluated TBK1 coprecipitation of TRAF3 in the presence or absence of the NY-1V Gn tail. Figure 4 indicates that TBK1 coprecipitation of TRAF3 was disrupted by coexpressing the NY-1V Gn tail. Consistent with the ability of the NY-1V Gn tail to inhibit TBK1-directed IFN-β transcriptional responses, this result indicates that the NY-1V Gn tail blocked TBK1 interactions with TRAF3, which are essential for IFN transcription. Although these findings do not define the means by which the Gn tail disrupts TBK1-TRAF3 complexes, these findings suggest that disruption of TBK1-TRAF3 interactions is a mechanism by which the NY-1V Gn tail blocks IFN induction.
FIG. 4.
Expression of the NY-1V Gn cytoplasmic tail disrupts TBK1-TRAF3 interactions. HEK 293 cells were cotransfected with TBK1 and TRAF3 expression vectors and either NY-1V Gn tail or GAL4 expression vector. Transfected cells were treated with MG132 (50 μM) for 6 h prior to cell lysis. Two days posttransfection, cells were lysed and TBK1 was immunoprecipitated (IP) using a monoclonal anti-myc antibody. Coprecipitated FLAG-TRAF3 was detected by anti-FLAG Western blotting (WB) (top panel). TBK1 and TRAF3 expression was determined by anti-myc or anti-FLAG Western blotting (middle and bottom panels). HC, IgG heavy chain.
Infection with NY-1V disrupts TBK1-TRAF3 complex formation.
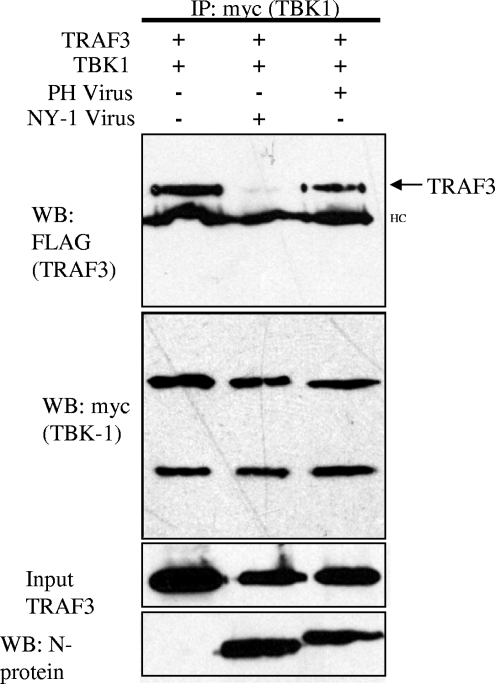
PHV strongly induces the transcription of ISGs 1 day postinfection while NY-1V regulates early IFN responses (1, 14, 26). Since expression of the NY-1V Gn tail disrupted TBK1-TRAF3 interactions, we assessed whether cells infected with NY-1V similarly disrupted TBK1-TRAF3 interactions. Cells were transfected with TBK1 and TRAF3 expression plasmids and subsequently infected with NY-1V or PHV. TBK1 coprecipitated TRAF3 from cell lysates in the absence of infection or following infection by PHV (Fig. 5). In contrast, cells infected with an identical amount of NY-1V dramatically reduced the ability of TBK1 to coprecipitate TRAF3 (Fig. 5). Thus, similar to expression of the NY-1V Gn tail, infection by NY-1V also disrupted TBK1 association with TRAF3. These experiments were performed in the absence of MG132, and since TRAF3 and TBK1 are abundantly expressed in the lysates, this indicated that NY-1V disrupted TBK1-TRAF3 complexes without TRAF3 degradation. However, this does not necessarily reflect whether endogenous TRAF3 is degraded following hantavirus infection. These findings suggest that pathogenic hantavirus regulation of host cell IFN-β induction and ISG expression is a consequence of Gn tail interactions with TRAF3, which block required TRAF3-TBK1 complex formation. However, the means by which TBK1-TRAF3 complexes are disrupted by hantavirus has yet to be defined.
FIG. 5.
NY-1V but not PHV infection disrupts TBK1-TRAF3 interactions. Vero E6 cells were transfected with TBK1 and TRAF3 expression plasmids, and 24 h posttransfection cells were infected with NY-1V or PHV or mock infected. One day postinfection cells were lysed and TBK1 was immunoprecipitated (IP) with anti-myc antibody. Coprecipitated TRAF3 was detected by anti-FLAG Western blotting (WB). Immunoprecipitated TBK1 and input TRAF3 were determined by Western blotting (center panels). NY-1V- and PHV-infected cells were determined by Western blotting for nucleocapsid protein (N-protein) 24 h postinfection by using an anti-N-protein rabbit polyclonal antiserum (bottom panel). HC, IgG heavy chain.
DISCUSSION
In response to viral infection cells rapidly induce IFN, an innate immune response that limits viral replication and spread. Many viruses counter innate immune responses by blocking pathways which direct IFN transcription (9, 51). Pathogenic hantaviruses inhibit IFN-β induction, and the ability to regulate innate immune responses distinguishes pathogenic hantaviruses from their nonpathogenic counterparts. The ability of pathogenic hantaviruses to inhibit IFN-β induction has been mapped to the long cytoplasmic tail of the hantavirus Gn protein (1). Expression of the pathogenic NY-1V Gn cytoplasmic tail inhibits transcriptional responses from IFN-stimulated response element and IFN-β promoters. This report demonstrates that the NY-1V Gn tail interacts with TRAF3 and prevents TBK1-TRAF3 complex formation, which is necessary for IFN-β transcription.
TRAF3 links upstream IFN signaling responses of IPS-1 (MAVS/Cardiff/VISA) to the TBK1-directed phosphorylation of IRF-3, and cells lacking TRAF3 are deficient in type I IFN responses (33). IPS-1 binds to the C-terminal domain of TRAF3 (TRAF-C) (41); however, requirements for TRAF3 binding to TBK1 have yet to be determined. Our data indicate that the TRAF3 N terminus is sufficient to form a complex with the NY-1V Gn tail and demonstrate that the NY-1V Gn tail blocks cellular TBK1-TRAF3 complex formation. These findings suggest that NY-1V Gn tail interactions with TRAF3 are discrete from TRAF3 domains required for IPS-1 binding.
TBK1 is known to bind TRAF3 and TRAF2, which direct IRF-3 phosphorylation and NF-κB activation, respectively (33, 38). Both of these responses are required for IFN-β transcription, and our findings indicate that the NY-1V Gn tail blocks the induction of IFN-stimulated response element and κB promoter responses which are directed by TBK1. The NY-1V Gn tail coprecipitates TRAF3, and the N-terminal domain of TRAF3 is sufficient to mediate this interaction. Interestingly the C-terminal domains of cellular TRAFs have been associated with pathway-specific responses, since N-terminal domains of TRAF3 and TRAF5 are reportedly interchangeable (7). Although further study is required, our findings suggest that the NY-1V Gn tail may regulate the function of several TRAFs through interactions with their N-terminal domains.
A cellular protein termed TNAP also reportedly binds TRAF3 and represses NF-κB activation, suggesting that the hantavirus Gn tail may mimic a normal cellular IFN regulatory mechanism (21). Additionally the cellular deubiquitinating protein DUBA has recently been shown to form a complex with TRAF3 that blocks TRAF3 ubiquitination and thereby recruitment and activation of TBK1 (24). Since the NY-1V Gn tail is proteasomally degraded, it is unclear whether its association with TRAF3 complexes stems from interactions with additional complex components that change TRAF3 ubiquitination or from activating Ub63 to proteasome-linked Ub48 ubiquitination or whether the Gn tail dissociates additional proteins required for TRAF3-TBK1 complex assembly (24). Although we have demonstrated that the Gn tail coprecipitates TRAF3, we have not defined the means by which the Gn tail inhibits TBK1-TRAF3 complex formation. Gn tail interactions could regulate many facets of the complex including altering TRAF3 trimerization or the recruitment of TBK1, TANK, E3 ubiquitin ligases, or deubiquitinating enzymes to TRAF3 complexes (24).
Hantaviruses are sensitive to the effects of IFN at early times postinfection and resistant to IFN addition at later times (1). Hantaviruses replicate normally when IFN is added ≥1 day postinfection, and pathogenic hantaviruses induce high-level ISG responses 2 to 4 days postinfection while persistently replicating within endothelial cells (1, 14, 26). These findings demonstrate the importance for hantaviruses of regulating early IFN responses and indicate that hantavirus regulation of IFN responses is likely to be a transient early inhibitory effect. These findings further imply that pathogenic hantaviruses have the ability to replicate in the presence of high-level ISG responses at later times postinfection (14, 26). One report suggests that hantaviruses regulate IFN receptor-directed STAT phosphorylation at later times postinfection, although these data are inconsistent with the reported high-level induction of STAT-dependent ISGs 2 to 4 days after hantavirus infection of endothelial cells (14, 47). Data presented here clearly demonstrate that the NY-1V Gn tail limits the induction of cellular IFN transcriptional responses. Since both NY-1 hantavirus infection and the expressed Gn tail block TBK1-TRAF3 complex formation, these findings suggest a mechanism by which pathogenic hantaviruses inhibit the early induction of IFN-β that would otherwise limit viral replication.
Hantaviruses express only four viral proteins, and the Gn protein plays multiple roles in the hantavirus life cycle. The Gn protein is an envelope protein assembled into virions, forms heterodimeric complexes with Gc, and may also function as a matrix protein during the viral budding process (53). Although viral assembly results in the accumulation of Gn into virions, the Gn tail of pathogenic hantaviruses also contains a C-terminal degron which directs proteasomal degradation (45). However, it is unclear whether there is a switch in Gn stability that accounts for early Gn degradation and subsequent Gn accumulation following assembly. Proteasomal degradation is also linked to TRAF3-directed ubiquitination (41). TRAF3 interactions direct the degradation of NF-κB inducing kinase, and deubiquitination of TRAF3 was recently implicated in the regulation of TBK1-directed IFN responses (17, 24). It remains to be determined whether TRAF3 interactions direct the ubiquitination and degradation of the NY-1V Gn tail or whether the Gn tail, like DUBA, alters the state of TRAF3 ubiquitination and determines whether TBK1-TRAF3 complexes are formed (24).
Viruses have evolved a variety of mechanisms to block IFN-β induction and antiviral immune responses. The hepatitis C virus NS3/4A protein inhibits RIG-I-directed IFN-β induction by cleaving IPS-1 from the mitochondria, while the VP35 protein of Ebola virus binds TBK1 and prevents IRF-3-directed IFN-β transcription (5, 11, 27-29, 31). The influenza virus NS1 protein is also an IFN antagonist with reported regulatory roles involving double-stranded RNA, RIG-I, and IPS-1 binding interactions (2, 5, 15, 25, 31, 34). Although we have previously reported that the NY-1V Gn tail also blocks TBK1-directed transcriptional responses, data presented here demonstrate that the Gn tail interacts with TRAF3 and disrupts TBK1-TRAF3 complex formation. The LMP1 protein of Epstein-Barr virus also binds TRAF3 through PxQxT motifs that reportedly regulate NF-κB activation, although there is no information on whether these interactions regulate IFN responses (50). The NY-1V Gn tail does not contain PxQxT motifs, and as a result Gn tail interactions with TRAF3 are discrete from those of LMP1. These findings suggest that the Gn tail blocks IFN-β transcription through unique TRAF3 interactions.
Some members of the Bunyaviridae family have been shown to encode a nonstructural NSs protein, which inhibits IFN-β transcription 50-fold (3, 4, 49). A recent report suggests that Tula and Puumala hantaviruses encode an NSs protein that reduces IFN-β transcription (22). However, Tula and Puumala NSs proteins reportedly reduced IFN-β transcriptional responses by only 30%, and it is unclear whether this level of inhibition contributes to the regulation of cellular IFN responses (22). Further, NSs proteins have not been identified during HTNV, ANDV, Sin Nombre virus, or NY-1V infection, and the open reading frame corresponding to the Tula virus NSs protein is disrupted by termination codons in NY-1V, HTNV, and ANDV. Although it is possible that hantaviruses have evolved redundant mechanisms for inhibiting IFN-β induction, the 40- to 50-fold reduction in IFN transcriptional responses directed by the NY-1V Gn tail (1) suggests that it plays a primary role in regulating early IFN responses during infection.
In this report we extend our previous findings regarding Gn tail regulation of IFN-β induction and define an IFN pathway-specific target bound by the pathogenic hantavirus Gn tail. We demonstrate that the recombinant expressed NY-1V Gn tail interacts with TRAF3 and disrupts TBK1-TRAF3 complex formation. Since infection of endothelial cells by a pathogenic hantavirus similarly disrupts TBK1-TRAF3 complex formation, our findings suggest a mechanism for hantavirus regulation of IFN transcriptional responses that is directed by Gn tail interactions with TRAF3. Collectively, our findings suggest that pathogenic hantaviruses disengage TBK1-TRAF3 complex formation at early times postinfection to prevent IFN-β induction.
Acknowledgments
We thank Varya Kirillov for technical assistance, David Goeddel for TRAF2 and TRAF3 plasmids, and David Baltimore for TBK1 plasmids used in these experiments. We also thank Lorenzo Bombardelli (Department of Pharmacology, Stony Brook University) for helpful discussions.
This work was supported by National Institutes of Health grants R01AI47873, PO1AI055621, and U54AI57158 (Northeast Biodefense Center-Lipkin) and by a Veterans Affairs Merit Award.
Footnotes
▿
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Alff, P. J., I. N. Gavrilovskaya, E. Gorbunova, K. Endriss, Y. Chong, E. Geimonen, N. Sen, N. C. Reich, and E. R. Mackow. 2006. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J. Virol. 809676-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 777945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 789798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakqori, G., S. Delhaye, M. Habjan, C. D. Blair, I. Sanchez-Vargas, K. E. Olson, G. Attarzadeh-Yazdi, R. Fragkoudis, A. Kohl, U. Kalinke, S. Weiss, T. Michiels, P. Staeheli, and F. Weber. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 814991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardenas, W. B., Y. M. Loo, M. Gale, Jr., A. L. Hartman, C. R. Kimberlin, L. Martinez-Sobrido, E. O. Saphire, and C. F. Basler. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 805168-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosgriff, T. M. 1991. Mechanisms of disease in Hantavirus infection: pathophysiology of hemorrhagic fever with renal syndrome. Rev. Infect. Dis. 1397-107. [DOI] [PubMed] [Google Scholar]
- 7.Dadgostar, H., and G. Cheng. 1998. An intact zinc ring finger is required for tumor necrosis factor receptor-associated factor-mediated nuclear factor-kappaB activation but is dispensable for c-Jun N-terminal kinase signaling. J. Biol. Chem. 27324775-24780. [DOI] [PubMed] [Google Scholar]
- 8.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43247-252. [DOI] [PubMed] [Google Scholar]
- 9.Finlay, B. B., and G. McFadden. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124767-782. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 11.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 3001145-1148. [DOI] [PubMed] [Google Scholar]
- 12.Geimonen, E., I. Fernandez, I. N. Gavrilovskaya, and E. R. Mackow. 2003. Tyrosine residues direct the ubiquitination and degradation of the NY-1 hantavirus G1 cytoplasmic tail. J. Virol. 7710760-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geimonen, E., R. LaMonica, K. Springer, Y. Farooqui, I. N. Gavrilovskaya, and E. R. Mackow. 2003. Hantavirus pulmonary syndrome-associated hantaviruses contain conserved and functional ITAM signaling elements. J. Virol. 771638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geimonen, E., S. Neff, T. Raymond, S. S. Kocer, I. N. Gavrilovskaya, and E. R. Mackow. 2002. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc. Natl. Acad. Sci. USA 9913837-13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, Z., L. M. Chen, H. Zeng, J. A. Gomez, J. Plowden, T. Fujita, J. M. Katz, R. O. Donis, and S. Sambhara. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36263-269. [DOI] [PubMed] [Google Scholar]
- 16.Hacker, H., V. Redecke, B. Blagoev, I. Kratchmarova, L. C. Hsu, G. G. Wang, M. P. Kamps, E. Raz, H. Wagner, G. Hacker, M. Mann, and M. Karin. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439204-207. [DOI] [PubMed] [Google Scholar]
- 17.He, J. Q., S. K. Saha, J. R. Kang, B. Zarnegar, and G. Cheng. 2007. Specificity of TRAF3 in its negative regulation of the noncanonical NF-kappa B pathway. J. Biol. Chem. 2823688-3694. [DOI] [PubMed] [Google Scholar]
- 18.Hiscott, J. 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 28215325-15329. [DOI] [PubMed] [Google Scholar]
- 19.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-kappaB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010237-248. [DOI] [PubMed] [Google Scholar]
- 20.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 21.Hu, W. H., X. M. Mo, W. M. Walters, R. Brambilla, and J. R. Bethea. 2004. TNAP, a novel repressor of NF-kappaB-inducing kinase, suppresses NF-kappaB activation. J. Biol. Chem. 27935975-35983. [DOI] [PubMed] [Google Scholar]
- 22.Jaaskelainen, K. M., P. Kaukinen, E. S. Minskaya, A. Plyusnina, O. Vapalahti, R. M. Elliott, F. Weber, A. Vaheri, and A. Plyusnin. 2007. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 791527-1536. [DOI] [PubMed] [Google Scholar]
- 23.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 24.Kayagaki, N., Q. Phung, S. Chan, R. Chaudhari, C. Quan, K. M. O'Rourke, M. Eby, E. Pietras, G. Cheng, J. F. Bazan, Z. Zhang, D. Arnott, and V. M. Dixit. 2007. DUBA: a deubiquitinase that regulates type I interferon production. Science 3181628-1632. [DOI] [PubMed] [Google Scholar]
- 25.Kochs, G., A. Garcia-Sastre, and L. Martinez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 817011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus, A. A., M. J. Raftery, T. Giese, R. Ulrich, R. Zawatzky, S. Hippenstiel, N. Suttorp, D. H. Kruger, and G. Schonrich. 2004. Differential antiviral response of endothelial cells after infection with pathogenic and nonpathogenic hantaviruses. J. Virol. 786143-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 10217717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 806072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 1036001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meylan, E., and J. Tschopp. 2006. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol. Cell 22561-569. [DOI] [PubMed] [Google Scholar]
- 31.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nolte, K. B., R. M. Feddersen, K. Foucar, S. R. Zaki, F. T. Koster, D. Madar, T. L. Merlin, P. J. McFeeley, E. T. Umland, and R. E. Zumwalt. 1995. Hantavirus pulmonary syndrome in the United States: a pathological description of a disease caused by a new agent. Hum. Pathol. 26110-120. [DOI] [PubMed] [Google Scholar]
- 33.Oganesyan, G., S. K. Saha, B. Guo, J. Q. He, A. Shahangian, B. Zarnegar, A. Perry, and G. Cheng. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439208-211. [DOI] [PubMed] [Google Scholar]
- 34.Opitz, B., A. Rejaibi, B. Dauber, J. Eckhard, M. Vinzing, B. Schmeck, S. Hippenstiel, N. Suttorp, and T. Wolff. 2007. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 9930-938. [DOI] [PubMed] [Google Scholar]
- 35.Panne, D., T. Maniatis, and S. C. Harrison. 2007. An atomic model of the interferon-beta enhanceosome. Cell 1291111-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panne, D., S. M. McWhirter, T. Maniatis, and S. C. Harrison. 2007. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J. Biol. Chem. 28222816-22822. [DOI] [PubMed] [Google Scholar]
- 37.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 38.Pomerantz, J. L., and D. Baltimore. 1999. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 186694-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raymond, T., E. Gorbunova, I. N. Gavrilovskaya, and E. R. Mackow. 2005. Pathogenic hantaviruses bind plexin-semaphorin-integrin domains present at the apex of inactive, bent alphavbeta3 integrin conformers. Proc. Natl. Acad. Sci. USA 1021163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothe, M., V. Sarma, V. M. Dixit, and D. V. Goeddel. 1995. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 2691424-1427. [DOI] [PubMed] [Google Scholar]
- 41.Saha, S. K., E. M. Pietras, J. Q. He, J. R. Kang, S. Y. Liu, G. Oganesyan, A. Shahangian, B. Zarnegar, T. L. Shiba, Y. Wang, and G. Cheng. 2006. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 253257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuel, C. E. 2007. Innate immunity minireview series: making biochemical sense of nucleic acid sensors that trigger antiviral innate immunity. J. Biol. Chem. 28215313-15314. [DOI] [PubMed] [Google Scholar]
- 43.Schmaljohn, C. 2001. Bunyaviridae and their replication, p. 1581-1602. In D. M. Knipe, P. M. Howley, D. E. Griffith, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 44.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 395-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen, N., A. Sen, and E. R. Mackow. 2007. Degrons at the C terminus of the pathogenic but not the nonpathogenic hantavirus G1 tail direct proteasomal degradation. J. Virol. 814323-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seth, R. B., L. Sun, and Z. J. Chen. 2006. Antiviral innate immunity pathways. Cell Res. 16141-147. [DOI] [PubMed] [Google Scholar]
- 47.Spiropoulou, C. F., C. G. Albarino, T. G. Ksiazek, and P. E. Rollin. 2007. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J. Virol. 812769-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 181359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 767949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, S., P. Xie, K. Welsh, C. Li, C. Z. Ni, X. Zhu, J. C. Reed, A. C. Satterthwait, G. A. Bishop, and K. R. Ely. 2005. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J. Biol. Chem. 28033620-33626. [DOI] [PubMed] [Google Scholar]
- 51.Yoneyama, M., and T. Fujita. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 28215315-15318. [DOI] [PubMed] [Google Scholar]
- 52.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 53.Zaki, S. R., P. W. Greer, L. M. Coffield, C. S. Goldsmith, K. B. Nolte, K. Foucar, R. M. Feddersen, R. E. Zumwalt, G. L. Miller, A. S. Khan, et al. 1995. Hantavirus pulmonary syndrome. Pathogenesis of an emerging infectious disease. Am. J. Pathol. 146552-579. [PMC free article] [PubMed] [Google Scholar]