Identification of 14 novel mutations in the long isoform of USH2A in Spanish patients with Usher syndrome type II (original) (raw)
Abstract
Mutations in USH2A gene have been shown to be responsible for Usher syndrome type II, an autosomal recessive disorder characterised by hearing loss and retinitis pigmentosa. USH2A was firstly described as consisting of 21 exons, but 52 novel exons at the 3' end of the gene were recently identified. In this report, a mutation analysis of the new 52 exons of USH2A gene was carried out in 32 unrelated patients in which both disease‐causing mutations could not be found after the screening of the first 21 exons of the USH2A gene. On analysing the new 52 exons, fourteen novel mutations were identified in 14 out of the 32 cases studied, including 7 missense, 5 frameshift, 1 duplication and a putative splice-site mutation.
Usher syndrome (USH, OMIM276901) is an autosomal recessive disorder, characterised by the association of retinitis pigmentosa, sensorineural hearing loss and, in some cases, vestibular dysfunction. It is thought to be responsible for more than half the cases of deaf‐blindness. The prevalence of USH ranges from 3.5 to 6.2 cases per 100 000.1 In Spain, the estimation is 4.2 per 100 000.2
Traditionally, three clinical types of USH have been distinguished—USH1, USH2 and USH3.3 Usher syndrome type I (USH1) is characterised by profound and congenital hearing loss, absent vestibular responses and retinitis pigmentosa with prepuberal onset. Patients with Usher syndrome type II (USH2) display moderate to severe hearing loss, normal vestibular function and postpuberal onset of retinitis pigmentosa. The traits of Usher syndrome type III (USH3) are variable onset of retinitis pigmentosa, progressive postlingual hearing loss and changes in vestibular responses in 50% of cases.
Three loci for USH2 have been mapped to date, but only the causative genes for USH2A (USH2A)4 and USH2C (VLGR1)5 have been isolated. USH2 is the most common form of Usher syndrome and the USH2A gene is thought to be involved in 74–90% of cases of USH2.2,6,7,8 Moreover, it is also responsible for the disease in patients described as having atypical Usher syndrome because they display progressive, rather than stable, hearing loss and/or vestibular arreflexia.9,10 In addition, this gene has been shown to be responsible for some recessive cases of non‐syndromic retinitis pigmentosa.10,11,12
The USH2A gene located in 1q4113 was first described as comprising 21 exons that expanded 259 kb of genomic DNA. The encoded protein was predicted to consist of 1546 amino acids containing laminin epidermal growth factor and fibronectin type III motifs, typical of extracellular matrix proteins.4,6 However, van Wijk et al14 identified 51 novel exons at the 3′ end of the USH2A gene, which indicated alternative splicing. A long open reading frame extends from exon 2 to 72, encoding a putative protein of 5202 amino acids that contains, in addition to the previously known extracellular domains, 2 laminin G and 28 fibronectin type III repeats, as well as a transmembrane region followed by an intracellular domain with a PDZ‐binding motif at the C‐terminal end. Additionally, Adato et al15 described a new alternatively spliced exon 71, which encodes a 24 amino‐acid peptide of the usherin cytoplasmic domain that is highly expressed in the murine inner ear and conserved throughout vertebrate evolution. The functional significance of the USH2A long isoform was shown by the presence of pathological mutations in several of the 51 novel exons in patients with USH2 syndrome.14 Furthermore, functional studies have shown that the PDZ‐binding domain of USH2A long isoform protein binds with harmonin,15,16 the defective protein in USH1C and whirlin,15,17 which is defective in the non‐syndromic hereditary deafness form DFNB31.
In previous studies of Spanish patients with USH2, atypical Usher, non‐syndromic retinal degeneration and non‐syndromic deafness, only one pathological mutation could be found in several cases, after screening exons 2–21 of the USH2A gene.10,18 Consequently, the aim of the present work was to screen for mutations in the 52 novel exons of the USH2A gene in these patients, to detect the second mutation responsible for the disease.
Key points
- Usher's syndrome type II (USH2) is the most common form of Usher syndrome, an autosomal recessive disorder characterised by hearing loss and retinitis pigmentosa. It has been shown that mutations in the USH2A gene are responsible not only for USH2 but also for atypical Usher syndrome and non‐syndromic retinitis pigmentosa.
- USH2A was first described as comprising 21 exons; however, 52 novel exons have recently been identified at the 3′ end of the gene. This paper reports a mutation analysis of the 52 new exons of the USH2A gene, carried out in 32 unrelated patients with Usher's syndrome, non‐syndromic retinal degeneration or non‐syndromic deafness. On screening the first 21 exons of the gene, patients were found not to be carrying both disease‐causing mutations.
- On analysing the 52 new exons, 14 novel mutations were identified in 14 of the 32 patients studied, including 7 mis‐sense, 5 frameshift, 1 duplication and 1 putative splice‐site mutation. The 14 patients were diagnosed with USH2 and 2 of them were found to carry both pathological variants in the 52 new USH2A exons.
Materials and methods
Subjects
Spanish patients with Usher syndrome, non‐syndromic retinal degeneration or non‐syndromic deafness were recruited from the Federacion de Asociaciones de Afectados de Retinosis Pigmentaria del Estado Español and from Ophthalmology and ENT Services of several Spanish hospitals as part of a large‐scale study on the genetics of Usher syndrome in the Spanish population.
The present study was carried out in 32 unrelated patients, all of whom had previously been screened for mutations in exons 2–21 of USH2A gene (unpublished data).10,18 Twenty nine of them were found to carry only one mutated allele. In another two patients a putative but unconfirmed pathological change was detected. The last patient was found not to carry any pathological variant, but intragenic SNPs showed homozygosity for this region. Consequently, in this patient a putative mutation in homozygotic state could be located in the 52 new exons of the gene.
On the basis of their clinical history and ophthalmological, audiological, neurophysiological and vestibular tests, 25 of these patients were clinically classified as having USH2, three displayed atypical Usher syndrome, another three were diagnosed with non‐syndromic retinal degeneration and one as having non‐syndromic deafness. Ophthalmological studies included determining visual acuity and visual field, fundus ophthalmoscopy and electroretinography. Audiometric tests included otoscopic exploration, pure‐tone audiometry and speech audiometry. Vestibular evaluation included electronystagmography, noting spontaneous, gaze and positional nystagmus, caloric testing and rotatory chair testing.
Fifty unrelated people of the Spanish population were screened as controls to evaluate the frequency of the mutations found in the patient sample.
Mutation screening
Genomic DNA from affected people and family members was extracted from leucocytes of peripheral blood samples using standard phenol‐choroform extraction procedures.
Exons 22–72 and their intron–exon boundaries of the USH2A gene were amplified using primers described by van Wijk et al.14 Primers 71CD and 71CR were designed to amplify the alternatively spliced exon 71 described by Adato et al.15 These primers and other additional primers designed to amplify some of the exons are available as an online supplementary table (http://jmg.bmjjournals.com/supplemental).
Samples from all affected people were analysed by single‐strand conformational polymorphism on polyacrylamide gel electrophoresis after polymerase chain reaction (PCR) amplification. The amplified DNA fragments were heat denatured, separated through 12% polyacrylamide gels at 18°C and silver stained.
Those PCR fragments showing different mobilities were analysed by direct sequencing on an automated sequencer (ABI‐PRISM, Applied Biosystems, California, USA, model 310).
Microsatellite‐marker analysis
Polymorphic markers from chromosome 1 were amplified using standard PCR conditions and analysed by polyacrylamide gel electrophoresis. Markers D1S1675, D1S199, D1S508 and D1S2734 were located in 1p. Markers D1S2141, D1S1602, D1S229 and D1S490 were located in 1q.
Prediction of splice scores
A Splice View program was used to assess how likely intron sequence variants were to create or exclude splice sites. This program is accessible at http://bioinfo.itb.cnr.it/oriel/splice‐view.html
Results
The mutation screening in our patients showed 14 different mutations, none of which had been reported previously (table 1). Thirteen of these changes were, as expected, found heterozygously and most of them were private.
Table 1 Novel mutations found in this study.
| Mutation | Exon | Nucleotide change | No of families |
|---|---|---|---|
| Mis‐sense | |||
| A2249D | 35 | 6746C→A | 1 |
| R2354H | 37 | 7061G→A | 1 |
| C3251R | 50 | 9751T→C | 1 |
| C3267R | 50 | 9799T→C | 1 |
| T3571M | 54 | 10712C→T | 2 |
| T4337M | 63 | 13010C→T | 1 |
| P4818L | 66 | 14453C→T | 1 |
| Frameshift | |||
| R2095GfsX2 | 32 | 6238delA | 1 |
| T2812MfsX17 | 42 | 8435_8438delCCTA | 1 |
| C3425FfsX4 | 52 | 10272_10273dupTT | 1 |
| Y3745fsX1 | 58 | 11234dupA | 1 |
| F4703HfsX6 | 64 | 14110_14111insA | 1 |
| Duplication | |||
| Y3472dup | 53 | 10414_10416dupTAT | 1 |
| Splicing | |||
| IVS26+1C→G | 1 |
The missense mutations, the in‐frame duplication and the splicing variant were not found in any of the 50 healthy controls. Segregation analysis was carried out in all cases except for families RP504, FRP60 and FRP37 (table 2).
Table 2 Clinical features of patients in whom both mutated alleles were detected.
| Family | Mutations | Sensorineural hearing loss | Onset of night blindness | Onset of visual field loss | Visual field | Visual acuity | Eye fundus | ERG | Cataracts |
|---|---|---|---|---|---|---|---|---|---|
| FRP13 | 2299delG/C3267R | Moderate, progressive | 16 | 16 | Marked concentric loss | RE: 0.4; LE: 0.2 | 1 | No response | BE |
| Moderate, progressive | 16 | 16 | Marked concentric loss | RE: 0.15; LE: 0.1 | 1 | No response | BE | ||
| FRP7 | 2299delG/11234dupA | Mild, progressive | 22 | 25 | Marked concentric loss | RE: 0.8; LE: 1 | 1 | No response | BE |
| Moderate, progressive | 11 | 16 | 2 | No response | BE | ||||
| RP504 | C759F/6238delA | Slight | 26 | 30 | Marked concentric loss | RE: 0.08; LE: 0.2 | 2 | No response | LE |
| FRP35 | 2299delG/P4818L | Moderate | 14 | 20 | Concentric loss, −10° | RE: 0.8; LE: 1 | 1 | No response | BE |
| FRP37 | 239_240insGTAC/10272_10273dupTT | Slight, progressive | 20 | 20 | Marked concentric loss | 1 | No response | LE | |
| Slight, progressive | 14 | 14 | Marked concentric loss | 1 | No response | BE | |||
| Slight, progressive | 17 | 20 | Marked concentric loss | 1 | No response | BE | |||
| FRP291 | 2299delG/14110_14111insA | Moderate | 10 | Marked concentric loss | RE: 0.8; LE: 1 | 1 | No response | BE | |
| FRP292 | 2299delG/A2249D | Moderate, progressive | 20 | Concentric loss, −5° | 1 | No response | BE | ||
| FRP293 | 2299delG/T3571M | Moderate | 10 | Marked concentric loss | RE: 1/3; LE: 1/2 | 1 | BE | ||
| FRP60 | 2299delG/C3251R | Moderate/severe | 20 | 25 | Slight concentric loss | RE: 0.9; LE: 0.9 | 1 | Iregular response | |
| FRP54 | T4337M (homozygotic) | Moderate/severe | 27 | 27 | Concentric loss,−10° | <0.1 | 1 | No response | No |
| FRP186 | G713R/Y3472dup/IVS26+1C→G | Slight | 18 | 17 | Marked concentric loss | 1 | No response | No | |
| FRP220 | 2299delG/T3571M | Mild | 13 | 13 | Marked concentric loss | RE: 0.5; LE: 0.5 | 1 | No response | BE |
| FRP229 | D778Y/R2354H | Moderate/severe | 20 | 22 | Marked concentric loss | RE: 0.6; LE: 0.8 | 1 | Altered | No |
| FRP232 | C759F//8435_8438delCCTA | Moderate | 30 | 33 | Marked concentric loss | 1 | No response | No |
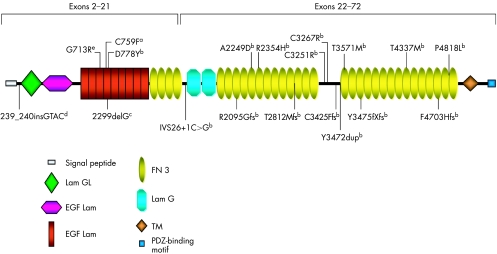
Figure 1 shows a schematic distribution of previously identified (exons 2–21) and novel (exons 22–72) mutations along the USH2A protein domains (appendix).
Figure 1 Schematic distribution of previously identified (exons 2–21) and novel (exons 22–72) mutations along USH2A protein domains. aMutation reported by Rivolta et al11; bpreviously unreported mutations; cmutation first reported by Eudy et al4 as 2314delG; dmutation previously reported by Najera et al18; emutation described by Dreyer et al7. EGF, epidermal growth factor; FN, fibronectin; GL, laminin G‐like; TM, transmembrane.
Only one mutation was found in more than one family. The amino acid change T3571M was detected in two families together with the 2299delG. To find out whether T3571M could have a common origin, intragenic haplotypes were constructed using the SNPs found in USH2A gene. Segregation analysis of the intragenic SNPs using healthy relatives showed that although both families shared the same haplotype for 2299delG it was not so for the novel mutation T3571M.
In another family, two additional potentially pathogenic changes were detected. IVS26+1C>G and Y3472dup were found together with the previously identified G713R change. Segregation analysis for the three mutations showed that the mother carried both the G713R and Y3472dup changes, whereas the IVS26+1C→G was carried by the father. An in silico analysis was carried out to predict whether IVS26+1C→G might affect the splicing of the mutant USH2A transcript. On analysis, the normal allele gave a score of 72 for the donor splice site, whereas for the mutant allele the splice sequence was not recognised.
One of the 14 novel mutations detected in this study, the amino acid change T4337M, was found homozygously in one patient. In this case, no mutation had been detected on screening the first 21 exons of the USH2A gene. This patient did not refer consanguinity, but intragenic haplotype showed homozygosity along the entire gene. Segregation analysis showed that T4337M was not homozygously carried by any of the healthy relatives studied. The patient's mother carried the mutation heterozygously, but there was no DNA sample available from the patient's father. A possible maternal uniparental disomy was discarded using microsatellite markers (see Materials and methods).
Moreover, 26 variants, presumed to be non‐pathological, were also found after screening the last 52 exons of USH2A. Table 3 summarizes these variants which were considered to be polymorphisms on the basis of their nature, frequency and segregation analysis.
Table 3 Presumed non‐pathological variants found in the last 52 exons of USH2A.
| Exon | Nucleotide change | Codon change | Allele frequency |
|---|---|---|---|
| 22 | 4714C→T | L1572F | 11/64 |
| 25 | 4994T→C | I1665T | 18/64 |
| 25 | 5013C→A | G1671G | 7/64 |
| IVS30+76A→T | — | 1/64 | |
| 32 | 6317T→C | I2106T | 15/64 |
| 34 | 6506T→C | I2169T | ND |
| 35 | 6713A→C | E2238A | 2/64 |
| IVS36+19A→G | — | 4/64 | |
| IVS38–65T→G | — | 3/64 | |
| 40 | 7506G→A | P2502P | 3/64 |
| IVS40+22C→T | — | 3/64 | |
| 43 | 8624G→A | R2875Q | 2/64 |
| 43 | 8656C→T | L2886F | ND |
| IVS44–52_53delTT | — | 2/64 | |
| 47 | 9296A→G | N3099S | 4/64 |
| 48 | 9430G→A | D3144N | 2/64 |
| 52 | 10232A→C | E3411A | 23/64 |
| IVS52–26T→C | — | ND | |
| IVS58+9A→T | — | 10/64 | |
| IVS59+98G→A | — | 8/64 | |
| 60 | 11602A→G | M3868V | 8/64 |
| 61 | 11736G→A | E3912E | 1/64 |
| 61 | 11907A→T | P3969P | 1/64 |
| 63 | 12666A→G | T4222T | 14/64 |
| 63 | 13191G→A | E4397E | 7/64 |
| IVS71–194A→T | — | 2/64 |
The 14 patients found to carry mutations in the last 52 exons of the USH2A gene were clinically diagnosed with USH2. In these patients, the age of onset of night blindness and visual field loss ranged from 10 to 30 years. The sensorineural hearing loss ranged from slight to moderate or severe and, in some cases, was subjectively progressive. Fundus ophthalmoscopy showed typical retinitis pigmentosa in all cases, but in two of them the macula was also affected, corresponding to the cases in which retinitis pigmentosa was more advanced. Most patients were found to have cataracts. Table 2 summarises the clinical data of these patients (appendix).
Discussion
The first 21 exons of the USH2A gene have been screened for mutations in many different populations,6,7,18,19,20 with most studies finding that several patients displayed only one mutated allele. In our Spanish patient sample, both pathogenic mutations were detected in 12 patients, whereas 24 patients displayed only one of the two expected mutations.10,18 This prompted us to screen the 52 novel exons of the USH2A gene to find the second mutation responsible for the disease.
A high diversity of mutations was found in our series: seven missense, five frameshift, one duplication and one putative splice‐site mutation. Although 2299delG, located at exon 13, is the most prevalent mutation found in the first part of the USH2A gene in several populations,4,6,7,19,21 no predominant mutation was detected for the Spanish population in the last part of the gene, as only one of the mutations was detected in two families and the remaining changes were private. These 14 novel mutations were found to be distributed along the whole new part of the USH2A gene, there being no indication of a hot spot for mutations in the new 52 exons. No mutation was found either in the transmembrane or in the intracellular domain of the protein. This fact is hardly surprising, as only the last 161 amino acids from a total of 5202 make up these regions. However, recent studies have shown that this intracytoplasmic domain can interact with harmonin and whirlin, defective proteins in USH1 and non‐syndromic sensorineural deafness, respectively.15,16,17 Therefore, mutations in the transmembrane and cytoplasmic domain of the USH2A protein could lead to a phenotype that is different from USH2. We also included some patients with non‐syndromic retinitis pigmentosa, non‐syndromic deafness and atypical Usher in this screening, but no mutations were detected.
Only the amino acid change T3571M was not found privately. This mutation was detected in two unrelated cases and in both the accompanying mutation was 2299delG. Segregation analysis using intragenic SNPs showed that the haplotype linked to 2299delG was identical in both cases, in agreement with the common origin indicated by Dreyer et al22 for this ancestral and widespread mutation. Conversely, a different haplotype linked to T3571M, suggesting a different origin of the mutation for these two families.
Here, we have described one patient with USH2, FRP186, with three potentially pathogenic changes in the USH2A gene: G713R (previously identified in exon 13), Y3472dup and the splicing variant IVS26+1C→G. With respect to IVS26+1C→G, no recognition of the splicing site was obtained using the Splice View program for the abnormal sequence, suggesting that this variant may affect USH2A transcript splicing. Segregation analysis showed that IVS26+1C→G was carried by the father, and both G713R and Y3472dup were carried by the mother. Although G713R was described as a pathological mutation by Dreyer et al,7 we were able to detect this change in 2 of 200 normal control chromosomes.18 Seyedahmadi et al23 showed that this variant did not segregate with the disease in some of their families. Furthermore, functional studies performed by Bhattacharya and Cosgrove24 showed that, as opposed to other missense mutations located in the LE domain of USH2A protein, G713R does not abolish usherin–fibronectin interactions. Consequently, we do not consider that there is convincing evidence for the pathogenic effect of G713R, but believe, rather, that it seems to be a polymorphism without clinical implications.
The amino acid change T4337M was found homozygously in patient FRP54. This change was heterozygously detected in the patient's mother, whereas paternal DNA was not available. No indication of maternal uniparental disomy was obtained. However, it is possible there is a deletion in heterozygotic state involving the USH2A region in this patient. Future studies using Southern blot, multiplex ligation probe assay or quantitative PCR might elucidate this question.
All the patients harbouring both mutated alleles displayed features that prompted us to classify them as USH2. The greatest clinical differences between affected people concerned the age of onset of retinitis pigmentosa, ranging from 10 to 30 years. Nevertheless, these data were obtained from personally interviewing the patients and are therefore clearly subjective. Also, differences in the degree of hearing loss must be mentioned. This ranged from slight to moderate or severe, and, in some cases, was slightly progressive. This progression is not as evident as it is in USH3 and, in any event, was subjectively qualified. We do, however; think that it is more important to remark on findings on hearing loss in that, in some cases, hearing loss was only mild or even slight. In these cases, USH2 could be misdiagnosed as non‐syndromic retinitis pigmentosa.
Evidently, a high proportion of mutated alleles for the USH2A gene are undetected, as the expected complement of two pathological mutations could not be found in a large number of patients. This could be explained by the fact that 52 exons of this gene were not screened in most previously published studies, and it is reasonable to expect the mutation detection rate to improve after screening this part of the gene.25 In this study, 22–72 USH2A exons were screened for mutations in 32 unrelated patients, in which the first 21 USH2A exons had previously been screened. Both mutations responsible for the disease were finally detected in 14 of them. All 14 novel mutations belonged to the patients with USH2. This means that 14 of 25 (56%) cases of USH2 were resolved by typing the additional 52 USH2A exons. However, we were still unable to identify the second mutation in 18 of the patients studied. Consequently, additional reasons for this finding must be postulated, such as sensitivity of the detection technique, mutations in non‐coding regions or the presence of other isoforms expressed in minimal amounts that have not yet been identified.
Acknowledgements
We thank the patients participating in the study and their family members, and also the FAARPEE for their help and cooperation. This work was supported by a grant from the Fondo de Investigaciones Sanitarias (PI04/0918), Redes Temáticas de Investigación Cooperativa (FIS G03/018 and G03/203), the Integrated Project EVI‐GenoRet (Contract No LSHG‐CT‐2005‐512036) and the ONCE. EA is recipient of a fellowship from Agència Valenciana de Ciència i Tecnologia (CTBPRB/2003/122). English text was corrected by F Barraclough.
Abbreviations
PCR - polymerase chain reaction
USH - Usher syndrome
Footnotes
Competing interests: None declared.
References
- 1.Keats B J, Corey D P. The Usher syndromes (review). Am J Med Genet 199989158–166. [PubMed] [Google Scholar]
- 2.Espinos C, Millan J M, Beneyto M, Najera C. Epidemiology of Usher syndrome in Valencia and Spain. Community Genet 19981223–228. [DOI] [PubMed] [Google Scholar]
- 3.Smith R J, Berlin C I, Hejtmancik J F, Keays B J, Kimberling W J, Lewis R A, Moller C G, Pelias M Z, Tranebjaerg L. Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am J Med Genet 19945032–38. [DOI] [PubMed] [Google Scholar]
- 4.Eudy J D, Weston M D, Yao S, Hoover D M, Rehm H L, Ma‐Edmonds M, Yan D, Ahmad I, Cheng J J, Ayuso C, Cremers C, Davenport S, Moller C, Talmadge C B, Beisel K W, Tamayo M, Morton C C, Swaroop A, Kimberling W J, Sumegi J. Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 19982801753–1757. [DOI] [PubMed] [Google Scholar]
- 5.Weston M D, Luijendijk H W, Humphrey K D, Moller C, Kimberling W J. Mutations in the VLGR1 gene implicate G‐protein signalling in the pathogenesis of Usher syndrome type II. Am J Hum Genet 200474357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weston M D, Eudy J D, Fujita S, Yao S, Usami S, Cremers C, Greenberg J, Ramesar R, Martini A, Moller C, Smith R J, Sumegi J, Kimberling W J. Genomic structure and identification of novel mutations in usherin, the gene responsible for Usher syndrome type IIa. Am J Hum Genet. 2000;66: 1199–210 (Erratum in, Am J Hum Genet 2000662020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreyer B, Tranebjaerg L, Rosenberg T, Weston M D, Kimberling W J, Nilssen O. Identification of novel USH2A mutations: implications for the structure of USH2A protein. Eur J Hum Genet 20008500–506. [DOI] [PubMed] [Google Scholar]
- 8.Pennings R J, Te Brinke H, Weston M D, Claassen A, Orten D J, Weekamp H, Van Aarem A, Huygen P L, Deutman A F, Hoefsloot L H, Cremers F P, Cremers C W, Kimberling W J, Kremer H. USH2A mutation analysis in 70 Dutch families with Usher syndrome type II. Hum Mutat 200424185. [DOI] [PubMed] [Google Scholar]
- 9.Liu X Z, Hope C, Liang C Y, Zou J M, Xu L R, Cole T, Mueller R F, Bundey S, Nance W, Steel K P, Brown S D. A mutation (2314delG) in the Usher syndrome type IIA gene: high prevalence and phenotypic variation. Am J Hum Genet 1999641221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aller E, Najera C, Millan J M, Oltra J S, Perez‐Garrigues H, Vilela C, Navea A, Beneyto M. Genetic analysis of 2299delG and C759F mutations (USH2A) in patients with visual and/or auditory impairments. Eur J Hum Genet 200412407–410. [DOI] [PubMed] [Google Scholar]
- 11.Rivolta C, Sweklo E A, Berson E L, Dryja T P. Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 2000661975–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernal S, Ayuso C, Antinolo G, Gimenez A, Borrego S, Trujillo M J, Marcos I, Calaf M, Del Rio E, Baiget M. Mutations in USH2A in Spanish patients with autosomal recessive retinitis pigmentosa: high prevalence and phenotypic variation. J Med Genet 200340e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimberling W J, Weston M D, Möller C, van Aarem, Cremers C WRJ, Sumegi J, Ing P S, Connolly C, Martini A, Milani M, Tamayo M L, Bernal J, Greenberg J, Ayuso C. Gene mapping of Usher syndrome type IIa: localization of the gene to a 2.1 –cM segment on chromosome 1q41. Am J Human Genet 199556216–223. [PMC free article] [PubMed] [Google Scholar]
- 14.van Wijk E, Pennings R J, te Brinke H, Claassen A, Yntema H G, Hoefsloot L H, Cremers F P, Cremers C W, Kremer H. Identification of 51 novel exons of the Usher syndrome type 2A (USH2A) gene that encode multiple conserved functional domains and that are mutated in patients with Usher syndrome type II. Am J Hum Genet 200474738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adato A, Lefevre G, Delprat B, Michel V, Michalski N, Chardenoux S, Weil D, El‐Amraoui A, Petit C. Usherin, the defective protein in Usher syndrome type IIA, is likely to be a component of interstereocilia ankle links in the inner ear sensory cells. Hum Mol Genet 2005143921–3932. [DOI] [PubMed] [Google Scholar]
- 16.Reiners J, van Wijk E, Marker T, Zimmermann U, Jurgens K, Te Brinke H, Overlack N, Roepman R, Knipper M, Kremer H, Wolfrum U. The scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum Mol Genet 2005143933–3943. [DOI] [PubMed] [Google Scholar]
- 17.van Wijk E, van der Zwaag B, Peters T, Zimmerman U, Te Brinke H, Kersten F F, Marker T, Aller E, Hoefsloot L H, Cremers C W, Cremers F P, Wolfrum U, Knipper M, Roepman R, Kremer H. The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum Mol Genet 200615751–765. [DOI] [PubMed] [Google Scholar]
- 18.Najera C, Beneyto M, Blanca J, Aller E, Fontcuberta A, Millan J M, Ayuso C. Mutations in myosin VIIA (MYO7A) and usherin (USH2A) in Spanish patients with Usher syndrome types I and II, respectively. Hum Mutat 20022076–77. [DOI] [PubMed] [Google Scholar]
- 19.Leroy B P, Aragon‐Martin J A, Weston M D, Bessant D A, Willis C, Webster A R, Bird A C, Kimberling W J, Payne A M, Bhattacharya S S. Spectrum of mutations in USH2A in British patients with Usher syndrome type II. Exp Eye Res 200172503–509. [DOI] [PubMed] [Google Scholar]
- 20.Ouyang X M, Hejtmancik J F, Jacobson S G, Li A R, Du L L, Angeli S, Kaiser M, Balkany T, Liu X Z. Mutational spectrum in Usher syndrome type II. Clin Genet. 2004;65: 288–93 (Erratum in, _Clin Genet_200465433. [DOI] [PubMed] [Google Scholar]
- 21.Beneyto M, Cuevas J M, Millan J M, Espinos C, Mateu E, Gonzalez‐Cabo P, Baiget M, Domenech M, Bernal S, Ayuso C, Garcia‐Sandoval B, Trujillo M J, Borrego S, Antinolo G, Carballo M, Najera C. Prevalence of 2314delG mutation in Spanish patients with Usher syndrome type II (USH2). Ophthalmic Genet 200021123–128. [PubMed] [Google Scholar]
- 22.Dreyer B, Tranebjaerg L, Brox V, Rosenberg T, Moller C, Beneyto M, Weston M D, Kimberling W J, Cremers C W, Liu X Z, Nilssen O. A common ancestral origin of the frequent and widespread 2299delG USH2A mutation. Am J Hum Genet. 2001;69: 228–34 (Erratum in, _Am J Hum Genet_200169922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seyedahmadi B J, Rivolta C, Keene J A, Berson E L, Dryja T P. Comprehensive screening of the USH2A gene in Usher syndrome type II and non‐syndromic recessive retinitis pigmentosa. Exp Eye Res 200479167–173. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya G, Cosgrove D. Evidence for functional importance of usherin/fibronectin interactions in retinal basement membranes. Biochemistry 20054411518–11524. [DOI] [PubMed] [Google Scholar]
- 25.Kimberling W J. Estimation of the frequency of occult mutations for an autosomal recessive disease in the presence of genetic heterogeneity: application to genetic hearing loss disorders. Hum Mutat 200526462–470. [DOI] [PubMed] [Google Scholar]