Multilayer three-dimensional super resolution imaging of thick biological samples (original) (raw)
Abstract
Recent advances in optical microscopy have enabled biological imaging beyond the diffraction limit at nanometer resolution. A general feature of most of the techniques based on photoactivated localization microscopy (PALM) or stochastic optical reconstruction microscopy (STORM) has been the use of thin biological samples in combination with total internal reflection, thus limiting the imaging depth to a fraction of an optical wavelength. However, to study whole cells or organelles that are typically up to 15 μm deep into the cell, the extension of these methods to a three-dimensional (3D) super resolution technique is required. Here, we report an advance in optical microscopy that enables imaging of protein distributions in cells with a lateral localization precision better than 50 nm at multiple imaging planes deep in biological samples. The approach is based on combining the lateral super resolution provided by PALM with two-photon temporal focusing that provides optical sectioning. We have generated super-resolution images over an axial range of ≈10 μm in both mitochondrially labeled fixed cells, and in the membranes of living S2 Drosophila cells.
Keywords: nanoscopy, PALM microscopy, 3D imaging, temporal focusing
Initial studies, based on defocusing (1), astigmatism (2), and others (3), have extended super resolution imaging (2, 4–13) to three dimensions (3D); however, only up to a few hundred nanometers in depth in biological samples. This is mainly due to the following: In photoactivated localization microscopy (PALM) (6, 12) and stochastic optical reconstruction microscopy (STORM) (8), a sufficient signal-to-noise ratio is required to discriminate single molecules from the background. In the defocusing approaches, background typically increases with imaging depth and the signal is further reduced as it is spread out across more pixels, making the signal to noise ratio insufficient for molecules that are more than a few hundred nanometers away from the focal plane. In addition, PALM is inherently a wide-field technique that is not well suited to point-scanning methods. The reason for this is 2-fold: First, in PALM, single molecules need to be imaged, that is, their emission has to be spread out across several pixels (6) and second, data are most efficiently collected when emissions from spatially segregated molecules are recorded in parallel—point-scanning methods are inherently serial, and are thus slower than wide-field methods (14).
Thus, super resolution imaging would benefit greatly by adopting a wide-field technique and combining it with optical sectioning for enhanced signal to noise ratio at depth.
In this article, we describe a method whereby a thin layer of photoactivatable fluorescent proteins (15) can selectively be activated in a location several micrometers deep in a cellular sample. This thin layer is then excited and imaged using the PALM technique with a demonstrated resolution of better than 50 nm. A succession of thin layer images can be combined to produce a volume image several micrometers in depth.
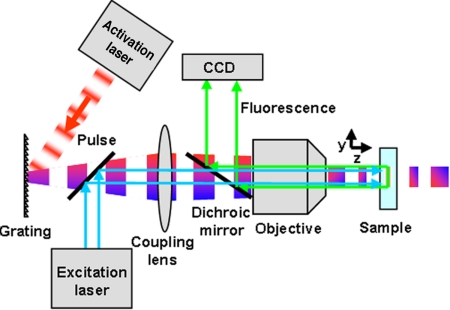
In a two-photon absorption process, the molecular excitation probability is proportional to the square of the intensity of the optical pulses. Thus, for constant pulse energy, the excitation probability in the focal plane can be increased by reducing the pulse width. In this approach, the molecular excitation is inversely proportional to pulse width squared and is called temporal focusing (16, 17). Temporal focusing is experimentally achieved by first broadening the pulse using a dispersive optical element such as a grating. The illuminated spot on the grating is then imaged onto the specimen plane using a telescope. This results in a pulse broadened everywhere in the sample except at the image plane, where the dispersion is compensated and the pulse reaches its minimum width (Fig. 1). Compared with an epi-fluorescence technique, this minimum width results in a depth of field that is orders of magnitude smaller (see supporting information (SI) Fig. S1 and SI Materials and Methods). Therefore, temporal focusing can be used for selective excitation of a thin layer of molecules in a biological sample.
Fig. 1.
Experimental setup for temporal focusing PALM. The activation light is provided by a two-photon temporally focused beam and the excitation light by a one-photon laser. Temporal focusing of the propagating pulse is achieved by imaging a spot on a grating onto the specimen via a two lens system (objective plus coupling lens). This 4f imaging configuration ensures that the pulse has the same (shortest) width in the specimen as at the grating, but the spatial and temporal dispersion caused by the grating generates a longer pulse outside the focal region.
The short depth of field of the temporal focusing and the fact that it is a wide-field technique makes it a suitable technique for subsurface layer PALM. Ideally, temporal focusing PALM would employ both two spatially overlapping two-photon, temporally focused beams, one at the two-photon excitation peak and the other at the two-photon activation peak of the photoactivatable protein. In this fashion, the two-photon activation of molecules would prevent all other molecules that are not in the same layer from unnecessary exposure and the two-photon excitation would lead to lower out-of-focus background. Temporal focusing PALM is also possible as long as sufficient overlap between the two-photon absorption spectrum of either the excitation or the activation of the desired photoactivatable fluorescent protein and the temporal focusing laser exists. In this case, the one of the beams can be implemented in a one-photon absorption wide-field configuration.
The localization precision provided by PALM is complemented when photoactivatable fluorescent proteins are used. These can be genetically expressed in a nonperturbative manner with high specificity in a target area that allows for functional cellular studies on the molecular level.
Results and Discussion
Depth of Field in Temporal Focusing.
In temporal focusing, the localization of excitation extends over the entire field of view and the intensity in the lateral plane drops off in the axial direction for a typical experimental configuration ≈ 75 times faster (16, 17) (Fig. S1) than a Gaussian beam with the same waist size. This gives the temporal focusing a depth of field and sectioning capability that is comparable with a confocal or two-photon microscope (18). We experimentally confirmed this by comparing the depth of focus of the temporal focusing in our setup with the depth of focus when the effect of the temporal focusing was removed (see SI Materials and Methods). The depth of field of the temporal focusing given our experimental parameters was measured to be 1.9 μm.
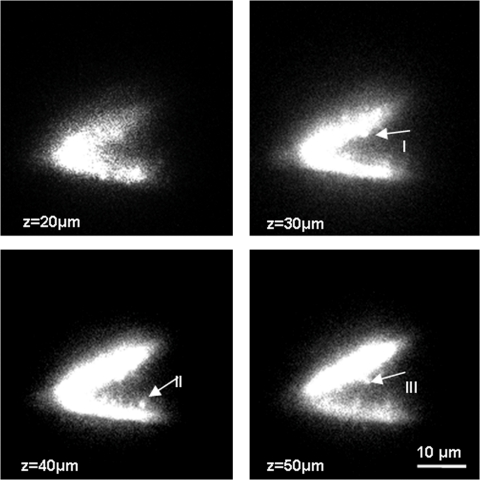
After confirming the sectioning performance of temporal focusing by axially scanning a thin fluorescent layer across the focal plane (see SI Materials and Methods), we next examined various biological samples. Acute transverse hippocampal slices (400 μm) from rat were prepared, microinjected with a fluorescent dye, and imaged in 1-μm steps over a range of 100 μm (see Movie S1). Four of the images at 10-μm intervals are shown in Fig. 2. Individual dendritic spines at different locations are visible in each frame. In acute slices, dendritic spines are typically only visible in a confocal or scanning two-photon microscope and to our knowledge have not been followed in 3D or in depth in a wide-field configuration.
Fig. 2.
Visualization of dendritic spines using temporal focusing. The depth of focus for a temporally focused beam with a spot size of ≈ 15 μm is shown. It is ≈ 1.9 μm, which is ≈ 75 times smaller than for the same spot size without temporal focusing (see SI Materials and Methods). Individual dendritic spines at different locations of a branched dendrite are visualized using temporal focusing. Top left image shows the membrane boundary of the two dendrites whereas the rest of images show the individual spines at I, II, and III coming in and going out of focus (see Movie S1 for the animation >100 μm). Images were taken at 10-μm steps in an acute hippocampal slice. Because the spines contained a lower concentration of dye compared with dendrites, the laser intensity was increased for the visualization of individual spines, which resulted in the saturation of the image at the dendrites.
Using Temporal Focusing for Super Resolution in 3D.
For temporal focusing PALM experiments, we used a two-photon activation, one-photon excitation approach and determined the suitability of several photoactivatable fluorescent proteins for this scheme. The two-photon activation responses of EosFP (19), Kikume (20), and Dronpa (21) were studied by using temporal focusing. We identified Dronpa as the molecule with the highest response to the temporal focusing activation light and chose it as the photoactivatable fluorescent protein for the following experiments. Under our experimental conditions, we collected an average of ≈ 600 photons from each Dronpa molecule before it switched off or bleached. Using other photoactivatable fluorescent proteins (such as EosFP), which emit more photons before bleaching and have a higher contrast ratio (ratio of the fluorescence intensity between the on and the off state), would likely allow a higher localization precision, lower background, and higher resolution (6). Efforts are underway in our laboratory to exploit these advantages by designing a tunable temporal focusing laser, thus allowing temporal focusing for both activation and excitation of many different photoactivatable fluorescent proteins.
The power for the activation laser was adjusted to activate just one molecule per diffraction limited area per frame within the acquisition time. The activated molecules were excited and imaged via a lens system, designed to fulfill the Nyquist-Shannon criterion (see ref. 22), onto an EMCCD camera until bleached. Subsequently, another subset of the molecules was photoactivated, excited, and bleached. This process was repeated for each layer until all molecules in that layer were bleached. Then, using a piezo element stage, the sample was axially translated by ≈ 1 μm and molecules in the next layer were imaged. We typically took PALM images from 5 to 8 different layers over a range of up to 10 μm within the cells.
Since in each layer the molecules within a single diffraction-limited volume are imaged at different times, their diffraction-limited emissions do not overlap. The centre of each molecular point spread function can thus be determined to a precision better than the diffraction limit, and assembling the aggregate position information from all molecules results in a super resolution image. The image processing was done using custom PALM analysis software (6). This routine searches for intensity peaks and fits them to 2D Gaussian profiles. As described in ref. 6, the Gaussian fit parameters, number of photons and background counts are used to determine the molecular positions and the corresponding localization uncertainties. The molecules are then rendered as Gaussian peaks with widths equal to these localization uncertainties so that the brightness of each pixel in the rendered PALM image can be interpreted as the probability distribution for finding the molecule at that location. However, in image processing there is a fundamental tradeoff between the number of molecules rendered and the image resolution; including fewer, but brighter, molecules results in higher localization and crisper images but at a reduced molecular density, giving less complete information about the spatial distribution of the target protein. In this article, we rendered all molecules that were localized to within 50 nm; this condition resulted in ≈ 80% of localized molecules being plotted.
Multilayer PALM of Mitochondrial Matrix and Drosophila S2 Cell Membrane.
As our first sample, we imaged fixed HFF1-cells that expressed Dronpa in the mitochondrial matrix (Fig. 3A). Mitochondrial expression was achieved by appending the mitochondrial matrix targeting sequence from cytochrome c oxidase VIII to the Dronpa fluorescent protein to mediate the import of Dronpa into the mitochondria (see SI Materials and Methods for details).
Fig. 3.
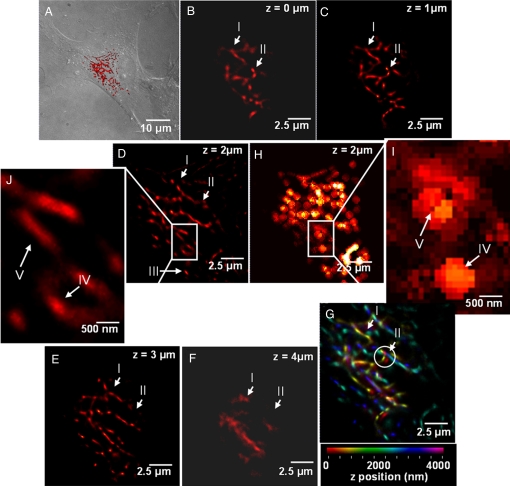
Multilayer super resolution imaging of mitochondrial network >5 μm in depth. (A) Overview of the region where the images were taken at 4× lower magnification than (B–G). The DIC images of the cell are overlaid with an epifluorescent image taken by a Xenon lamp and subsequently overlaid with (D) (red region). (B–F) PALM images of mitochondrial networks in HFF1-cell labeled with Dronpa taken in 1-μm steps. Different axial regions of the same mitochondria (region marked between I and II in B–E) and individual mitochondria (III in D) can be seen at different focal depths. (D and H) Comparison of the PALM image and the sum of diffraction-limited images for the same layer. (J and I) Five-fold magnification of a selected region from D and H, respectively. Several features seen as individual structures in J (IV and V) are clearly below the resolution limit in at the respective locations in I. (G) Superposition of (B–F) where the axial position is represented using a color map. (G) The 3D topology of the mitochondrial network. Interestingly, there are several features which are laterally as close as ≈ 200 nm but originate from planes that are axially 3 μm apart (structures within the circle in G) The volume information in (G) was further used to create a 3D animation (See SI Movie S2).
Fig. 3 B–F shows a series of these images taken in 1-μm steps from Dronpa-labeled reticular mitochondrial networks. Each PALM image contains structural details about the location of individual molecules and their relative locations, information that is not evident in the matching diffraction-limited images (Fig. 3 H and I). Moreover, the fact that images Fig. 3 B–F clearly show different mitochondrial structures in each layer demonstrates two points. First, it shows the effectiveness of the sectioning capability of temporal focusing for activating a single layer within the cell, thereby preventing activation of molecules in other layers. Such extraneous activation in other layers would result in lower axial sectioning and higher background. Second, it shows temporal focusing can be used to activate photoactivatable fluorescent proteins deep inside the cell and thus allows the collection of super resolution images at multiple axial locations within the sample. This has not been possible thus far with PALM and STORM, because these techniques have been limited to areas within 1 μm of the surface.
Fig. 3G shows the superposition of Fig. 3 B–F with colors coding for the axial position. Although the axial resolution in temporal focusing is significantly lower than the lateral resolution, individual PALM images at 1-μm steps can be combined to create a volume and rendered in three-dimensions. The 3D reconstruction of the images presented in Fig. 3 extends axially over a region of ≈5 μm and laterally over ≈ 15 μm. It clearly shows the relative spatial relation of different mitochondrial structures in three dimensions. Although the strength of temporal focusing PALM primary lies in the sectioning capability over large axial regions and the resulting capability for super resolution at depth, the super resolution images from individual layers can also be combined to create a three-dimensional rendering allowing the visualization of relative location of biological structures in a volume (see animation in Movie S2). The lacking super resolution in the third dimension in this approach can be improved by combining temporal focusing PALM with a recently developed technique which allows super resolution imaging in all three dimensions, but only over an axial length of 0.25 μm. (Shtengel G., Galbraith J.A., Galbraith C.G., Lippincott-Schwartz J., Gillette J.M., Manley S., Sougrat R., Waterman C.M., Kanchanawong P., Davidson M.W., Fetter R.D., Hess H.F., personal communication). This combination would then enable three-dimensional isotropic super resolution imaging in whole cells.
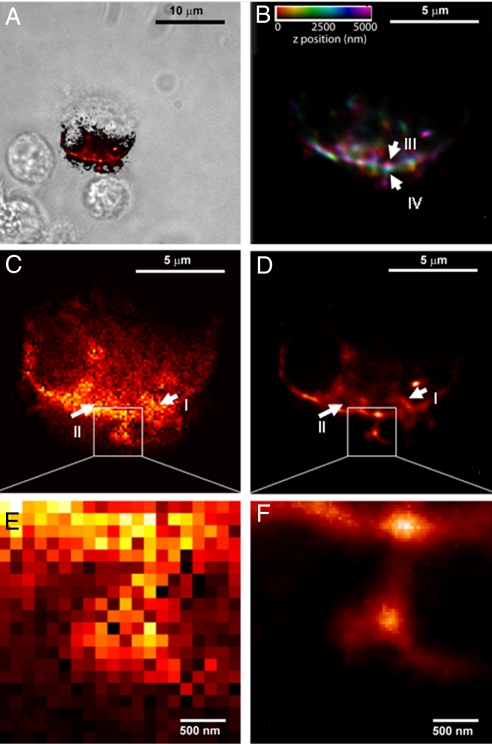
As a second example, we have demonstrated temporal focusing PALM in live Drosophila S2 cells expressing membrane targeted Dronpa (Fig. 4A) (Pfeiffer B., Rubin G.M., personal communication). PALM images were taken at six consecutive axial layers each separated by 1 μm; a representative layer and its close up are shown in Fig. 4 D and F. Fig. 4 C and D and their respective close ups, Fig. 4 E and F, show the comparison between the PALM image of a membrane region and the respective diffraction limited image both taken at the same axial depth. The PALM image in Fig. 4D shows details on the membrane region and some regions closer to the centre of the image (I and II) which are clearly below the resolution of the diffraction-limited image, Fig. 4C. This is more evident in the close up images Fig. 4 E and F. As in Fig. 3, the axial position of each PALM image was further used for color coding, and the images were subsequently superimposed, Fig. 4B. This color coded image displays information about the axial structure of the cell membrane, showing a sharp edge of ≈ 2.5 μm. Areas marked as III and IV that are laterally within 200 nm are axially separated by ≈ 2.5 μm. The axial topology of the membrane is thus revealed by using temporal focusing PALM.
Fig. 4.
Multilayer PALM of Drosophila S2 cells. (A) Stack of PALM images at six axial layers separated by 1 μm each are taken from membrane labeled Drosophila S2 cells. The axial position of each layer is used for color coding and the superimposed images are shown in (B). The color information clearly shows the axial structure of the cell membrane showing a sharp axial edge between III and IV of ≈ 2.5 μm over a lateral extension of less than ≈ 200 nm. (C and D) Comparison between PALM and diffraction-limited images for one of the layers. Although the area marked as I is resolved in the PALM image (D), it is clearly below the diffraction limit in (C). The area II in (D) shows a ring type structure that is not resolved in (C). Close-ups (E and F) show protrusions from the membrane. These are individually resolved in (F) and are below the diffraction limit in (E). (A) is a DIC/PALM overlay, showing the S2 cell and the region PALM-imaged in (B–F).
Outlook.
Using temporal focusing to generate PALM images in a volume has several key advantages. First, the optical sectioning characteristics provided by temporal focusing results in a higher signal to noise ratio over a larger axial range than previous techniques. In this sense, the ultimate imaging depth with temporal focusing can be expected to be comparable to a two-photon scanning microscope and is ultimately limited by scattering and aberrations*. Second, by using a slightly modified configuration and by using an objective with a higher numerical aperture, the axial resolution of temporal focusing could be improved to ≈ 0.8 μm. As mentioned earlier, the temporal focusing could then be combined with existing methods for further improving resolution in 3D and over larger axial extensions. Finally, the temporal focusing setup is relatively simple, stable and can be built by using noncustomized components.
In summary we have demonstrated super resolution and 3D imaging up ≈ 10-μm depths in different biological systems. This was achieved by combing the characteristics of photoactivatable fluorescent proteins with temporal focusing PALM. Given the power of temporal focusing making optical sectioning over a range of many tens of micrometers possible, this technique can be expected to become a method of choice when single-molecule resolution in larger volumes is required. Areas that could benefit from temporal focusing PALM include developmental biology on the molecular level or in neuroscience studies using Drosophila where questions of neural circuitry could be addressed on the level of whole brains.
Materials and Methods
Instrumentation.
To demonstrate PALM in layers deep within the cell, we used temporal focusing to activate ≈ 1.9-μm thick layers of Dronpa-tagged proteins that were subsequently excited by a diode laser at 471 nm and imaged onto an electron multiplying CCD (EMCCD) camera. The Dronpa molecules in each layer were activated by two-photon absorption from a near IR femtosecond pulsed laser source. The light was directed to an uncoated gold diffraction grating with a spatial period of 100 grooves per mm, which was used to spread the spectral components of the pulse in space. The light from the first diffraction order of the grating was passed through a coupling lens, which together with a 60× water-immersion objective (NA = 1.2) imaged the ≈ 3-mm spot on the grating onto the specimen plane. This resulted in a field of view of the temporal focusing of ≈ 15 μm. The excitation laser was expanded to match the size of the activation region. Both beams were combined after the grating by using a long pass dichroic mirror (Fig. 1).
In the multilayer 3D PALM experiments, temporal focusing was used to activate ≈ 1.9-μm thick layers of Dronpa-tagged proteins that were subsequently excited by a diode laser and imaged onto an EMCCD camera. A 10 femtosecond-pulsed laser source (FemtoLasers) with a central wavelength at 795 nm and a spectral full width half maximum (FWHM) of ≈ 105 nm was used as the source for TF. The beam freely propagated to a spot size of ≈ 3 mm and was diffracted by an uncoated gold diffraction grating with a groove density of 100 lines per mm and a blazing angle of 2.3° toward the imaging system. The first order diffracted beam was next passed through an achromatic lens (Linos) with a focal length f = 60 cm, which together with the 60× microscope objective (Olympus UPLSAPO60XW) imaged the spot on the grating onto the specimen plane. The objective was housed in an inverted microscope (Olympus IX-71) in Epi-fluorescence configuration. The TF light was reflected by a customized dichroic mirror (Chroma) that was designed to have two reflection bands [590–920 nm, covering the spectrum of the femtosecond laser and 400–490 nm for reflecting a 471-nm laser (required for excitation of Dronpa)] and one transmission band with FWHM of ≈ 50 nm centered at 535 nm (for Dronpa emissions). The specimen plane was imaged (after passing through the 60× objective) via a 1.6× magnification coupling lens onto a back illuminated EM-CCD camera (Andor iXon DU-888, Andor Technology) with a pixel size of 13 μm. The magnification was chosen to satisfy the Nyquist criteria (22), which requires that the sampling frequency be at least twice as high as the highest frequency present in the signal. In our case, it meant that the magnification had to be designed so that the standard deviation in the PSF is approximately equal to one pixel. The excitation light for PALM was provided by a 471-nm diode laser (Spectra Physics) that was combined with the TF light after the grating via a long pass dichroic mirror (Semrock). Typical frame acquisition times for the PALM images were ≈ 0.1 s, and for each image layer ≈ 10,000 frames were acquired. The excitation power was typically attenuated to ≈ 4 μW and the TF activation light power was ≈ 4 mW before the microscope objective. While the excitation laser was on during the entire acquisition time, the activation laser was switched on for ≈ 2 s following an approximate 8-s off period.
Cover Slip Preparation.
Twenty-five-mm glass cover slips were (i) incubated in 5:1:1 Milli-Q filtered H2O: ammonium hydroxide: hydrogen peroxide for ≈ 12 h at 90°C; (ii) serially rinsed in H2O and methanol; (iii) flamed; (iv) transferred to 35-mm cell culture plates; (v) coated overnight with 10 μg/ml fibronectin-1× PBS at 4°C; (vi) suctioned to remove fibronectin solution; and (vii) incubated with 2 ml of growth medium under recommended growth conditions per cell line.
Plasmid Construction.
Dronpa mitochondria.
To produce the mitochondrially targeted Dronpa, the plasmid tdEos-mito containing the mitochondria targeting sequence of cytochrome c oxidase VIII was digested with _Bam_HI and _Not_I to remove the tdEos fluorescent protein. This was replaced with a Dronpa-containing insert that was isolated from a similarly digested plasmid Dronpa-Paxillin. The tdEos-Mito and Dronpa-Paxillin plasmids were a gift from Michael W. Davidson (National High Magnetic Field Laboratory, Florida State University, Tallahassee, Florida).
S2 Dronpa membrane tagged.
A membrane-tagged Drosophila codon optimized Dronpa protein was used in S2 cell assays (Pfeiffer B., Rubin G.M., personal communication). The Dronpa plasmids for S2 cells were a gift from Barret Pfeifer (Howard Hughes Medical Institute, Janelia Farm research Campus, Ashburn, VA).
Cell Culture and Transfection.
Dronpa mitochondria.
HFF-1 cells (ATCC, SCRC-1041) passage 19, were grown in DMEM-HG (without phenol red) containing 15% FBS at 37°C in 5% CO2 to 75% confluency. Trypsinized cells were then transiently transfected at 8 × 105 cells per shuttle well using the Nucleofector 96-well shuttle system (Amaxa Biosystems) using Cell Line Nucleofector Kit SE, program DS-137, and 1.0 μg per shuttle well of plasmid DNA.
Nucleofected cells were (i) transferred to the cover slips in 35-mm culture plates; (ii) incubated at 37°C in 5% CO2 for 24–36 h; (iii) fixed for ≈ 15 min with 2% paraformaldehyde in PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mMMgCl2, pH 6.9); and (iv) rinsed 3× with PHEM.
S2 Dronpa membrane tagged.
S2 (DGRC) passage JFRC + 14 were grown in Schneider's Drosophila Medium containing 10% heat-inactivated FBS.
Cells were (i) cotransfected with 150 ng Ub-GAL4 and 250 ng of pJFRC-Dronpa using Effectene (Qiagen); (ii) transferred to the cover slips in 35-mm culture plates; (iii) incubated at 25°C for ≈ 24–36 h; (iv) fixed for ≈ 15 min with 2% paraformaldehyde in PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mMMgCl2, pH 6.9); and (v) rinsed 3× with PHEM.
Acute hippocampal slices.
Transverse hippocampal slices (400 μm) were prepared from 6 to 12-week-old Sprague Dawley rats. According to methods approved by the Howard Hughes Medical Institute, Janelia Farm Research Campus Institutional Animal Care and Use Committee, animals were anesthetized by a lethal dose of isoflurane inhalation and perfused through the heart with ice-cold cutting solution containing (mM): 234 sucrose, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 0.5 CaCl2, 7 MgCl2, and 7 dextrose, bubbled with 95% O2–5% CO2 at ≈ 0°C (pH 7.4). Brains were rapidly removed after decapitation and placed in a cold oxygenated cutting solution. Slices were prepared using a vibratome (Vibratome), and subsequently kept in artificial cerebrospinal fluid (ACSF) [containing (in mM): 125 NaCl, 3 KCl, 25 NaHCO3, 1.25 NaH2PO4, 1.3 CaCl2, 1 MgCl2, 25 glucose, 3 Na-pyruvate, and 1 ascorbic acid], as normal external solution, for 30 min at 37°C, and then at room temperature. Experiments were conducted from the soma or apical trunk of CA1 pyramidal cells.
Patch pipettes (5–8 MΩ) were pulled from borosilicate glass and filled with an internal solution containing (mM): 140 CsCl, 0.5 EGTA, 4 NaCl, 0.3 CaCl2, 4 Mg2ATP, 0.3 Tris2GTP, 14 phosphocreatine, and 10 Hepes (pH 7.25) Pipette solution additionally contained 50 μM Alexa Fluor 488 hydrazide (Molecular Probes) to visualize and identify the cell with two-photon microscopy.
All neurons had resting potentials between −55 and −70 mV. Series resistances from dendritic whole-cell recordings were between 10 and 40 MΩ.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Kevin McGowan and Barret Pfeiffer for plasmid design and construction, Helen White and Alma Arnold for sample preparation and fixation, Judit Makara for help with brain slices, Eric Betzig and Harald Hess for many helpful discussions and for the use of the PALM code and Gleb Shtengel for help in image processing. This work was supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*
The ultimate imaging depth in temporal focusing when using ultra short pulses (i.e., ≈10 fs) is determined by two competing processes; the spectral width (or pulse duration) and dispersion of the ultra short pulses in the sample. In temporal focusing, different frequency components within the pulse enter the back focal plane of the objective at different locations. Therefore, the only location in the sample where these frequency components overlap and create two-photon excitation is the focal plane. In contrast, in a conventional two-photon microscope, all frequency components are distributed equally in each lateral plane. As a result, although in the conventional two-photon microscope, the axial integration over the out-of-focus fluorescent light from each lateral plane is one of the factors limiting the ultimate imaging depth, this is not the case in the temporal focusing configuration. For two-photon absorption, the sum energy of the two photons has to match the optical transition energy. Therefore, the separation of spectral components in the lateral plane guarantees that the axial integration over the out of focus volume does not contribute to a two-photon absorption process. This effect is more pronounced for shorter pulses because in the frequency domain they represent a wider spectral range and therefore in the focusing process, a wider separation of the individual frequency components. However, shorter pulses are more susceptible to dispersion as they propagate through a sample that in turn increases the pulse width. In this regard the suitability of ultra short pulses for imaging in depth and its comparison with the widely used 100-fs pulses calls for a systematic experimental study.
References
- 1.Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 2.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt R, et al. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 4.Gustafsson MGL, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustafsson MGL. Super-resolution light microscopy goes live. Nat Methods. 2008;5:385–387. doi: 10.1038/nmeth0508-385. [DOI] [PubMed] [Google Scholar]
- 6.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y-l, Hahn KM, Murphy RF, Horwitz AF. From imaging to understanding: Frontiers in live cell imaging, Bethesda, MD, April 19–21, 2006. J Cell Biol. 2006;174:481–484. doi: 10.1083/jcb.200607097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 10.Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 11.Donnert G, et al. Macromolecular-scale resolution in biological fluorescence microscopy. PNAS. 2006;103:11440–11445. doi: 10.1073/pnas.0604965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess ST, Girirajan TPK, Mason MD. Ultra-High resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fölling J., et al. Photochromic rhodamines provide nanoscopy with optical sectioning 13. Angew Chem Int Ed. 2007;46:6266–6270. doi: 10.1002/anie.200702167. [DOI] [PubMed] [Google Scholar]
- 15.Wiedenmann J, Nienhaus GU. Live-cell imaging with EosFP and other photoactivatable marker proteins of the GFP family. Expert Rev Proteomics. 2006;3:361–374. doi: 10.1586/14789450.3.3.361. [DOI] [PubMed] [Google Scholar]
- 16.Oron D, Tal E, Silberberg Y. Scanningless depth-resolved microscopy. (Translated from English) Optics Express. 2005;13:1468–1476. doi: 10.1364/opex.13.001468. in English. [DOI] [PubMed] [Google Scholar]
- 17.Zhu GH, van Howe J, Durst M, Zipfel W, Xu C. Simultaneous spatial and temporal focusing of femtosecond pulses. Optics Express. 2005;13:2153–2159. doi: 10.1364/opex.13.002153. [DOI] [PubMed] [Google Scholar]
- 18.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 19.Wiedenmann J, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miywaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habuchi S, et al. From The Cover: Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci USA. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon CE. Communication in the presence of noise. Proc IRE. 1949;37:10–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information