Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes (original) (raw)
Abstract
Advances in molecular phylogeny of eukaryotes have suggested a tree composed of a small number of supergroups. Phylogenomics recently established the relationships between some of these large assemblages, yet the deepest nodes are still unresolved. Here, we investigate early evolution among the major eukaryotic supergroups using the broadest multigene dataset to date (65 species, 135 genes). Our analyses provide strong support for the clustering of plants, chromalveolates, rhizarians, haptophytes and cryptomonads, thus linking nearly all photosynthetic lineages and raising the question of a possible unique origin of plastids. At its deepest level, the tree of eukaryotes now receives strong support for two monophyletic megagroups comprising most of the eukaryotic diversity.
Keywords: eukaryote evolution, deep phylogeny, phylogenomics, endosymbiosis, root, megagroup
Abbreviations: HC; monophyletic grouping of haptophytes and cryptomonads; SAR; monophyletic grouping of stramenopiles, alveolates and Rhizaria
1. Introduction
Resolving the global tree of eukaryotes is one of the most important goals in evolutionary biology. Molecular phylogenies, morphology and biochemical characteristics have allowed the division of the majority of eukaryotic diversity into five or six putative supergroups (reviewed in Keeling et al. (2005) and Lane & Archibald (2008)); these comprise the opisthokonts and Amoebozoa (united as ‘unikonts’; Cavalier-Smith 2002), Plantae (or Archaeplastida), Excavata, Chromalveolata and Rhizaria (often considered as members of the so-called ‘bikonts’; Stechmann & Cavalier-Smith (2003_a_)). Recent phylogenomic reconstructions based on large sequence datasets have been used to infer the relationships between some of these large assemblages, and notably Rhizaria have been shown to share a common origin with members of the chromalveolates (Burki et al. 2007; Hackett et al. 2007; Rodríguez-Ezpeleta et al. 2007_a_). However, the order of divergence among the deepest nodes remains uncertain, particularly the relationships between plants, chromalveolates and other photosynthetic lineages (haptophytes and cryptomonads). In order to investigate early evolution among eukaryotic supergroups, we have assembled the broadest dataset to date (65 species, 135 genes representing 31 921 amino acids) and show that the eukaryotes can be divided into two highly supported monophyletic megagroups and a few less diversified lineages related to the excavates.
2. Material and methods
Our multigene dataset was assembled according to a custom pipeline, as follows: (i) construction of databases made of all existing sequences for species specifically selected for their broad taxonomic distribution and availability of genomic sequences (downloaded from http://www.ncbi.nlm.nih.gov/ and http://amoebidia.bcm.umontreal.ca/pepdb/searches/welcome.php), (ii) BLAST searches against these databases using as queries the single-gene sequences composing our previously described multiple alignments (Burki et al. 2007), (iii) retrieval (with a stringent _e_-value cut-off at 10−50) and addition of the new homologous copies to the existing single-gene alignments, (iv) automatic alignments using MAFFT (Katoh et al. 2002), followed by manual inspection to extract unambiguously aligned positions, (v) testing the orthology, in particular possible lateral or endosymbiotic gene transfer, for each of the selected genes by performing single-gene maximum-likelihood (ML) reconstructions using Treefinder Whelan and Goldman (WAG, four gamma categories; Jobb et al. 2004), and (vi) the final concatenation of all single-gene alignments was done using SCaFoS (Roure et al. 2007). Owing to the limited data for certain groups and to maximize the number of genes by taxonomic assemblage, some lineages were represented by different closely related species always belonging to the same genus (electronic supplementary material). Potential interesting species with full genomes available, such as the excavates Giardia and Trichomonas or the red algae Cyanidioschyzon, have been discarded from our taxon sampling owing to their extreme rate of sequence evolution or their demonstrated tendency to lead to systematic errors in phylogenies (Rodríguez-Ezpeleta et al. 2007_b_).
The concatenated alignment was analysed using both bayesian (BI) and ML frameworks, with Phylobayes v. 2.3 (Lartillot & Philippe 2004) and RAxML-VI-HPC v. 2.2.3 (Stamatakis 2006), respectively. Phylobayes was run using the site-heterogeneous mixture CAT model and two independent Markov chains with a total length of 10 000 cycles, discarding the first 4000 points as burn-in and calculating the posterior consensus on the remaining 6000 trees. The convergence between the two chains was checked and always led to the exact same tree, except for uncertainties of the order of divergence between the glaucophytes, the red algae and haptophytes+cryptomonads (HC). In order to reduce mixing problems of the chains, the constant sites were removed from the alignment in a subsequent analysis. The convergence was in this case much quicker, after only 5000 cycles (burn-in of 1000), and HC was unambiguously positioned as sister to the Plantae. RAxML was used in combination with the WAG amino acid replacement matrix and stationary amino acid frequencies estimated from the dataset. The best ML tree was determined with the PROTMIX implementation, in a multiple inferences using 20 randomized maximum parsimony (MP) starting trees. Statistical support was evaluated with 100 bootstrap replicates. Two independent runs were performed on each replicate, using a different starting tree (MP and the best ML tree), in order to prevent the analysis from getting trapped in a local maximum. The tree with the best log likelihood was selected for each replicate, and the 100 resulting trees were used to calculate the bootstraps proportions. To save computational burden, the PROTMIX solution was chosen with 25 distinct rate categories. To minimize potential systematic errors associated with saturation and homoplasy, the fast-evolving sites were identified using PAML (Yang 1997), given the 20 topologies obtained in the ML analysis. Sites were classified according to their mean site-wise rates and ML bootstrap values were computed from shorter concatenated alignments with sites corresponding to categories 7 and 6+7 removed.
3. Results
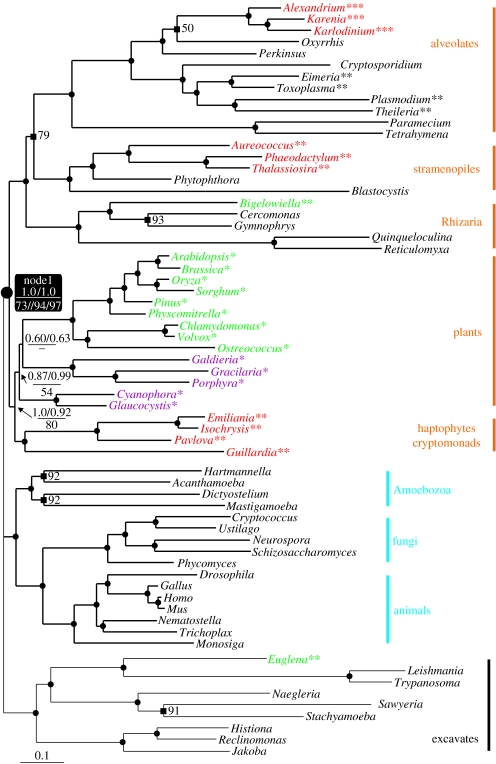
We first performed a bayesian analysis on a species-rich dataset, using the powerful CAT model that has been developed to overcome systematic errors due to homoplasy (Lartillot & Philippe 2004; Lartillot et al. 2007; figure 1). The tree obtained is in agreement with previously published studies; it strongly supports monophyletic groupings of unikonts (Amoebozoa, fungi and animals), excavates, plants, stramenopiles+alveolates+Rhizaria (SAR) and HC. This latter group appears as sister to plants, with 1.0 Bayesian posterior probability (PP) when the constant sites were removed and 0.92 PP with the full-length alignment. Remarkably, the plants+HC clade form a strongly supported monophyletic megagroup with the SAR assemblage (1.0 PP, node 1), revealing an ancient split in eukaryote evolution and almost entirely resolving the relationships within most ‘bikont’ supergroups.
Figure 1.
Bayesian unrooted phylogeny of eukaryotes, with a basal trichotomy representing uncertainties in the relationships between the three groups. The tree was obtained from the consensus between two independent Markov chains, run under the CAT model implemented in Phylobayes. The species colour code corresponds to the type of plastid pigments, as follows: purple, chlorophyll a; green, chlorophyll a+b; and red, chlorophyll a+c. The asterisks represent primary, secondary or tertiary endosymbiosis. Underlined numbers at nodes represent PP of the analysis performed with the constant sites removed/analysis performed with all sites; other numbers represent the result of the ML bootstrap analysis (BS)—Node 1 below the line: ML analysis of the full-length alignment//ML analysis with category 7 removed/ML analysis with catergories 6+7 removed. Black dots correspond to 1.0 PP and 100% BS; black squares correspond to 1.0 PP and the specified values of BS. The scale bar represents the estimated number of amino acid substitutions per site.
This new megagroup received relatively low support (73% bootstrap support, BS) in the ML analysis of the complete dataset (figure 1). However, because we are investigating relationships deriving from very ancient splits in the eukaryotic tree, it is probable that multiple substitutions occurred at several sites in our alignment, decreasing the true phylogenetic signal and rendering standard site-homogeneous models based on empirical matrices of amino acid replacement (such as WAG) less accurate. To test this further, we investigated the effect of the exclusion of the fastest evolving sites, which are more likely to be saturated and thus be the cause of model violations (Rodríguez-Ezpeleta et al. 2007_b_). Not surprisingly, the removal of the noisiest positions led to a drastic increase in the statistical support for the new megagroup (94 and 97% BS when categories 7 and 6+7 were removed, respectively; figure 1).
4. Discussion
At its deepest level, the tree of eukaryotes presented here displays only three stems, i.e. the two highly supported megagroups, enclosing the vast majority of eukaryotic species, and the excavates. If the monophyly of excavates is further confirmed and strong support is found for their possible sister position to the new megagroup, we may well be able to provide independent evidence (based on phylogenetic reconstructions) for the concept of the two primary clades of eukaryotes—unikonts and bikonts (Stechmann & Cavalier-Smith 2003_b_; Richards & Cavalier-Smith 2005). This model, however, would need to be modified as the widely used dihydrofolate reductase-thymidylate synthase gene fusion is questionable for several reasons (see discussion in Kim et al. 2006). Of course, this does not rule out the possibility that some protists, such as Telonemia or the centrohelid heliozoans that have not yet been placed with confidence (Shalchian-Tabrizi et al. 2006; Sakaguchi et al. 2007), might represent additional independent lineages. But generally, we believe that most eukaryotes fall into one of these megagroups.
As we are getting closer to a fully resolved phylogeny for the eukaryotes, an obvious question of crucial importance is the position for the root. We chose, however, to show an unrooted tree as the absence of compelling information leaves the rooting of the eukaryotic tree an open question. Over the past few years, independent data proposed a root lying either between unikonts and bikonts (Stechmann & Cavalier-Smith 2003_b_) or within excavates, e.g. basal to jakobids (Rodríguez-Ezpeleta et al. 2007_a_) or on the branch leading to diplomonads/parabasalids (Arisue et al. 2005). In the absence of evidence for rooting the eukaryotes within the plants+HC+SAR megagroup, the plausible rooting scenarios together with our tree consistently suggest that this assemblage is holophyletic.
Our results bring convincing support for the clustering of almost all photosynthetic groups in a unique clade (with the notable exception of the second-hand green plastids in Euglenozoa, belonging to the excavates), and sustain a single primary endosymbiotic event as also suggested by gene-based models of the import machinery (McFadden & van Dooren 2004). The strongest scenario to date for the evolution of primary plastid-containing species is that a unique endosymbiosis involving a cyanobacterium took place in the last common ancestor of Plantae (see Bhattacharya et al. 2007). The trees presented here allow the possibility that the primary plastid was established even earlier in one of the ancestors of the new megagroup, and was subsequently lost and independently replaced by plastids of secondary origin in several lineages (HC, Rhizaria, alveolates and stramenopiles), corroborating the hypothesis of an early chloroplast acquisition in eukaryotes based on the phylogeny of the 6-phosphogluconate dehydrogenase gene (Andersson & Roger 2002; see also Nosaki (2005) for a more general discussion). We speculate that the high observable diversity of plastids within the new megagroup can be traced back to its last common ancestor, and is the consequence of an increased capability of all its members to accept and keep plastids or plastid-bearing cells.
Acknowledgments
The authors would like to thank the Vital-it server (www.vital-it.ch) and the Bioportal platform (www.bioportal.uio.no) for allowing us to perform phylogenomic analyses. They are also very grateful to the Canadian consortium PEP that has made publicly available several EST projects through its database (http://tbestdb.bcm.umontreal.ca/searches/welcome.php). This research was supported by the Swiss National Science Foundation grant 3100A0-112645 (J.P.).
Supplementary Material
Additional material
Table S1 OTU (Operational Taxonomic Unit) names and chimera constructions. Table listing the full species names used to build the concatenated alignment, as well as specifying the species used to construct chimera. The new species in our alignment, compared to our previously published dataset (Burki et al. 2007), are marked in bold.
Additional material
Table S2 Percentage of missing data per species and per genes. Detailed list of the 135 genes used, precising the amount of missing data per species Burki, F., Shalchian-Tabrizi, K., Minge, M., Skjaeveland, A., Nikolaev, S. I., Jakobsen, K. S. & Pawlowski, J. 2007 Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 2, e790.
References
- Andersson J.O, Roger A.J. A cyanobacterial gene in nonphotosynthetic protists—an early chloroplast acquisition in eukaryotes? Curr. Biol. 2002;12:115–119. doi: 10.1016/s0960-9822(01)00649-2. doi:10.1016/S0960-9822(01)00649-2 [DOI] [PubMed] [Google Scholar]
- Arisue N, Hasegawa M, Hashimoto T. Root of the eukaryota tree as inferred from combined maximum likelihood analyses of multiple molecular sequence data. Mol. Biol. Evol. 2005;22:409–420. doi: 10.1093/molbev/msi023. doi:10.1093/molbev/msi023 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Archibald J.M, Weber A.P, Reyes-Prieto A. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays. 2007;29:1239–1246. doi: 10.1002/bies.20671. doi:10.1002/bies.20671 [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev S.I, Jakobsen K.S, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One. 2007;2:e790. doi: 10.1371/journal.pone.0000790. doi:10.1371/journal.pone.0000790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Hackett J, Yoon H, Li S, Reyes-Prieto A, Rümmele S, Bhattacharya D. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. doi:10.1093/molbev/msm089 [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. Treefinder: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. doi:10.1186/1471-2148-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT version 5.25: multiple sequence alignment program. Nucl. Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. doi:10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P.J, Burger G, Durnford D.G, Lang B.F, Lee R.W, Pearlman R.E, Roger A.J, Gray M.W. The tree of eukaryotes. Trends Ecol. Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. doi:10.1016/j.tree.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Kim E, Simpson A.G, Graham L.E. Evolutionary relationships of apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol. Biol. Evol. 2006;23:2455–2466. doi: 10.1093/molbev/msl120. doi:10.1093/molbev/msl120 [DOI] [PubMed] [Google Scholar]
- Lane C.E, Archibald J.M. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. doi:10.1016/j.tree.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. doi:10.1093/molbev/msh112 [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 2007;7(Suppl. 1):S4. doi: 10.1186/1471-2148-7-S1-S4. doi:10.1186/1471-2148-7-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G.I, van Dooren G.G. Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. doi:10.1016/j.cub.2004.06.041 [DOI] [PubMed] [Google Scholar]
- Nosaki H. A new scenario of plastid evolution: plastid primary endosymbiosis before the divergence of the “Plantae”, emended. J. Plant Res. 2005;111:247–255. doi: 10.1007/s10265-005-0219-1. doi:10.1007/s10265-005-0219-1 [DOI] [PubMed] [Google Scholar]
- Richards T.A, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. doi:10.1038/nature03949 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burger G, Roger A.J, Gray M.W, Philippe H, Lang B.F. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr. Biol. 2007a;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. doi:10.1016/j.cub.2007.07.036 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang B.F, Philippe H. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 2007b;56:389–399. doi: 10.1080/10635150701397643. doi:10.1080/10635150701397643 [DOI] [PubMed] [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol. Biol. 2007;7(Suppl. 1):S2. doi: 10.1186/1471-2148-7-S1-S2. doi:10.1186/1471-2148-7-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M, Inagaki Y, Hashimoto T. Centrohelida is still searching for a phylogenetic home: analyses of seven Raphidiophrys contractilis genes. Gene. 2007;405:47–54. doi: 10.1016/j.gene.2007.09.003. doi:10.1016/j.gene.2007.09.003 [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, et al. Telonemia, a new protist phylum with affinity to chromist lineages. Proc. R. Soc. B. 2006;273:1833–1842. doi: 10.1098/rspb.2006.3515. doi:10.1098/rspb.2006.3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. doi:10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J. Mol. Evol. 2003a;57:408–419. doi: 10.1007/s00239-003-2490-x. doi:10.1007/s00239-003-2490-x [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr. Biol. 2003b;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. doi:10.1016/S0960-9822(03)00602-x [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional material
Table S1 OTU (Operational Taxonomic Unit) names and chimera constructions. Table listing the full species names used to build the concatenated alignment, as well as specifying the species used to construct chimera. The new species in our alignment, compared to our previously published dataset (Burki et al. 2007), are marked in bold.
Additional material
Table S2 Percentage of missing data per species and per genes. Detailed list of the 135 genes used, precising the amount of missing data per species Burki, F., Shalchian-Tabrizi, K., Minge, M., Skjaeveland, A., Nikolaev, S. I., Jakobsen, K. S. & Pawlowski, J. 2007 Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 2, e790.