Quantitative Proteomics Reveals the Function of Unconventional Ubiquitin Chains in Proteasomal Degradation (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 3.
Summary
All seven lysine residues in ubiquitin contribute to the synthesis of polyubiquitin chains on protein substrates. Whereas K48-linked chains are well established as mediators of proteasomal degradation, and K63-linked chains act in nonproteolytic events, the roles of unconventional polyubiquitin chains linked through K6, K11, K27, K29, or K33 are not well understood. Here we report that the unconventional linkages are abundant in vivo, and all non-K63 linkages may target proteins for degradation. Ubiquitin with K48 as the single lysine cannot support yeast viability, and different linkages have partially redundant functions. By profiling both the entire yeast proteome and ubiquitinated proteins in wild-type and ubiquitin K11R mutant strains using mass spectrometry, we identified K11 linkage-specific substrates, including Ubc6, a ubiquitin conjugating enzyme involved in endoplasmic reticulum-associated degradation (ERAD). Ubc6 primarily synthesizes K11-linked chains, and K11 linkages function in the ERAD pathway. Thus, unconventional polyubiquitin chains are critical for ubiquitin-proteasome system function.
Keywords: ubiquitin, polyubiquitin chains, K11 linkages, protein degradation, proteasome, Ubc6, mass spectrometry, proteomics, SILAC
Introduction
Protein ubiquitination is a highly conserved modification that controls a variety of cellular processes (Kerscher et al., 2006). Ubiquitin (Ub) is covalently attached to protein substrates by a cascade of enzymatic reactions involving activating enzymes (E1), conjugating enzymes (E2), and ligases (E3). These reactions generally form an isopeptide bond between the carboxyl group of the C-terminus of Ub (G76) and the ε-amino group of a lysine residue within the substrates. All seven lysines in Ub contribute to the assembly of polyUb chains, thereby producing a variety of structures with diverse lengths and linkages (Peng et al., 2003b; Pickart and Fushman, 2004). Furthermore, ubiquitination is reversible through the action of deubiquitinating enzymes (DUBs). The specificity of Ub signaling is mediated by substrate recognition via the E3 ligases, the interaction of Ub moieties (monoUb and polyUb) with Ub receptors, and Ub removal by specific DUBs. More recently, the regulation of Ub pathways by Ub chains bearing distinct polyUb linkages has emerged as an important mechanism.
The synthesis of particular polyUb chain linkages appears to be a function of specific E2s and the E2-E3 combinations involved (Jin et al., 2008; Kim et al., 2007; Kirkpatrick et al., 2006). For example, the Cdc34 E2 generates predominantly K48-linked polyUb products, and its specificity is dependent on an acidic loop region of Cdc34 (Petroski and Deshaies, 2005). The Mms2/Ubc13 E2 complex preferentially catalyzes the formation of polyUb with K63 linkages, because the structure of Mms2 allows for selective insertion of the K63 side chain of Ub into the Ubc13 active site during chain elongation (Eddins et al., 2006). The interaction between Ub enzymes and specific surface residues adjacent to the lysine sites in Ub may be important for selection of the docking site during chain assembly (Wang et al., 2006). Indeed, a “TEK-box” in Ub facilitates the formation of K11-linked polyUb by the APC E3 and UbcH10 E2 (Jin et al., 2008).
Ubiquitinated substrates may be sorted into different pathways based on diverse polyUb structures (Pickart and Fushman, 2004). Canonical K48-linked polyUb chains are believed to be the principal signal for targeting substrates for degradation by the 26S proteasome, whereas K63-linked chains act in a range of other processes, including protein trafficking, DNA repair, and inflammation. Monoubiquitination also functions as a nonproteolytic signal. However, the prevalence of other polyUb topologies and their roles in biological processes are not well understood, in spite of a few intriguing observations. K6 linkages catalyzed by the BRCA1/BARD1 E3 might regulate DNA repair (Nishikawa et al., 2004). K11 linkages were suggested to support proteasomal degradation of certain protein targets (Baboshina and Haas, 1996; Jin et al., 2008; Kim et al., 2007; Kirkpatrick et al., 2006). Both K27 and K33 linkages may be assembled by U-box-type E3 ligases during stress response (Hatakeyama et al., 2001). Finally, K29-linked chains may participate in Ub-fusion degradation (Johnson et al., 1995).
In the present study, we combined genetic and proteomic approaches to systematically study the function of different polyUb linkages in the yeast S. cerevisiae. Our data revealed unexpectedly high levels of unconventional polyUb linkages on a broad range of substrates in cells. We found that proteasomal degradation is mediated by all non-K63 polyUb linkages in vivo. K11 linkages preferentially modify a subset of protein substrates and play an important role in endoplasmic reticulum-associated degradation (ERAD).
Results
Mass Spectrometry Reveals Abundant Non-K48 Linkages in PolyUb Chains
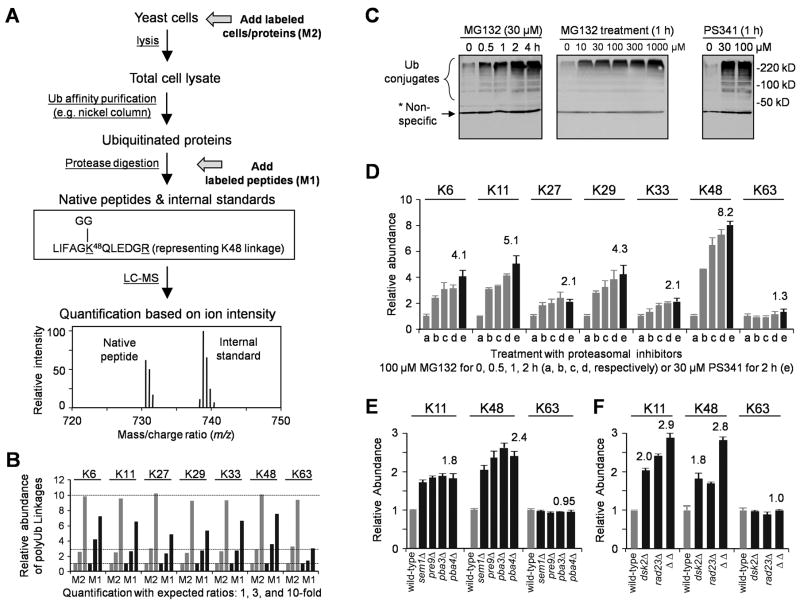
To analyze the level of polyUb linkages by mass spectrometry (MS), we employed an isotope dilution method (Kirkpatrick et al., 2006; Xu et al., 2006), which uses heavy isotope-labeled peptides as internal standards for lighter versions of unlabeled, native linkage-specific peptides that are generated during trypsin digestion of Ub polymers (Figure 1A, Table S1). Ubiquitinated proteins are usually isolated from yeast cells by affinity purification and digested by trypsin, during which Ub is trimmed to a tag of two Gly residues (GG, monoisotopic mass of 114.0429 Da, Figure 1A) attached to the modified Lys side chains of remnant peptides of Ub itself or non-Ub proteins (Peng et al., 2003b). Therefore, the abundance of specific Ub-Ub linkages is represented by the level of the corresponding GG-tagged tryptic peptides. The chemical properties of the labeled standards are indistinguishable from those of native peptides, but, being different in mass, the labeled and unlabeled peptides are resolved by MS, allowing for quantitation of the native peptides. Using chemically synthesized, heavy isotope-labeled, GG-tagged peptides as references, we found that the concentration of K48 linkages is 7.3 ± 0.3 picomoles per milligram protein in log-phase yeast cell lysate. There is a striking and unexpectedly high abundance of the unconventional non-K48 linkages, especially K11 linkages. Based on the absolute quantification of all seven linkages (a sum of 100%), the percent abundance of individual linkages is 10.9 ± 1.9% (K6), 28.0 ± 1.4% (K11), 9.0 ± 0.1% (K27), 3.2 ± 0.1% (K29), 3.5 ± 0.1% (K33), 29.1 ± 1.9% (K48) and 16.3 ± 0.2% (K63). Thus, unconventional Ub chains form a major component of the conjugated Ub pool.
Figure 1.
Inhibition of the Ub-proteasome system increases the level of all non-K63 linked polyUb chains.
(A) Schematic of quantitative MS. In Method 1 (M1), isotope-labeled labeled peptides are used as standards that are added during trypsin digestion. In Method 2 (M2), labeled cells/proteins are spiked in after cell harvest, but before cell lysis, minimizing variations in sample processing. In addition, labeled proteins can be added in any steps between cell lysis and trypsin digestion as internal standards.
(B) Using labeled cells/proteins (M2, grey) instead of peptides (M1, black) reduced quantitative variations. Three yeast lysates with different amounts of polyUb chains (1x, 3x and 10x) were processed in parallel, and the data were normalized to the result of the 1× sample.
(C) Proteasome inhibitor treatment caused accumulation of Ub conjugates in a dose- and time-dependent manner. Strain JMP001 (_pdr5_Δ) expressing His-myc-Ub was treated and harvested for immunoblotting with myc antibodies.
(D) Distinct polyUb chain linkages were measured by MS, shown as mean ± SEM.
(E–F) Yeast strains with mutations in Ub-proteasome system raised K11 and K48 linkages but not K63-linked chains. Data are represented as mean and SEM.
We further developed a more precise method to quantify polyUb linkages using stable-isotope labeled cells/proteins instead of peptides (Figure 1A, 1B), and found that levels of monoUb and all seven Ub-Ub linkages are essentially unchanged as a function of cell growth stage (Table S2). When monoUb pool was augmented ~10-fold by induction in yeast, all seven polyUb linkages were increased, but to varying levels (3- to 12-fold, Figure S1, supplemental Data), suggesting that the seven polyUb linkages display different kinetics of synthesis and disassembly.
Proteasomal Degradation is Mediated by Non-K63 PolyUb Linkages
To investigate the potential physiological roles of the abundant unconventional polyUb chains compared with K48-linked chains, we analyzed the change in these linkages during proteasomal inhibition. After treatment with MG132 proteasome inhibitor, bulk Ub conjugates accumulated, leveling off after ~2 hr (Figure 1C). When the MG132 concentration was increased from 10 to 1000 μM, ubiquitinated species accumulated in a dose-dependent manner, almost reaching saturation at ~100 μM. The Ub conjugate levels also markedly increased in the presence of 30 μM of PS341, a more potent and specific proteasome inhibitor. MS analyses indicated that MG132 (100 μM) and PS341 (30 μM) had similar effects on the level of polyUb linkages (Figure 1D, Figure S2, Table S2). After 2 hr treatment, K48 linkages increased ~8-fold; K6, K11, and K29 linkages 4–5-fold; and K27 and K33 linkages ~2-fold. In contrast, K63 linkages did not change significantly. When MG132 was administrated at a level (30 μM) causing incomplete proteasomal inhibition, all non-K63 linkages also accumulated, though to a lesser degree (Table S2). These data indicate that the levels of all non-K63 polyUb chains are inversely correlated with the proteolytic activity of the proteasome. Substrates modified by such chains might therefore be targeted to the proteasome.
To test the apparent involvement of unconventional polyUb chains in proteasomal targeting, we further measured levels of polyUb linkages in yeast strains carrying mutations in components of the Ub-proteasome system. We first developed a method that used total cell lysate directly, rather than Ub conjugates purified by nickel affinity chromatography, because these strains only express untagged Ub (Figure 1A). With this method, we were able to detect only the three most abundant linkages (K11, K48, and K63), the other linkages being undetectable because of their lower concentrations and the greater protein complexity in total cell lysate versus purified Ub conjugates. Single-gene deletions of proteasomal subunits (_sem1_Δ or _pre9_Δ) or proteasome-assembly factors (_pba3_Δ or _pba4_Δ) caused comparable increases of K11 linkages (~1.8-fold) and K48 linkages (~2.4-fold, Figure 1E, Table S2). Similarly, deletion of Ub-receptors Dsk2 or Rad23, both of which deliver Ub-conjugates to the proteasome, raised the level of K11 (~2.0-fold) and K48 (~1.8-fold) linkages, and the double deletion had a more dramatic effect (K11, 2.9-fold; K48, 2.8-fold; Figure 1F). In all these mutant strains, K63 linkages remained constant. The data strongly support the possibility that both K11-and K48-linked chains are targeting signals for the proteasome.
In addition, we analyzed the abundance of K11, K48 and K63 linkages in yeast strains containing mutations in DUB genes (Figure 2A). Consistent with previous results that deletion of proteasome-associated Ubp6 caused little accumulation of high molecular weight ubiquitinated proteins (Amerik et al., 2000a), we did not observe large changes in the three linkages. In contrast, in _doa4_Δ strain, K11 and K48 linkages were markedly decreased (5-fold and 2.5-fold, respectively), probably due to Ub depletion (Swaminathan et al., 1999), whereas K63 linkages showed a small increase (1.3-fold) despite the decrease of overall Ub levels, in agreement with the role of Doa4 in membrane trafficking that involves K63-linked polyUb chains (Amerik et al., 2000b). Loss of the UBP2 gene, encoding another DUB that has preference for K63 linkages and regulates protein sorting efficiency (Kee et al., 2006), raised the level of K63 linkages (1.8-fold), but also increased K48 linkages (1.9-fold). Loss of UBP14, a DUB that cleaves unanchored polyUb chains with various linkages (Amerik et al., 2000a), resulted in higher amount of all three linkages (1.2 to 1.7-fold). These results suggest that distinct DUBs have specific Ub-Ub linkage preferences in vivo. The patterns of polyUb linkages in the DUB mutants are also different from that in the above proteasome-defective cells, supporting the nondegradative role for K63-linked chains and the function of other linkages in degradation.
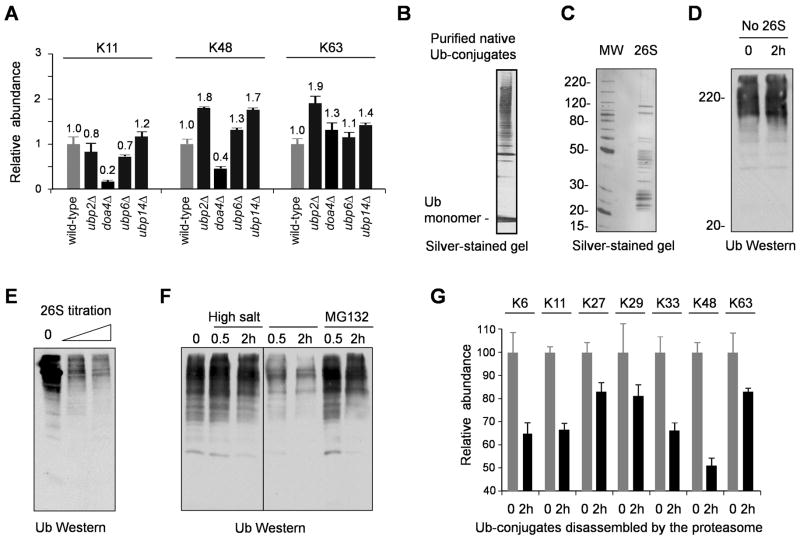
Figure 2.
PolyUb chains with distinct linkages are processed by the 26S proteasome.
(A) Yeast DUB mutations have distinct effects on the composition of polyUb linkages in vivo. The data are normalized to the amount of linkages in isogenic wild-type strains and shown as mean ± SEM.
(B–C) Purified His-myc-Ub conjugates and 26S proteasome analyzed by gel electrophoresis.
(D) The Ub conjugates were not contaminated by active DUBs, shown by immunoblotting (myc Ab).
(E) The 26S (0.1 and 1 μg) deubiquitinated native Ub conjugates (0.3 μg) in a 2-h reaction.
(F) The proteasome-associated DUB activity was sensitive to high salt and MG132 (100 μM) treatment. The proteasome (0.3 μg) was incubated with the Ub conjugates (0.3 μg). For high-salt treatment, the proteasome was incubated for 30 min with 250 mM NaCl and then diluted to 50 mM NaCl before the reaction.
(G) The disassembly of polyUb linkages (2 μg of native Ub conjugates) by the 26S (2 μg), analyzed by MS using heavy isotope-labeled Ub conjugates as internal standards. Data are represented as mean and SEM.
If the unconventional polyUb chains direct proteins for proteasomal degradation, the Ub in these chains could be recycled by deubiquitinating activities associated with the proteasome complex. To test this idea, we set up an in vitro deubiquitinating assay using purified native Ub conjugates (Figure 2B) and 26S proteasome (Figure 2C). The purified Ub-conjugates were not disassembled without the addition of proteasome, indicating that the Ub-conjugates had little contamination by active DUBs or other proteases (Figure 2D). When the conjugates were incubated with the 26S in the presence of ATP, Ub-conjugate levels were dramatically decreased (Figure 2E), suggesting that polyUb chains were disassembled by the action of the 26S proteasome. This process was obstructed by the addition of MG132 (Figure 2F), probably because it inhibits the proteasome core particle and indirectly interferes with the deubiquitinating activity of a proteasome subunit Rpn11 (Jin et al., 2008). In addition, this deubiquitination was also sensitive to high-salt treatment (Figure 2F), which inactivates the 26S-associated DUBs, such as Rpn11 (Verma et al., 2002) and USP14 (a mammalian homolog of yeast Ubp6) (Leggett et al., 2002). Moreover, we found that all seven linkages were cleaved by proteasome-associated DUBs, but to different extents, with K48 linkages hydrolyzed most readily (Figure 2G). All these data further confirm that unconventional polyUb linkages can be recognized and disassembled in the 26S proteasome.
Phenotypic Effects of Combining Multiple Substitutions in Chain-forming Lys Residues
Although these unconventional polyUb chains are abundant in the cell and appear to function as signals for proteasomal degradation, only the K48R substitution in Ub is lethal in yeast, and the K27R mutation causes a small growth defect (Spence et al., 1995). Therefore, we asked whether polyUb chains with unconventional linkages have redundant functions by combining mutations in non-K48 lysine residues and testing yeast survival. We generated a series of yeast Ub-mutant strains, including single (R11 or R27), double (R11R63), triple (R11R27R63), and quadruple (R6R11R27R63) lysine substitutions (Figure S3A). We also attempted to make a strain expressing a sextuple Ub mutant (R6R11R27R29R33R63) with K48 as the single lysine but failed in spite of extensive screening (Figure S3B), suggesting that Ub with K48 alone cannot support viability in yeast.
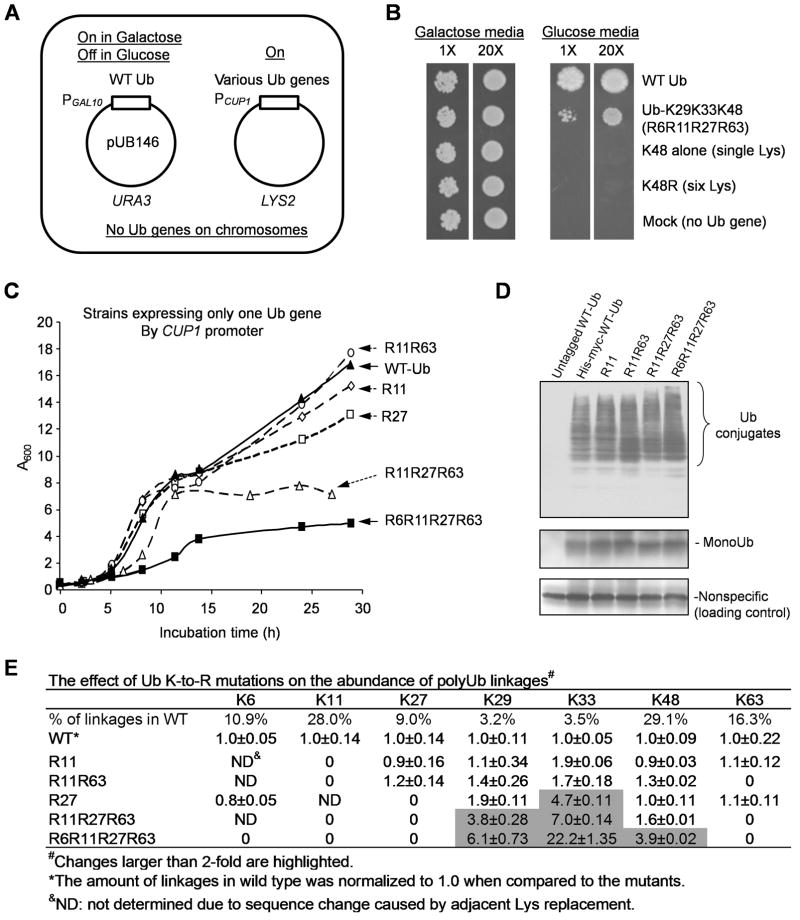
To confirm that Ub with K48 as the lone lysine is not sufficient for yeast survival, we used a strategy to switch expression from wild-type to mutant Ub genes in yeast and then examined cell growth (Figure 3A). We constructed yeast strains that maintained two plasmids: one expressing wild-type Ub from galactose-inducible GAL10 promoter, and the other expressing individual Ub mutants under the constitutive CUP1 promoter (Figure 3A). In synthetic galactose media, Ub genes on both plasmids were actively transcribed. Switching to glucose media suppressed the expression of wild-type Ub, whereas the mutant gene was still expressed as the only source of ubiquitin. When Ub-K48R was introduced into the strain, it could not support growth (Figure 3B), confirming that Ub-K48 is an essential residue (Finley et al., 1994). More importantly, expressing the single-lysine Ub (K48 alone) also resulted in a complete deficiency in cell growth (Figure 3B), and the culture’s growth curve was indistinguishable from that of the Ub-K48R strain (Figure S3D). These data indicate that Ub with K48 as its only lysine is not sufficient to sustain yeast growth, underscoring the physiological significance of non-K48 lysine residues.
Figure 3.
Ub with K48 alone cannot support yeast viability and cumulative K to R substitutions lead to growth defects.
(A) The strategy for switching Ub expression in yeast.
(B) Expression of Ub-K48 as the only Ub source resulted in lethality (1X = ~100 cells).
(C) Growth curves of yeast strains expressing a single Ub gene under the P_CUP1_ promoter YPD medium.
(D) Comparison of His-myc-Ub monomer and conjugated forms in yeast strains. Total cell lysates (10 μg) were blotted with anti-myc antibodies.
(E) Quantification of polyUb linkages in yeast strains by MS. All values are normalized according to the levels in the wild-type strain and shown as mean and SEM.
To rescue viability of the K48-only Ub mutant, restitution of K29 and K33 (R6R11R27R63 strain) is sufficient. However, this mutant exhibited severely retarded growth (Figure 3B). The growth curves of various lysine mutant strains in rich medium (YPD) were further compared (Figure 3C). The order of growth rates was WT = R11 = R11R63 > R27 > R11R27R63 > R6R11R27R63, suggesting that the mutations have a cumulative effect on cell proliferation. To eliminate the possibility that the mutations affected the stability of ubiquitin, thereby reducing the availability of Ub for conjugation, we examined the level of Ub and its conjugates in all strains by Western blotting. In spite of a slight increase in Ub-conjugates in the quadruple mutant, all strains showed comparable levels of Ub monomer and conjugates (Figure 3D). Thus, the lethal effect of mutating combinations of non-K48 lysines is unlikely resulted from a general defect in conjugate formation. However, this does not rule out subtle functional defects not related to the formation of specific chains.
To examine potential interdependence of the different Ub-Ub linkages, we used MS to measure the changes in the polyUb linkage levels in the Ub mutants (Figure 3E). Neither R11 nor R11R63 affected cell growth, and they had little effect on the abundance of other linkages (< 2-fold). K27R replacement influenced linkages only at the nearby lysines K29 (1.9-fold) and K33 (4.7-fold). The triple substitution mutant (R11R27R63) dramatically reduced cell growth and increased K48 (1.6-fold), K29 (3.8-fold), and K33 (7.0-fold) linkages. Additional mutation of K6 resulted in a further increase in the utilization of the remaining sites (K48, 3.9-fold; K29, 6.1-fold; and K33, 22.2-fold). These results indicate that the physiological roles of these lysine residues are partially redundant, and thus the growth defects are cumulative when more lysine residues are mutated. Conversely, concomitant increases of polyUb linkages at the remaining lysines occur but do not support wild-type growth rate. This suggests that different polyUb linkages may modify specific substrates and have unique functions (unnaturally high levels of alternative Ub-Ub linkages might be deleterious as well; see Discussion).
K11 Linkages Modify Specific Substrates Revealed by Quantitative MS
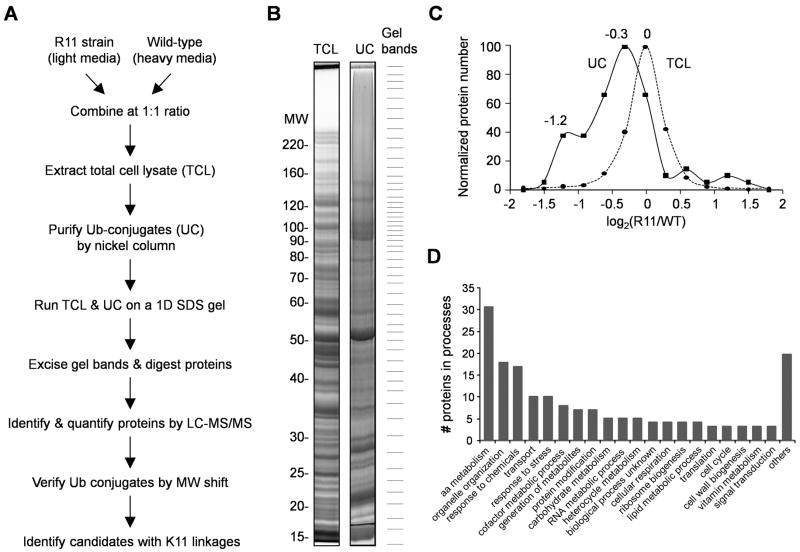
Since K11 Ub-Ub isopeptide bonds are as abundant as K48 linkages in vivo, but are not nearly as well characterized, we sought to identify K11 polyUb chain-linked substrates by comparing R11 and wild-type strains using a quantitative MS strategy, termed stable isotope labeling with amino acids in cell culture (SILAC) (Ong et al., 2002). R11 and wild-type cells were differentially labeled in light and heavy stable isotope media, mixed after harvesting, and then used for the purification of Ub conjugates under denaturing conditions (Figure 4A). Proteins from total cell lysate and Ub conjugates were both resolved on an SDS gel, excised from the gel, digested by trypsin and analyzed by reverse phase liquid chromatography coupled with tandem mass spectrometry (LC/MS/MS, Figure 4B). This resulted in the quantification of 1,576 proteins in the total lysate (Table S3), and 347 Ub conjugate candidates with at least two assigned peptides. Because samples enriched for Ub conjugates can still be contaminated by unmodified proteins, we developed an algorithm to construct “virtual Western blot” images of every identified protein (Figure 5C, 5D) to evaluate whether the protein is likely to be ubiquitinated based on the large mobility shift expected from the polyUb tag (Seyfried et al., 2008). Ultimately, out of the 347 candidates, we accepted 75 Ub conjugates, 37 (49%) of which were also quantified in the total cell lysate (Table S4). Comparing the accepted Ub conjugates to the proteins in the total cell lysate revealed that the K11R substitution caused a small, global decrease in ubiquitination (main peak for the Ub conjugates in Figure 4C) and markedly inhibited the ubiquitination of a subset of proteins (shoulder in the histogram of Ub conjugates; Figure 4C). Moreover, out of the 1,576 proteins in the cell lysate, 91 proteins showed statistically significant changes in abundance (absolute log2 ratio at least 0.8 and signal-to-noise ratio > 1.5, Figure S4E, Table S3). Gene ontology classification (Ashburner et al., 2000) indicates that the K11R mutation perturbs the yeast proteome in a profound manner, suggesting that K11 linkages are involved in a broad range of cellular processes (Figure 4D, table S5).
Figure 4.
Large-scale protein profiling of the wild-type and Ub-R11 strains to identify linkage-specific substrates.
(A) Outline of the SILAC method for comparing total cell lysate (TCL) and purified Ub conjugates (UC) in the two strains.
(B) Comparison of the TCL and the UC by SDS-PAGE. Both samples were resolved on a 6–12% gel, stained with Coomassie Blue, excised into ~50 gel bands, digested by trypsin, and analyzed by LC/MS/MS.
(C) Histograms of log abundance ratios of quantified proteins in the TCL (n = 1,576) and in the UC (n = 75).
(D) GO categories of biological processes of 91 proteins, the levels of which in the TCL were significantly altered by the Ub-K11R mutation.
Figure 5.
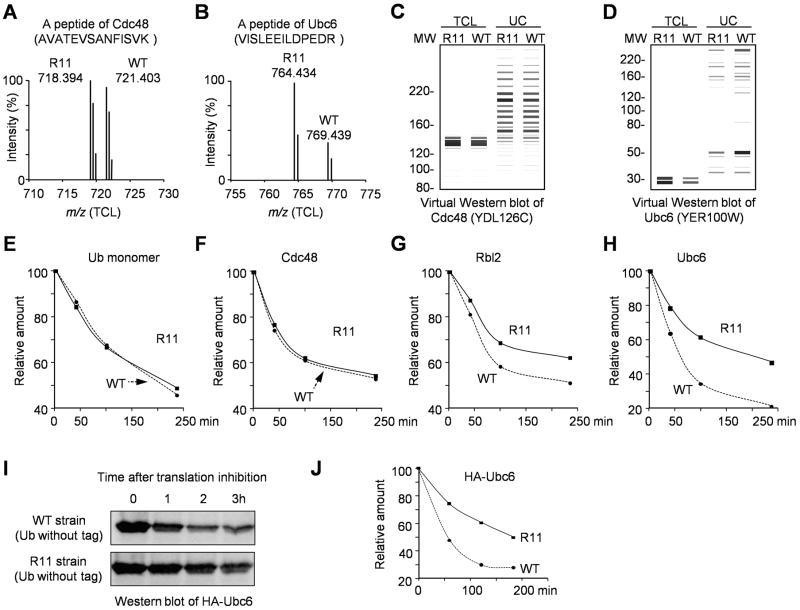
Validation of K11 linkage-specific substrates by virtual Western blots and protein turnover analyses.
(A–B) Representative isotope-labeled peptide pairs of Cdc48 and Ubc6 in the total cell lysate (TCL). The light and heavy labeled peptides were distinguished by different mass-to-charge ratios (m/z).
(C–D) Virtual Western blots reconstructed from proteomics data for Cdc48 and Ubc6, reflecting quantitative data in all gel bands in the TCL and the Ub-conjugates (UC). The protein abundance was represented by the darkness and thickness of the bands; and the molecular weight information was extracted from the 1D SDS gel.
(E–H) Protein half-life analyzed by cycloheximide chase and quantitative MS. The experiment was repeated, and the relative standard errors were under 10%.
(I–J) Protein half-life analysis in yeast strains expressing untagged WT or R11 Ub. The cells were treated with cycloheximide to inhibit translation. The degradation of HA-Ubc6 was examined by Western blotting (HA Ab).
Among the 37 proteins identified in both the total cell lysate and the ubiquitinated proteome, we searched for potential K11 linkage targets using two criteria: (i) the protein’s level was elevated in the total cell lysate, and (ii) the level of the ubiquitinated forms was reduced. Two potential substrates were found: Ubc6, an E2 Ub-conjugating enzyme (Chen et al., 1993; Sommer and Jentsch, 1993), and Rbl2, a tubulin-folding cofactor (Lopez-Fanarraga et al., 2001). In protein half-life assays, the lack of K11 in Ub significantly reduced the turnover rate of these two proteins, but did not stabilize two negative control proteins (Ub and Cdc48, Figure 5A-H). The reduced degradation of Ubc6 by Ub-K11R mutation was further validated in different yeast strains expressing untagged Ub (Figure 5I-J). These data support the inference that the degradation of a specific subset of proteins is dependent on K11-linked polyUb chains.
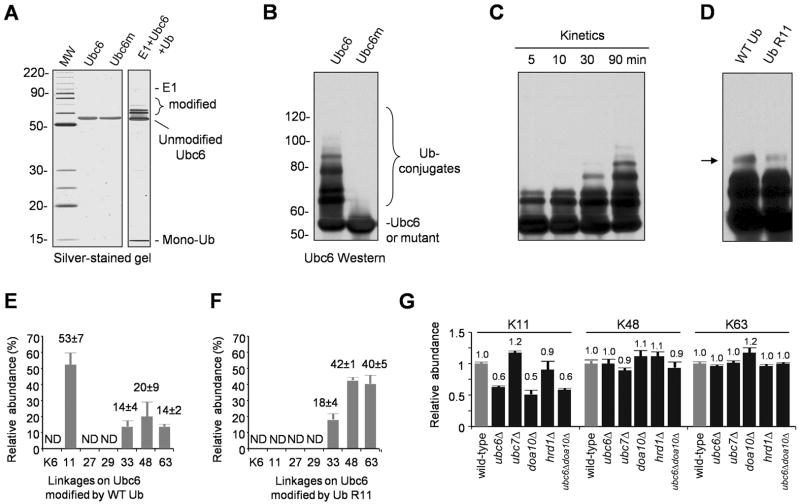
To confirm that Ubc6 is a genuine substrate modified by K11-linked polyUb chains, we reconstituted an in vitro ubiquitination reaction using recombinant yeast E1 and Ubc6. The recombinant Ubc6 lacked the C-terminal transmembrane domain (M233-K250) to promote its solubility. In this reaction, Ubc6 modified itself with multiple Ub molecules, causing a mass shift of up to 40 kDa (~4–5 Ub tags), whereas no free Ub polymers were generated during the reaction (Figure 6A). The autoubiquitination of Ubc6 was abolished by substituting Cys87 with Ser in the catalytic site (Figure 6B). After a 30-min reaction (a time point prior to the reaction plateau, Figure 6C), the K11R mutation in Ub caused a small, but visible defect in the formation of higher order Ubc6-linked Ub-conjugates (Figure 6D). Further MS analysis (Figure 6E) indicated that Ubc6 was modified mainly by K11-linked polyUb (53%), but also by other polyUb chains assembled through K33 (14%), K48 (20%), and K63 (14%). Ub-K11R could still form polyUb chains on Ubc6 in vitro through increased use of other lysines: K33 (18%), K48 (42%) and K63 (40%) sites (Figure 6F). More interestingly, deletion of the UBC6 gene in yeast cells reduced the total cellular K11 polyUb linkages by ~40%, but had no impact on K48 or K63 chains (Figure 6G). These results reveal that Ubc6 is not only a substrate modified by K11-linked polyUb chains, but is also one of the primary E2s contributing to the synthesis of K11 linkages in vivo.
Figure 6.
Ubc6 and Doa10 contribute to the synthesis of K11 linkages.
(A) Purified GST-Ubc6 and a catalytically inactive mutant (GST-Ubc6m, C87S). When incubated with E1, and Ub for 2 h, Ubc6 was self-ubiquitinated. No free Ub polymers (e.g. dimers, trimers, and etc.) were observed.
(B) The Cys87 residue in Ubc6 is essential for its activity (immunoblotting with GST Ab).
(C) Time course of Ubc6 in vitro ubiquitination reaction.
(D) Comparison of Ubc6 self-ubiquitination (30 min) by WT or R11 Ub.
(E) MS measurement of polyUb linkages on Ubc6 (2 h reaction). The polyUb-Ubc6 was resolved on a SDS gel, excised and analyzed by MS using synthetic peptides as internal standards. Total amount of polyUb linkages was normalized to 100%. ND: not determined.
(F) MS analysis of Ub-R11-Ubc6 conjugates.
(G) Deletion of UBC6, DOA10, or both genes in yeast reduced the global level of K11 linkages. Data in panel E–G are represented as mean and SEM.
K11 Linkages Function in Endoplasmic Reticulum Associated Degradation (ERAD)
Ubc6 is a component of the ERAD pathway, a quality control pathway in which misfolded or improperly assembled proteins of the ER are ubiquitinated, translocated to the cytosol if necessary, and degraded by the proteasome (Ravid et al., 2006; Sommer and Jentsch, 1993). Thus we tested whether polyUb chains with K11 linkages impact the ERAD in any way. There are two main E3 ligases involved in the ERAD: Doa10, which requires two different E2s, Ubc6 and Ubc7 (Ravid et al., 2006), and Hrd1, which acts primarily with Ubc7, an E2 that assembles K48-linked polyUb chains (Bazirgan and Hampton, 2008). Consistent with the connection between Doa10 and Ubc6, we found a selective decrease of K11 linkages in _doa10_Δ and _ubc6_Δ_doa10_Δ double mutant cells, an effect comparable to that measured in the _ubc6_Δ strain; in contrast, _ubc7_Δ or _hrd1_Δ did not affect K11 linkages (Figure 6G). Thus, Ubc6 and Doa10 define a pathway for synthesizing K11-linked polyUb chains in vivo.
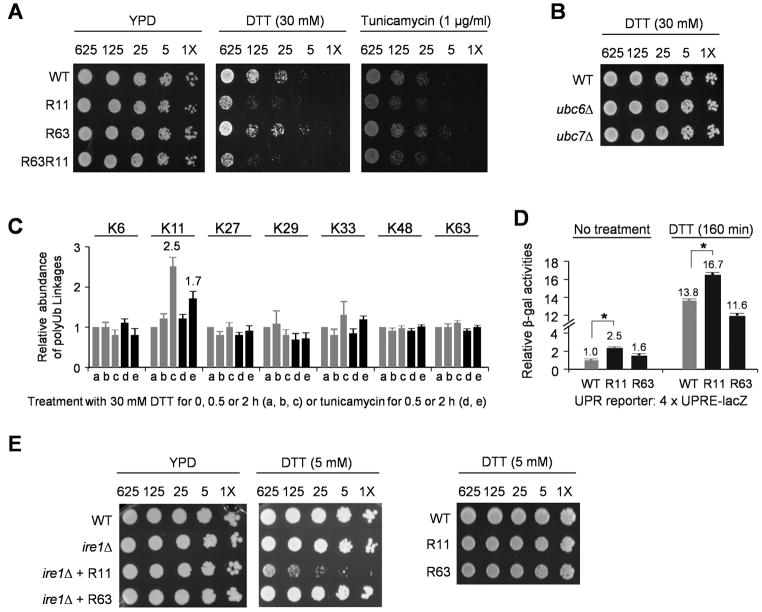
Given the link between K11 polyUb chains and ERAD, we examined whether the R11 mutant had a higher sensitivity to ER stress-inducing reagents. Indeed, the R11 strain grew more poorly than the wild-type in the presence of high levels of dithiothreitol (30 mM DTT, a thiol reductant that interferes with disulfide bond formation) or tunicamycin (an antibiotic that blocks the synthesis of N-linked glycoproteins in the ER) (Figure 7A). In contrast, the Ub-R63 mutation had no obvious effect on cell proliferation under the same conditions, and a strain expressing the doubly mutated Ub (R11R63) displayed defects similar to those of the R11 single mutant (Figure 7A). However, no hypersensitivity to ER stress was observed in _ubc6_Δ or _ubc7_Δ cells (Figure 7B), indicating that _ubc6_Δ is not as sensitive to ER stress as the Ub-K11R mutant. These findings are consistent with the fact that Ubc6 only catalyzes formation of ~40% of cellular K11 Ub-Ub linkages.
Figure 7.
Ub-K11 linkages function in the ER stress response.
(A) The Ub-K11R but not the K63R substitution affected cell growth under ER stress. The PDR5 gene was deleted from the strains to increase drug sensitivity (1X = ~30 cells). Cells were grown on YPD plates at 24°C, and recorded (control cells after 4 days; treated cells after 5 days).
(B) Under high concentration of DTT (30 mM), _ubc6_Δ or _ubc7_Δ strains had no growth defect.
(C) ER stress with DTT (30 mM) or tunicamycin (1 μg/ml) specifically raised the levels of K11 linkages (mean ± SEM) in the WT strain.
(D) The Ub-R11 strain had a higher level of UPR activation than wild-type and Ub-R63 cells. Yeast cells were transformed with a UPR reporter that expresses lacZ gene under the control of UPR element. The basal and induced (30 mM DTT) levels β-galactosidase activity were measured. Data are shown as mean and SEM, and the asterisks indicate p value < 0.01 (student _t_-test).
(E) Deletion of UPR gene (IRE1) had synthetic defects with the Ub-R11 mutation under low concentration of DTT (5 mM).
Furthermore, we measured the level of all seven polyUb linkages in wild-type yeast treated with DTT or tunicamycin. The treatment selectively stimulated formation of K11 linkages, and DTT had a stronger effect than tunicamycin (2.5-fold vs. 1.7-fold after 2 hr, Figure 7C), consistent with their relative potency in growth inhibition (Figure 7A). There was no detectable change in the levels of K48 linkages, either because formation of K48-linked polyUb chains, such as those made by Ubc7, is already occurring at maximal rates in unstressed cells, or because K48 chains are used more widely than K11 chains in processes other than ER homeostasis. The global levels of other polyUb linkages did not change either (Figure 7C). The selective accumulation of K11 Ub-Ub linkages during ER stress strongly supports the role for such polyUb chains in the ER stress response.
ER stress induces the unfolded protein response (UPR), a homeostatic mechanism initiated by activation of the ER-resident Ire1 kinase (Ron and Walter, 2007). We measured the UPR induction in the WT, R11 and R63 strains using a UPR reporter, consisting of the E. coli lacZ gene under the control of a promoter containing 4 copies of the UPR element. Indeed, the R11 but not R63 mutation caused a weak but reproducible increase in both the basal and DTT-induced levels of the UPR (Figure 7D, p<0.01). Notably, when we deleted IRE1 gene in our yeast strains, we observed a striking synthetic growth defect in combination with Ub-R11, but not Ub-R63, in the presence of low concentrations of DTT (5 mM, Figure 7E). These results demonstrate that K11-linked polyUb chains function in ubiquitination events related to the ER stress pathway.
Discussion
The Complexity of PolyUb Linkages in Cells
The use of all seven lysine residues in Ub as conjugation acceptor sites permits an enormous variety of theoretical configurations of polyUb chains (e.g., seven forms of di-Ub and 70 forms of tri-Ub, Figure S5). In some cases, the N-terminal α-amino group of Ub provides an additional acceptor site for polyUb assembly (Kirisako et al., 2006). The αN-linked linear Ub-Ub linkages were not observed in our yeast strains, which may be due to the interference of N-terminal tagging or their low abundance in yeast. The structural diversity of Ub-protein modification becomes even greater when multiple residues in a given substrate are modified. Previous MS measurements implied that K48, K63, and K11 are used more frequently than other lysine residues in yeast (Peng et al., 2003b) and mammals (Bennett et al., 2007), suggesting that the formation of diverse polyUb chains is a highly conserved mechanism.
Function of Diverse PolyUb Chains in Proteasomal Degradation
Our work provides several lines of evidence that many different non-K63-linked chains are involved in proteasomal degradation. K63 linkage levels do not change significantly when the proteasome is inhibited chemically or genetically, consistent with their well-known roles in nonproteolytic mechanisms (Pickart and Fushman, 2004). However, in mammalian cells, K63 linked polyUb chains were reported to accumulate after MG132 treatment, although their accumulation was slower than that of either K48 or K11 chains (Bennett et al., 2007; Meierhofer et al., 2008). This could have several explanations. First, formation of K63 linkages may be part of a stress response to inhibition of proteasome activity or depletion of free ubiquitin in mammalian cells. Second, global analysis of polyUb linkages may not accurately reflect the behavior of individual proteins. We cannot rule out the possibility that a subset of K63-linked substrates is degraded by the proteasome. The capacity of K63 Ub linkages to target proteins for degradation by the proteasome system has been suggested in some recent in vitro studies (Crosas et al., 2006; Kim et al., 2007; Kirkpatrick et al., 2006). The contrasting observations in yeast and mammals could also be accounted for if K63-linked substrates for degradation are more abundant in mammals.
A role in proteasomal proteolytic targeting for non-K63 polyUb chains is further supported by the analysis of polyUb linkages that increase in yeast strains with mutations in the proteasome, proteasome-binding Ub receptors, or DUBs. The proteasome is capable of cleaving mixed endogenous polyUb chains and has different activities toward the various linkages. Interestingly, the kinetics of disassembling different linkages (K48 > K6, K11, K33 > K27, K29, K63, Figure 2G) is roughly correlated with their accumulation during treatment with proteasomal inhibitors (K48 > K6, K11, K29 > K27, K33 > K63, Figure 1D). Proteasome-associated DUB activity is sensitive to MG132 treatment because deubiquitination by Rpn11/POH1, a major DUB in the 26S proteasome, is tightly coupled with protein degradation. Moreover, deletion of multiple other DUBs, such as Ubp2, Doa4, Ubp6 and Ubp14, had highly distinct effects on the pattern of polyUb linkages (Figure 2A). These data suggest that the polyUb accumulation by proteasomal inhibitor treatment in yeast is largely a consequence of inhibiting proteasome-associated DUB activities.
Since all non-K63 polyUb linkages accumulate upon proteasome inhibition and may therefore be able to mediate proteasomal protein degradation, one would predict that these alternative linkages might show some functional redundancy, which is what we observed in Ub mutagenesis analysis. Simultaneous replacement of multiple lysine residues led to increasingly severe defects in the strains of R11R27R63 and R6R11R27R63, and mutation of all six non-K48 lysine residues resulted in lethality. Further MS analysis revealed that the cells appear to compensate for mutations by using the remaining lysine sites more frequently for chain formation, consistent with the functional importance of unconventional linkages. For instance, K33 linkages are increased 22-fold in the quadruple mutant (R6R11R27R63) compared with the wild-type strain.
When multiple K-to-R mutations force the assembly of unconventional chains, it is conceivable that the high level of unconventional linkages become toxic to cells because proteins bearing such chains are not degraded or deubiquitinated with appropriate kinetics. To determine whether increased unconventional Ub-Ub linkages might cause a general block of proteasomal activity, e.g., by failing to be cleared efficiently from the proteasome, we examined the degradation of ornithine decarboxylase, a well-characterized Ub-independent proteasome substrate (Coffino, 2001). We found no major change in the global activity of the proteasome in the set of Ub mutants (Figure S6). The results suggest that the proteasome activity per se is not reduced, but instead suggest that proteolysis of particular substrates requires specific polyUb linkages. This view is bolstered by our detailed analysis of K11 linkages.
The Role of Abundant K11 Linkages in Degradation and the ER stress response
The large-scale proteomic comparison of wild-type and K11R mutant cells indicates that this Ub mutation has a global impact on both the bulk yeast proteome and the ubiquitinated proteome. More significantly, the role of K11 linkages in protein degradation in vivo is directly demonstrated by the characterization of Ubc6 as a K11 Ub-chain-modified proteolytic substrate. This finding further led to the hypothesis that K11 linkages function in the ERAD pathway. Recently, systematic analysis of UBX protein interactomes also revealed more prominent K11 Ub-Ub linkages than K48 linkages in immunoprecipitated complexes (Alexandru et al., 2008). As UBX proteins target ubiquitinated proteins to Cdc48/p97, a key component in the ERAD (and other pathways) in both yeast and mammalian cells, the results implicate a conserved function for K11 linkages in the ER stress response.
Interestingly, when we asked whether two known Ubc6/Doa10 substrates, _Deg1_-βgal and _Deg1_-VP (Ravid et al., 2006), were degraded more slowly in the Ub-K11R strain, we found that this was not the case (Figure S7 and data not shown). This may be explained by several compensatory mechanisms in the mutant cells: (i) upregulation of Ubc6 (~1.8-fold), (ii) complementary use of other Lys residues for polyUb assembly in the absence of K11 (Figure 6), and (iii) the activation of UPR to increase the level of proteins involved in ERAD. Doa10 also functions with the Ubc7 E2, which synthesizes K48-linked polyUb chains, so substrates of Ubc6 and Doa10 may not necessarily get modified by homogeneous K11 chains. Finally, it is also possible that K11-linked polyUb chains have additional nonproteolytic regulatory roles in the ER-stress response, which could account for the mutational effects on ER stress being stronger than those on ERAD. Proteins modified by K11-modified polyUb chains but not dependent on such linkages for degradation were excluded from our analysis.
K11-linked polyUb chains have been suggested to target individual proteins to the proteasome in vitro (Baboshina and Haas, 1996; Kirkpatrick et al., 2006). More recently, K11 linkages have been reported to regulate the cell cycle in Xenopus in vivo, but the responsible E2 (UbcH10) is not present in budding yeast (Jin et al., 2008). Consistent with this, we saw no cell proliferation defect in the yeast K11R Ub mutant. Perhaps the contribution of K11 linkages to cell-cycle control evolved only in higher organisms. Nevertheless, K11 linkages are abundant in yeast and play important roles in specialized degradation pathways, such as the ERAD. In addition, based on the spectrum of proteins altered in the K11R mutant, K11 linkages are likely involved in a broad range of physiological processes.
Our studies suggest that K11 polyUb linkages are commonly assembled on substrates together with other linkages. Despite the high abundance of K11 linkages, we found that the K11R mutation did not completely eliminate the ubiquitination of any substrates. Similar to Ubc6, which is modified by chains with distinct Ub-Ub linkages (possibly, but not necessarily, in the same polyUb chain), cyclin B1 is known to be modified by K11, K48, and K63 linkages, and mixed chains appeared competent to mediate the degradation of cyclin B1 in vitro and in vivo (Jin et al., 2008; Kirkpatrick et al., 2006). Furthermore, mixed linkage polyUb chains may be synthesized on one substrate by the sequential actions of multiple E2s with different linkage preferences (Rodrigo-Brenni and Morgan, 2007), and subsequently edited by the DUBs (Crosas et al., 2006; Newton et al., 2008). K48 and K63 linkage-specific antibodies were recently developed and allowed the immunoisolation of specific Ub-conjugates, but mass spectrometric analysis still revealed a mixture of linkages (Newton et al., 2008). Such complex polyUb chain topologies may provide a high density of Ub to increase the binding affinity for subsequent targeting events. Alternatively, various polyUb linkages may result in distinct signaling events.
Contribution of PolyUb Linkages to Ubiquitin Signaling Specificity
The diversity of polyUb structures has suggested the hypothesis that Ub polymers with different Ub-Ub linkages allow for specificity in downstream Ub-mediated events by binding to distinct Ub receptors or by regulation through linkage-specific DUBs (Pickart and Fushman, 2004). Our work reveals diverse kinetics in the synthesis and deubiquitination of different polyUb linkages under various conditions (e.g., with increased monoUb pools, proteasome inhibition, and by in vitro DUB assays). These kinetic differences may also be exploited as a means to regulate Ub signaling, leading to distinct biological outcomes.
Experimental Procedures
Yeast Strains
Yeast strains (Table S6) and protocols are described in the Supplemental Data.
Purification of Recombinant Proteins, 26S Proteasome, and Ub Conjugates
Ubc6 and its mutant (C87S) were expressed as GST-fusion proteins in E. coli using pET21a expression system and purified to near homogeneity. The 26S proteasome was purified from bovine red blood cells (Liu et al., 2006), which contained three DUBs: Rpn11/POH1, Uch37 and Usp14.
His-myc-tagged Ub conjugates were purified from yeast under either denaturing conditions (Peng et al., 2003b) or native conditions. For native conditions, cells were harvested during log phase and lysed in buffer A (50 mM Na2HPO4, pH 8.0, 500 mM NaCl, 0.01% SDS, 5% glycerol, 5 mM imidazole, 10 mM iodoacetamide). The clarified lysate was loaded onto a nickel column followed by washing with buffer A and then buffer B (50 mM NH4HCO3, 5 mM imidazole, and 10 mM iodoacetamide). Bound Ub conjugates were step-eluted by the increase of imidazole (10, 20, 30, 40, 500 mM). Whereas most of the monoUb was eluted prior to 40 mM, polyubiquitinated species were eluted at 40 mM and 500 mM of imidazole.
During the purification of Ub-conjugates, iodoacetamide is commonly used to alkylate cysteine residues to inhibit endogenous DUB activities, but it can also modify lysine residues to produce a monoisotopic mass tag of 114.0429 Da, which is indistinguishable from the GG-tag resulting from ubiquitination (Nielsen et al., 2008). This side reaction was highly dependent on the temperature of incubation. Under our experimental conditions (4°C or 21°C), the spurious lysine alkylation had no detectable effect on the quantification results (see Supplemental Data for details on comparison of iodoacetamide, chloroacetamide and N-ethylmaleimide).
Quantitative Mass Spectrometry of Selected Proteins and Modifications
The quantification of Ub, polyUb linkages, Cdc48, Rbl2, and Ubc6 was performed using synthetic peptides or metabolically labeled cells/proteins as internal standards. The stable isotope-labeled GG-linked Ub peptides were synthesized, quantified by amino acid analysis and used for absolute quantification, whereas labeled cells were used as internal standards for relative quantification. Moreover, Ub-conjugates purified from labeled cells were also used as standards, when analyzing the in vitro DUB activity in the 26S proteasome. The MS instrument was operated in selective reaction monitoring (SRM) mode to increase sensitivity (Table S1 and Supplemental Data).
Proteomics Analysis by SILAC
The wild type and the Ub-K11R strain (both with deletion of auxotrophic LYS2 and ARG4 genes, Table S6) were cultured for ~8 generations to reach A600 of 0.7 in synthetic media (0.7% Difco yeast nitrogen base, 2% dextrose, supplemented with adenine, uracil, and amino acids). The light medium contained Arg (12 mg/liter) and Lys (18 mg/liter), and the heavy medium contained equal molar amount of [13C615N4] arginine (+10.0083 Da) and [13C6] lysine (+6.0201 Da) from Cambridge Isotope Laboratories. Differentially labeled cells were equally mixed, lysed and subjected to the enrichment of Ub-conjugates. Total cell lysate (~150 μg) and purified Ub conjugates (~100 μg) were run on a 6–12% gradient gel, stained with Coomassie blue, and cut into ~50 slices. After in-gel digestion by trypsin, the samples were analyzed by LC-MS/MS (75 μm i.d. column, 2-hr elution gradient) on an LTQ-Orbitrap mass spectrometer (Thermo Finnigan).
Proteins were identified by MS/MS spectra in a composite target/decoy yeast database search (Elias and Gygi, 2007; Peng et al., 2003a). The assigned peptides were filtered until all decoy matches were discarded, and we only accepted proteins with at least two peptide matches to further minimize false discoveries. The identified candidate Ub-conjugates were filtered by virtual Western blot images using a dynamic threshold that includes the mass of ubiquitin and experimental variations (10 kDa + 3 moving standard deviations) (Seyfried et al., 2008).
Protein quantification was performed using an in-home program, in which peptide ion peaks in survey scans were first defined to include peak intensity, area intensity, and signal-to-noise ratio. The heavy and light peaks were matched using predicted m/z difference (±6 ppm). After subtracting the noise, the peak intensity of matched peptides was compared to obtain abundance ratios. More details in SILAC protocols were also described in Supplemental Data.
In Vitro Ubiquitination Assays
GST-Ubc6 or the C87S mutant (500 nM) was incubated with E1 (25 nM) and human Ub (5 μM) in the presence of 2 mM ATP and buffer (50 mM Tris-HCl, pH 7.5, 20 mM KCl, 5 mM MgCl2, 1 mM DTT) at 30°C for the indicated time points. Reactions were terminated by the addition of SDS Laemmli loading buffer containing 10 mM DTT and boiled for 5 min to dissociate Ub thiolester. The samples were analyzed by SDS-PAGE followed by silver staining, immunoblotting or MS.
In Vitro Deubiquitination Assays
Native Ub conjugates purified from yeast were treated with 10 mM DTT to quench trace amounts of iodoacetamide. Disassembly of polyUb chains was initiated by the addition of 26S proteasomes (with 1 mM ATP, 50 mM Tris, pH 7.5, 1 mM EDTA, 5 mM MgCl2), usually in 20-μl reactions at 37°C for the indicated times, terminated by SDS Laemmli loading buffer, and followed by Western blotting or MS.
Supplementary Material
1. Supplemental Data.
Supplemental data include Supplemental Experimental Procedures and Discussion, six tables, and eight figures.
2
Acknowledgments
We thank P. Coffino, A. Corbett, G. Crouse, R. Cohen, E. Dammer, J. Fridovich-Keil, M. Hoyt, M. Jonikas, C. Kahana, C.-W. Liu, J. Mullally, P. Philippsen, D. Reines, J. Roelofs, R. Rothstein, T. Sommer, Y. Tone, P. Walter, J. Weissman, K. Wilkinson, Q. Xia and T. Yao for reagents and discussion. We also thank D. Kirkpatrick and K. Wilkinson for critical reading, H. Boldt and C. Strauss for editing. This work was funded by NIH grants (GM046904 to M.H.; GM043601 to D.F.; DK069580, CA126222 and AG025688 to J.P.).
Abbreviations
MS
mass spectrometry
ERAD
endoplasmic reticulum-associated degradation
SILAC
stable isotope labeling with amino acids in cell culture
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000a;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000b;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem. 1996;271:2823–2831. doi: 10.1074/jbc.271.5.2823. [DOI] [PubMed] [Google Scholar]
- Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Coffino P. Regulation of cellular polyamines by antizyme. Nat Rev Mol Cell Biol. 2001;2:188–194. doi: 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kee Y, Munoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem. 2006;281:36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Fanarraga M, Avila J, Guasch A, Coll M, Zabala JC. Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J Struct Biol. 2001;135:219–229. doi: 10.1006/jsbi.2001.4386. [DOI] [PubMed] [Google Scholar]
- Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Ooka S, Sato K, Arima K, Okamoto J, Klevit RE, Fukuda M, Ohta T. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279:3916–3924. doi: 10.1074/jbc.M308540200. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003a;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003b;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Seyfried NT, Xu P, Duong DM, Cheng D, Hanfelt J, Peng J. Systematic approach for validating the ubiquitinated proteome. Anal Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Cheng D, Duong DM, Rush J, Roelofs J, Finley D, Peng J. A proteomic strategy for quantifying polyubiquitin chain topologies. Israel J Chem. 2006;46:171–182. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Supplemental Data.
Supplemental data include Supplemental Experimental Procedures and Discussion, six tables, and eight figures.
2