JAK mutations in high-risk childhood acute lymphoblastic leukemia (original) (raw)
Abstract
Pediatric acute lymphoblastic leukemia (ALL) is a heterogeneous disease consisting of distinct clinical and biological subtypes that are characterized by specific chromosomal abnormalities or gene mutations. Mutation of genes encoding tyrosine kinases is uncommon in ALL, with the exception of Philadelphia chromosome-positive ALL, where the t(9,22)(q34;q11) translocation encodes the constitutively active BCR-ABL1 tyrosine kinase. We recently identified a poor prognostic subgroup of pediatric _BCR-ABL1_-negative ALL patients characterized by deletion of IKZF1 (encoding the lymphoid transcription factor IKAROS) and a gene expression signature similar to _BCR-ABL1_-positive ALL, raising the possibility of activated tyrosine kinase signaling within this leukemia subtype. Here, we report activating mutations in the Janus kinases JAK1 (n = 3), JAK2 (n = 16), and JAK3 (n = 1) in 20 (10.7%) of 187 _BCR-ABL1_-negative, high-risk pediatric ALL cases. The JAK1 and JAK2 mutations involved highly conserved residues in the kinase and pseudokinase domains and resulted in constitutive JAK-STAT activation and growth factor independence of Ba/F3-EpoR cells. The presence of JAK mutations was significantly associated with alteration of IKZF1 (70% of all JAK-mutated cases and 87.5% of cases with JAK2 mutations; P = 0.001) and deletion of CDKN2A/B (70% of all _JAK_-mutated cases and 68.9% of _JAK2_-mutated cases). The JAK-mutated cases had a gene expression signature similar to BCR-ABL1 pediatric ALL, and they had a poor outcome. These results suggest that inhibition of JAK signaling is a logical target for therapeutic intervention in JAK mutated ALL.
Keywords: IKAROS, kinase, mutation
Acute lymphoblastic leukemia (ALL) is the most common pediatric cancer, and despite high overall cure rates (1), ALL remains the second leading cause of cancer death in children. To improve outcome, it is necessary to identify high-risk patients at the time of diagnosis and then tailor therapy toward the genetic lesions driving their leukemia.
Recent genome-wide analyses have identified common genetic alterations in childhood ALL that contribute to leukemogenesis (2, 3). To identify genetic lesions predictive of poor outcome in childhood ALL, we recently performed genome-wide analysis of DNA copy number alterations, transcriptional profiling, and gene resequencing in a cohort of 221 children with B progenitor ALL predicted to be at high risk for relapse based on age and presentation leukocyte count (4). These patients were treated on the Children's Oncology Group P9906 trial by using an augmented reinduction/reconsolidation strategy (“Berlin–Frankfurt–Münster” regimen) (5, 6). This cohort excluded patients with known good (ETV6-RUNX1 or trisomies 4 and 10) or very poor (hypodiploid, BCR-ABL1) risk sentinel genetic lesions, and it represents ≈12% of noninfant B precursor ALL cases (Table S1). Alteration of the lymphoid transcription factor IKZF1 (IKAROS) was associated with poor outcome and a leukemic cell gene expression signature highly similar to that of BCR-ABL1 pediatric ALL (4). Furthermore, hierarchical clustering of gene expression profiling data identified a subset of 24 cases with poor outcome (4-year incidence of relapse, death, or second malignancy: 79.1%; 95% C.I., 58.6–99.6%) and expression of outlier genes similar to those seen in BCR-ABL1 ALL. Together, these observations suggested that the poor-outcome, _IKZF1_-deleted, _BCR-ABL1_-negative cases might harbor activating tyrosine kinase mutations. The JAK-STAT pathway may mediate BCR-ABL1 signaling and transformation (7, 8), and JAK1 and JAK2 are mutated in myeloproliferative diseases (9), Down syndrome-associated ALL (DS-ALL), and T lineage ALL (10–12). Here, we have performed genomic resequencing of JAK1, JAK2, JAK3, and TYK2 in 187 diagnostic samples from this high risk B-progenitor ALL cohort that had available DNA and gene expression profiling data. This identified mutations in JAK1, JAK2, and JAK3 in 20 patients (10.7%). The _JAK_-mutated cases had a high frequency of concomitant deletion of IKZF1 (IKAROS) and CDKN2A/B, a gene expression profile similar to BCR-ABL1 ALL, and extremely poor outcome.
Results
JAK1, JAK2, and JAK3 Mutations in High-Risk Pediatric ALL.
Genomic resequencing of JAK1, JAK2, JAK3, and TYK was performed for 187 cases in the P9906 cohort that had available DNA, single-nucleotide polymorphism array, and gene expression profiling data. This identified 20 pediatric ALL patients (10.7%) with 20 heterozygous, somatic mutations of JAK1, JAK2, and JAK3 (Fig. 1, Tables S2 and S3, and Fig. S1). All patients with JAK mutations lacked known common chromosomal translocations.
Fig. 1.
Primary structure of JAK1, JAK2, and JAK3 showing the location of missense (▼) and insertion/deletion (▲) mutations. FERM, band 4.1 ezrin, radixin, and moesin domain; SH2, src-homology domain; JH2, pseudokinase domain; and JH1, kinase domain.
A total of 16 cases had JAK2 mutations, with 13 located in the pseudokinase domain (R683G, n = 10; R683S, n = 1; I682F, n = 1; and QGinsR683, n = 1) and 3 within the kinase domain (R867Q, D873N, and P933R). Three previously undescribed missense or in-frame deletion mutations were also identified in the pseudokinase domain of JAK1 (L624_R629>W, S646F, and V658F), as well as a single JAK3 mutation, S789P. A total of 2 of the 9 DS-ALL cases in our cohort harbored JAK2 mutations (QGinsR683 and R683G), with the remaining 18 JAK mutations occurring in non-DS-ALL patients (Tables S2 and S3). With the exception of JAK3 S789P, each mutation was located in highly conserved residues in either the pseudokinase or kinase JAK domains (Fig. S2). Mutation of JAK2 R683 and JAK1 V658F (which is homologous to the JAK2 V617F mutation common in myeloproliferative disease) (13–16) results in cytokine-independent in vitro growth of Ba/F3-Epo-R or Ba/F3 cells (10, 11, 17).
Concomitant Genomic Abnormalities in JAK-Mutated ALL.
The presence of JAK mutations in this cohort was significantly associated with alterations of IKZF1 and CDKN2A/CDKN2B (Table S2). IKZF1 deletions or mutations were present in 14 (70%) _JAK_-mutated cases (and in 14 of 16 cases with JAK2 mutations) but in only 25.7% of cases that lacked a JAK mutation (P = 0.0001). JAK mutations were also associated with CDKN2A/B deletion (70% vs. 47%, P = 0.06). An increased frequency of copy number alterations at or flanking the IL3RA/CSF2RA/CRLF2 locus at the pseudoautosomal region of Xp22.3/Yp11.3 was also observed in _JAK_-mutated cases (45.0% vs. 4.2%; P < 0.0001). A trend to a significantly higher presenting leukocyte count in JAK-mutated cases was observed (158 × 109/L vs. 101 × 109/L; P = 0.06), but there was no difference in age of presentation.
Structural Modeling of JAK Mutations.
The JAK pseudokinase domain is thought to negatively regulate activity of the kinase domain (18) and may mediate protein–protein interactions (19–21). JAK2 I682 and R683 map to the junction between the N and C lobes of the pseudokinase domain (Fig. S3_A_). All 4 pseudokinase domain mutations identified affect these residues and are predicted to influence the structure and dynamics of the loops that pack together at the interlobe interface, and this may result in a loss of the inhibitory activity of the pseudokinase domain. Accordingly, the R683G and R683S mutations result in the activation of the tyrosine kinase activity of JAK2 (10, 11). R867Q and D873N map to the β2-β3 loop of the kinase domain and are predicted to alter surface electrostatic properties of this region (Fig. S3_B_). The P933 residue lies in the JAK2 kinase hinge region, adjacent to the ATP-binding site (22), and is thought to impart rigidity to this hinge that may be important for catalytic activity. These data suggest that the kinase mutations may lead to enhanced kinase activity.
In Vitro Analysis of JAK Mutations.
To examine the functional consequences of the JAK variants, we transduced murine pro-B Ba/F3 cells expressing the erythropoietin receptor (Ba/F3-EpoR cells) with retroviral constructs expressing wild-type or mutant murine Jak1 or Jak2 alleles. Each Jak mutation examined conferred growth factor independence to Ba/F3-EpoR cells (Fig. 2 A and B) and resulted in constitutive Jak-Stat activation, as assessed by western blotting (Fig. 2E), and by phosphoflow cytometry analysis of Jak2 and Stat5 phosphorylation after serum and cytokine starvation and subsequent erythropoietin or pervanadate stimulation (Fig. 3 A and C). Interestingly, expression of the Jak2 pseudokinase domain mutants resulted in higher growth rates and Jak-Stat phosphorylation than that observed for the Jak2 kinase domain mutants (Figs. 2 A and E and 3 A and C).
Fig. 2.
Functional effects of JAK mutations. (A) Ba/F3-EpoR cells were transduced with retroviruses expressing wild-type or mutant Jak2 alleles and cultured in the absence of cytokine. Each Jak2 mutant examined resulted in cytokine-independent growth. Untransduced cells and cells transduced with wild-type mJak2 remained cytokine-dependent. Mean ± SDs of triplicates are shown. (B) Transduction of Ba/F3-EpoR cells with retrovirus expressing mJak1 S646F resulted in factor independence. (C) Ba/F3-EpoR cells transduced with wild-type or mutant Jak2 were cultured without cytokine in the presence of increasing concentrations of Jak inhibitor I. Each mutation was sensitive to Jak inhibition. The _BCR-ABL1_-positive cell line K562 is shown as a control. (D) Growth of Ba/F3 cells transduced with S646F was inhibited by Jak inhibitor I. (E) Western blots showing activation of JAK-STAT signaling in Ba/F3-EpoR cells transduced with each mutant Jak allele. Cells were cultured without erythropoietin for 15 h and then harvested for blotting before and after 15 min of erythropoietin at 5 units/mL. Each mutation resulted in constitutive Jak2 and Stat5 phosphorylation that was augmented by pulsed erythropoietin (shown for Jak2 617F and 683G). The Jak2 kinase domain mutations showed less constitutive Jak-Stat activation than the pseudokinase domain mutations. Epo, erythropoietin; WT, wild type. (F) Western blots demonstrating abrogation of Jak-Stat activation by Jak inhibitor I. Ba/F3-EpoR cells transduced with each Jak allele were grown in the absence of cytokine, then harvested after 5 h of exposure to 5 mM Jak inhibitor I or vehicle (DMSO).
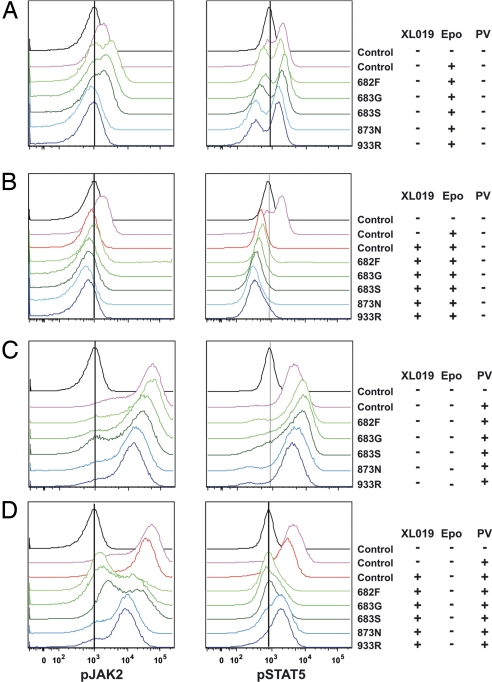
Fig. 3.
Phosphoflow cytometry analysis of Jak-Stat activation in Ba/F3-EpoR cells transduced with Jak2 retroviral constructs. Transduced cells were serum-starved and cytokine-starved and then stimulated either with erythropoietin (Epo; A and B) or pervanadate (PV; C and D), either without pharmacologic Jak inhibition (A and C) or after administration of the Jak2 inhibitor XL019 (B and D). (A) Activation of Jak-Stat phosphorylation with erythropoietin stimulation. Notably, Jak2 phosphorylation was evident for Jak2 pseudokinase mutant alleles but not the kinase domain mutants. (B) Signaling was abrogated in control and mutants treated with XL019 with subsequent erythropoietin stimulation. (C) Marked Jak2 and Stat5 phosphorylation was observed for each mutant after pervanadate stimulation. (D) Jak2 and Stat5 signaling was preferentially abrogated in mutants treated with XL019 with subsequent pervanadate stimulation.
This transformation was abrogated by the pan-Jak-specific inhibitor Jak inhibitor I (Fig. 2 C, D, and F). The Jak2 inhibitor XL019 abrogated ligand-induced Jak-Stat activation induced by all tested Jak2 mutants (Fig. 3B). Treatment of the cells with the tyrosine phosphatase inhibitor pervanadate led to greater levels of Jak2 and Stat5 phosphorylation (Fig. 3C), which was more completely inhibited for mutations involving the pseudokinase domain than the kinase domain (Fig. 3D). The basis of this variable inhibition is unknown but raises the possibility of differences in the mechanism of transformation induced by each JAK mutation.
Similarity of the Gene Expression Profiles of _JAK_-Mutated and _BCR-ABL1_-Positive ALL.
The similarity in gene expression signatures between _IKZF1_-deleted _BCR-ABL1_-positive and _BCR-ABL1_-negative ALL suggested the possibility of activated tyrosine kinase signaling in the _BCR-ABL1_-negative cases (4). As expected, the JAK-mutated cases exhibited a _BCR-ABL1_-like gene expression signature (Fig. 4 A and B). Notably, additional _IKZF1_-mutated cases that lacked JAK mutations also showed enrichment of the _BCR-ABL1_-like signature (Fig. 4B), suggesting that these cases may harbor additional tyrosine kinase or JAK-STAT-activating mutations.
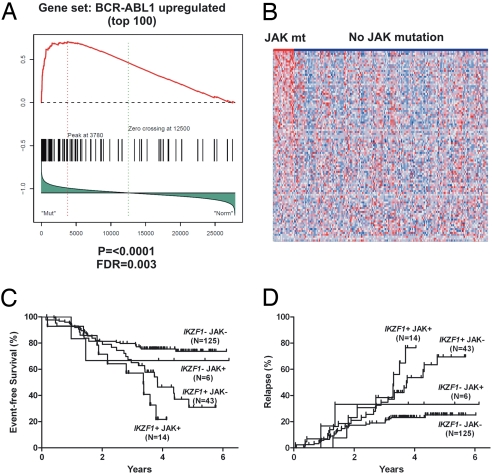
Fig. 4.
Gene expression profile and outcome of JAK-mutated B-progenitor ALL. (A) Gene set enrichment analysis demonstrates significant enrichment of the BCR-ABL1 gene expression signature in JAK-mutated ALL. (B) Heatmap of the enriched BCR-ABL1 up-regulated gene set in the P9906 cohort, showing overexpression of BCR-ABL1 up-regulated genes in JAK-mutated ALL. Notably, several cases lacking JAK mutations also have a BCR-ABL1 signature, suggesting the presence of additional kinase mutations in these cases. (C and D) JAK mutation and IKZF1 alteration are associated with a high incidence of events (C) and relapse (D).
JAK Mutations and IKZF1 Alteration Are Associated with Poor Outcome in Pediatric ALL.
We observed highly significant associations between IKZF1 and JAK lesions and outcome. The 4-year cumulative incidence of events (relapse, death, or second malignancy) was 78.2% for patients with both a JAK mutation and IKZF1 alteration, compared with 54.4% for IKZF1 alteration only, 33.3% for JAK mutation only, and 24.3% for neither lesion (P = 0.0002; Fig. 4C). This was primarily attributable to differences in the risk of relapse. The 4-year cumulative incidence of relapse was 76.6% for patients with both a JAK mutation and IKZF1 alteration, compared with 53.6% for IKZF1 alteration only, 33.3% for JAK mutation only, and 23.2% for neither lesion (P = 0.0004; Fig. 4D). In multivariable analyses incorporating clinical and laboratory variables, there was a trend toward an association between JAK mutations and increased risk of events or relapse (Table S4). However, no independent association was observed after incorporation of IKZF1 status in the model (Table S5). This is in part due to the highly significant correlation of JAK mutations and IKZF1 alterations. Moreover, additional _IKZF1_-mutated, JAK wild-type cases also have a “_BCR-ABL1_-like” signature and poor outcome, suggesting additional unidentified kinase-activating lesions in these cases.
Discussion
These results demonstrate that JAK kinase mutations are not limited to patients with DS-ALL, but also occur in about 10% of high-risk pediatric B-progenitor ALL patients. Notably, the P9906 cohort studied here is not an unselected series of childhood ALL cases, but comprises patients with high white blood cell counts and/or older age that were predicted to have a poor outcome. Patients with high hyperdiploidy, hypodiploidy, ETV6-RUNX1, or BCR-ABL1 were not included; however, the cohort did include TCF3-PBX1 (n = 22) and _MLL_-rearranged (n = 18) B-progenitor cases, none of whom had JAK mutations. These differences in cohort composition provide a potential explanation as to why JAK mutations have not been detected more frequently in non-DS-ALL in other studies (10, 11). Future studies including larger numbers of patients with recurring cytogenetic alterations will be of interest to determine whether JAK mutations occur predominantly among ALL patients lacking known translocations and aneuploidy.
JAK mutations were associated with concomitant IKZF1 and CDKN2A/B alterations, suggesting that genetic lesions targeting multiple cellular pathways, including lymphoid development (IKZF1), tumor suppression (CDKN2A/B), and activation of tyrosine kinases (BCR-ABL1, JAK, or other kinase mutations) cooperate to induce aggressive lymphoid leukemia in both DS-ALL and non-DS-ALL that is resistant to conventional ALL therapy. The majority of the identified JAK mutations occur in the pseudokinase domain of JAK2 in a region (R683) distinct from the predominant mutation (V617F) seen in polycythemia vera and related myeloproliferative diseases (9). It has been hypothesized that the nature of the JAK mutation plays a direct role in establishing the disease phenotype (9, 23, 24), and mutations at R683 have been identified almost exclusively in DS-ALL-related ALL (10, 11). Experiments testing the in vivo-transforming activity of the JAK mutations and the cooperative effect of concomitant genetic lesions, such as alteration of IKZF1, should provide valuable insights into how these lesions contribute to leukemogenesis and treatment resistance. Notably, both DS-ALL cases with JAK mutations in this study had concomitant alterations of IKZF1 and CDKN2A/B, suggesting that the cooccurrence of these lesions is important in the pathogenesis of DS and non-DS high-risk ALL. The identification of JAK mutations in a subset of the _IKZF1_-mutated, poor-outcome group raises the possibility that inhibition of JAK activity will be a logical therapeutic approach in these patients. Indeed, our data demonstrate impressive inhibition of the Jak-Stat activation induced by Jak pseudokinase domain mutations by the JAK2 inhibitor XL019, a drug currently in early-phase trials for myeloproliferative disorders. Finally, the absence of JAK mutations in additional cases with a _BCR-ABL1_-like expression signature suggests that efforts to identify the causes of activated kinase signaling in these cases should identify additional therapeutic targets in high-risk pediatric ALL.
Materials and Methods
Patients and Treatment.
Patients were enrolled in the Children's Oncology Group P9906 trial and treated with an augmented reinduction/reconsolidation strategy (5). All patients were high-risk based on the presence of central nervous system or testicular disease, MLL rearrangement, or based on age, sex, and presentation leukocyte count (25). BCR-ABL1 and hypodiploid ALL, as well as cases of primary induction failure were excluded. The cohort is described further in the SI Methods.
Genomic Resequencing and Structural Modeling of JAK2 Mutations.
Resequencing of the coding exons of JAK1, JAK2, JAK3, and TYK2 was performed by Agencourt Biosciences. Sequencing, sequence analysis, structural modeling, and homology alignment of JAK mutations are described in the SI Methods.
Functional Assays of JAK Mutants.
The JAK1 S646F and JAK2 V617F, I682F, R683G, R683S, D873N, and P933R mutations were introduced into the bicistronic MSCV-IRES-GFP retroviral vector encoding either murine Jak1 or Jak2 containing the C-terminal HA tag (26) by site-directed mutagenesis (QuikChange XL II; Stratagene). Retroviral supernatants were produced by using ecotropic Phoenix packaging cells (G.P. Nolan; www.stanford.edu/group/nolan/). Murine pro-B Ba/F3 cells were transduced with MSCV-EpoR-IRES-puro, and after puromycin selection they were transduced with wild-type or mutant Jak retroviral supernatants. Transduced cells were purified by flow sorting for GFP and were maintained in RPMI-1640 with 10% FCS (HyClone) penicillin-streptomycin, l-glutamine, and 5 units/mL erythropoietin. To assess growth factor independence, cells were washed 3 times and were plated at 500,000 cells per milliliter in media without cytokine, with or without JAK inhibitor I (Calbiochem), and growth was monitored daily by using a ViCell cell counter (Beckman Coulter).
For Western blotting, Jak-transduced Ba/F3-EpoR cells were cultured for 15 h without erythropoietin, followed by 15 min of treatment with erythropoietin at 5 units/mL or vehicle (DMSO). Whole-cell lysates were blotted and probed with anti-Jak2, anti-phospho-Jak2 (Tyr 1007–1008), anti-Stat5, and anti-phospho-Stat5 (Cell Signaling Technology), and with anti-PCNA (Santa Cruz Biotechnology).
Cytokine stimulation and intracellular phosphoprotein analysis using flow cytometry was performed as described previously (27). Ba/F3-EpoR cells were serum- and cytokine-starved for 30 min, then incubated with the JAK2 inhibitor XL019 (Exelixis) at a concentration of 5 μM for 30 min. Control and XL019-treated cells were subsequently stimulated with 5 ng/mL murine IL-3, 2 units/mL human erythropoietin, or 125 μM pervanadate for 15 min. Cells were fixed, permeabilized, rehydrated overnight, and then stained with anti-phospho-Stat5-Alexa 647 (Tyr-694; BD Biosciences), anti-phospho-Jak2 (Tyr 1007–1008), and phycoerythrin-conjugated donkey anti-rabbit IgG secondary antibody (Jackson ImmunoResearch). Samples were analyzed on an LSRII flow cytometer (BD Biosciences), and data were collected and analyzed by using DIVA (BD Biosciences) and FlowJo (Tree Star).
Gene Set Enrichment Analysis (GSEA).
GSEA (28) was performed as described previously (2, 4) by using the collection of publicly available gene sets (www.broad.mit.edu/gsea/msigdb/) and gene sets derived from the top up- and down-regulated genes of BCR-ABL1 de novo pediatric ALL (29, 30).
Statistical Analysis.
Associations between clinical, laboratory, and genetic variables and outcome (event-free survival and relapse) were performed as described previously (4). Cumulative incidence of relapse according to IKZF1 and JAK status was analyzed by using Gray's test (31). Associations with event-free survival were examined by using the methods of Kaplan and Meier and the Mantel–Haenszel test (32). Multivariable analyses of event-free survival were performed by using the EFS-PHREG procedure in SAS version 9.1.3 (SAS Institute); multivariable analyses of relapse were performed by using the Fine and Gray method (33) in S-Plus version 7.0.6 (Insightful).
Supplementary Material
Supporting Information
Acknowledgments.
We thank E. Parganas and J. Ihle (St. Jude Children's Research Hospital, Memphis, TN) for murine Jak and EpoR-puro retroviral constructs, and D. Clary (Exelixis) for providing XL019. The correlative biology studies described in this manuscript were funded by grants, funds from the National Institutes of Health (NIH), and philanthropic funds of the Children's Oncology Group, and not by a commercial entity. This work was supported by funds provided as a supplement to the Children's Oncology Group Chair's Award CA098543 (to S.P.H.); National Cancer Institute (NCI) Strategic Partnering to Evaluate Cancer Signatures (SPECS) Program Award CA114762 (to W.L.C., I.-M.C., R.C.H., and C.L.W.); NIH Cancer Center Core Grant 21765 (to J.R.D. and C.G.M.); NCI Grant U10 CA98543 supporting the TARGET initiative, the Children's Oncology Group, and U10 CA98413 supporting the Statistical Center (to G.H.R); Leukemia and Lymphoma Society Specialized Center of Research Grant 7388-06 (to C.L.W.); NCI Grant P30 CA118100 (to C.L.W) supporting the University of New Mexico Cancer Center Shared Resources; CureSearch; St. Baldrick's Foundation (M.L.L.); a National Health and Medical Research Council (Australia) CJ Martin Traveling Fellowship (to C.G.M.); and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital. B.A.S. is an investigator of the Howard Hughes Medical Institute. S.P.H. is the Ergen Family Chair in Pediatric Cancer. The sequencing was funded with federal funds from the National Cancer Institute, National Institutes of Health, Contract N01-C0-12400.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 3.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 4.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nachman JB, et al. Augmented post-induction therapy for children with high-risk acute lymphoblastic leukemia and a slow response to initial therapy. N Engl J Med. 1998;338:1663–1671. doi: 10.1056/NEJM199806043382304. [DOI] [PubMed] [Google Scholar]
- 6.Borowitz MJ, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: A Children's Oncology Group study. Blood. 2008;111:5477–5485. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samanta AK, et al. Janus kinase 2: A critical target in chronic myelogenous leukemia. Cancer Res. 2006;66:6468–6472. doi: 10.1158/0008-5472.CAN-06-0025. [DOI] [PubMed] [Google Scholar]
- 8.Xie S, et al. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene. 2001;20:6188–6195. doi: 10.1038/sj.onc.1204834. [DOI] [PubMed] [Google Scholar]
- 9.Vainchenker W, Dusa A, Constantinescu SN. JAKs in pathology: Role of Janus kinases in hematopoietic malignancies and immunodeficiencies. Semin Cell Dev Biol. 2008;19:385–393. doi: 10.1016/j.semcdb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Bercovich D, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372:1484–1492. doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- 11.Kearney L, et al. A specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukemia. Blood. 2008;113:646–648. doi: 10.1182/blood-2008-08-170928. [DOI] [PubMed] [Google Scholar]
- 12.Flex E, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 14.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 15.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Baxter EJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 17.Staerk J, et al. JAK1 and Tyk2 activation by the homologous polycythemia vera JAK2 V617F mutation: Cross-talk with IGF1 receptor. J Biol Chem. 2005;280:41893–41899. doi: 10.1074/jbc.C500358200. [DOI] [PubMed] [Google Scholar]
- 18.Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- 19.Russo AA, et al. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 20.Russo AA, et al. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 21.Brotherton DH, et al. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395:244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- 22.Lucet IS, et al. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–183. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 23.Levine RL, Pardanani A, Tefferi A, Gilliland DG. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer. 2007;7:673–683. doi: 10.1038/nrc2210. [DOI] [PubMed] [Google Scholar]
- 24.Levine RL, Gilliland DG. Myeloproliferative disorders. Blood. 2008;112:2190–2198. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuster JJ, et al. Identification of newly diagnosed children with acute lymphocytic leukemia at high risk for relapse. Cancer Res Ther Control. 1999;9:101–107. [Google Scholar]
- 26.Funakoshi-Tago M, et al. Jak2 FERM domain interaction with the erythropoietin receptor regulates Jak2 kinase activity. Mol Cell Biol. 2008;28:1792–1801. doi: 10.1128/MCB.01447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotecha N, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14:335–343. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeoh EJ, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 30.Ross ME, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 31.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–1154. [Google Scholar]
- 32.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 33.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information