Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N (original) (raw)
. Author manuscript; available in PMC: 2010 Jan 1.
Published in final edited form as: Nat Cell Biol. 2009 Jun 21;11(7):896–902. doi: 10.1038/ncb1899
Abstract
Centromeres are specialized chromosomal domains that direct kinetochore assembly during mitosis. CENP-A, a histone H3-variant present exclusively in centromeric nucleosomes, is thought to act as an epigenetic mark that specifies centromere identity. Here we identify the essential centromere protein CENP-N as the first protein to selectively bind CENP-A nucleosomes but not H3 nucleosomes. CENP-N bound CENP-A nucleosomes in a DNA-sequence independent manner but did not bind soluble CENP-A/H4 tetramers. Mutations in CENP-N that reduced the affinity of CENP-N for CENP-A nucleosomes caused defects in CENP-N localization and had dominant effects on the recruitment of CENP-H, CENP-I and CENP-K to centromeres. Depletion of CENP-N with siRNA’s led to similar centromere assembly defects and resulted in reduced assembly of nascent CENP-A into centromeric chromatin. These data suggest that CENP-N interprets the information encoded within CENP-A nucleosomes and recruits to centromeric chromatin other proteins required for centromere function and propagation.
Accurate chromosome segregation during mitosis is essential for the maintenance of genome integrity. Eukaryotic cells have evolved complex machinery to ensure the fidelity of chromosome segregation. Each chromosome directs the assembly of a kinetochore that mediates attachment to the mitotic spindle and is required for microtubule-dependent chromosome movement during mitosis. Kinetochores also act as signaling centers for the mitotic checkpoint, which delays the initiation of anaphase in response to improper chromosome attachment to the mitotic spindle (reviewed in 1). The centromere is the region of the chromosome upon which the kinetochore is assembled. Human centromeric DNA is composed of repetitive α-satellite sequences but centromeres are thought to be epigenetically specified as centromeric DNA is not well conserved between species and no DNA sequences have been identified that are necessary or sufficient for kinetochore function in vertebrates (reviewed in 2). Centromeric chromatin in all eukaryotes contains specialized nucleosomes in which histone H3 is replaced by a histone H3-variant called centromere protein A (CENP-A) (reviewed in 3). CENP-A is currently the best candidate for the epigenetic mark that specifies centromere identity. CENP-A is essential for the recruitment to the centromere of most other proteins required for kinetochore function but the molecular basis for recognition of CENP-A-containing chromatin as the site of kinetochore assembly is poorly understood. Furthermore, the mechanisms that target newly synthesized CENP-A/H4 to established centromeric chromatin to maintain centromere identity have not been determined.
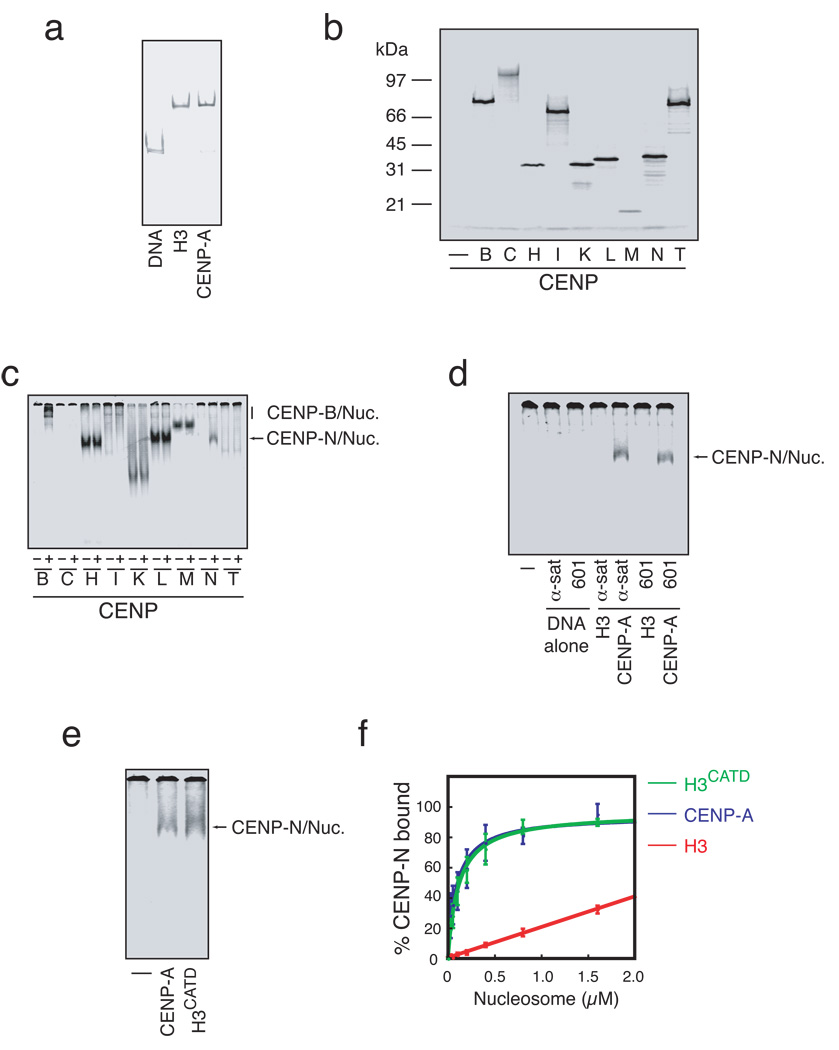
A fundamental limitation in understanding centromere assembly is the lack of well-defined biochemical assays for studying this process. In no case has a direct and specific interaction between CENP-A nucleosomes and any of the >75 proteins that make up a vertebrate mitotic kinetochore been demonstrated. In order to identify CENP-A-nucleosome interacting proteins we developed a simple and rapid binding assay using reconstituted mononucleosomes that contained α-satellite DNA derived from human centromeres and either histone H3 or CENP-A (Figure 1a). Potential CENP-A-nucleosome interacting proteins were selected for testing in this assay based on the analysis of kinetochore assembly in several organisms. We focused on the constitutive centromere associated network (CCAN) of proteins, which includes centromere protein (CENP)-C, H, I, and K through U, because the CCAN proteins are localized to centromeres during interphase and are required for the assembly of functional kinetochores in mitosis. Moreover, the localization by immuno-electron microscopy of CENP-C to the inner kinetochore plate and the recent demonstration that several of these proteins co-purify with CENP-A mononucleosomes suggests that CCAN proteins are likely to be the chromatin proximal elements of kinetochores4–7. We also included CENP-B in our analysis because it has previously been shown to bind directly to a conserved 17-nucleotide motif called the CENP-B box present in α-satellite DNA 8. We expressed and 35S-labeled CENP-B, C, H, I, and K through U by coupled in vitro transcription and translation (Figure 1b, S1), incubated the labeled proteins with or without CENP-A mononucleosomes, and then resolved the mixtures by native gel electrophoresis. Both CENP-B and CENP-N displayed an increased relative migration in the presence of CENP-A nucleosomes assembled with α-satellite DNA suggesting that CENP-B and CENP-N bind directly to CENP-A nucleosomes (Figure 1c). We did not detect any change in the migration of CENP-C, H, I, K, L, M or O through U (Figure 1c, S1a). We also tested hMis18α, hMis18β and M18BP1/hKNL-2 that have been implicated in CENP-A assembly but we did not detect any interaction with CENP-A nucleosomes in this assay (Figure S1b)9, 10.
Figure 1. CENP-N binds CENP-A nucleosomes.
a) Reconstitution of conventional and centromeric nucleosomes. Free 186bp human alpha satellite DNA (DNA) or mononucleosomes containing either histone H3 or CENP-A were assembled by salt dialysis and resolved by native gel electrophoresis. b) In vitro expression of centromere proteins. Centromere proteins to be used as substrates in nucleosome binding assays were expressed in coupled transcription and translation reactions containing with 35S-methionine. c) CENP-A nucleosome binding assay. 35S-labeled centromere proteins (~1 nM) were incubated in the presence (+) or absence (−) of reconstituted CENP-A nucleosomes (50 nM) and separated by native gel electrophoresis. Both CENP-B and CENP-N exhibit CENP-A nucleosome dependent changes in migration. d) CENP-N binding to nucleosomes depends upon CENP-A and not DNA sequence. α-satellite DNA (α-sat) or 601 DNA (601), or H3-or CENP-A-containing nucleosomes reconstituted with α-satellite or 601 DNA were incubated with 35S-labeled CENP-N and resolved by native gel electrophoresis. The faster migrating band (arrow) indicates nucleosome bound CENP-N. No nucleosome or DNA control (−). e) Binding of CENP-N to CENP-A nucleosomes occurs through the CATD region of CENP-A. No nucleosome control (−) and CENP-A or H3CATD nucleosomes reconstituted with α-satellite DNA were bound to 35S-CENP-N and assayed as in a. f) CENP-N binds with equal affinity to CENP-A and H3CATD nucleosomes. CENP-N binding was assayed as in (a) with increasing nucleosome concentration and quantified based upon the 35S-CENP-N signal in the gel (N=3, error bars represent SEM).
We determined the contribution of DNA sequence and histone protein composition to CENP-B and CENP-N binding of CENP-A nucleosomes by alternately exchanging the nucleosomal DNA and histones. As expected, CENP-B bound to naked α-satellite DNA as well as both H3 and CENP-A nucleosomes that contained α-satellite DNA (Figure S1c). CENP-B did not bind the synthetically derived 601 nucleosome positioning sequence or nucleosomes that contained the 601 DNA sequence. In contrast, CENP-N bound equally well to CENP-A nucleosomes that contained either the α-satellite DNA or the 601 DNA (Figure 1d). CENP-N did not bind the α-satellite or 601 DNA fragment or histone H3 nucleosomes assembled on either DNA fragment. CENP-N also bound to CENP-A/H4 tetrasomes reconstituted with α-satellite DNA that lacked histone H2A and H2B but did not specifically bind to soluble CENP-A/H4 tetramers (Figure S1d,e,f). Thus, CENP-N binds specifically to CENP-A-associated chromatin.
Domain transfer experiments have previously suggested that a contiguous portion of the loop I and helix II region within the histone fold of CENP-A, called the CENP-A targeting domain (CATD), is sufficient for CENP-A function in vivo11, 12. We tested whether the CATD was sufficient for CENP-N binding using reconstituted nucleosomes that contained the histone H3CATD chimera. CENP-N bound efficiently to H3CATD-containing nucleosomes (Figure 1e), and dose-response experiments revealed that the affinity of CENP-N for H3CATD nucleosomes was indistinguishable from the affinity of CENP-N for wildtype CENP-A nucleosomes (apparent Kd =163 nM ±60 vs. 169 nM ±70, respectively) (Figure 1f, Table S1). The CATD imparts structural differences to CENP-A nucleosomes, in comparison to H3 nucleosomes, that have been suggested to act as an epigenetic mark within chromatin to specify centromere identity13. Our data suggest that CENP-N recognizes the unique structural information encoded by the CATD within CENP-A nucleosomes.
Depletion of CENP-N with siRNA causes defects in kinetochore assembly and chromosome congression during metaphase, and results in the loss from the centromere of most other CCAN components5, 14. However, the depletion of other CCAN subunits, including CENP-H, CENP-I, and CENP-K, results in mitotic phenotypes similar to those caused by CENP-N depletion4, 5, 14, 15. Furthermore, CENP-N co-purifies with CENP-H, CENP-I, CENP-M, CENP-K, CENP-L and CENP-T, which are all likely to be interdependent for centromere localization based on pairwise dependency relationships4, 5, 14, 15. Accordingly, a specific function for CENP-N in recognizing centromeric chromatin in vivo cannot be inferred from siRNA-based studies.
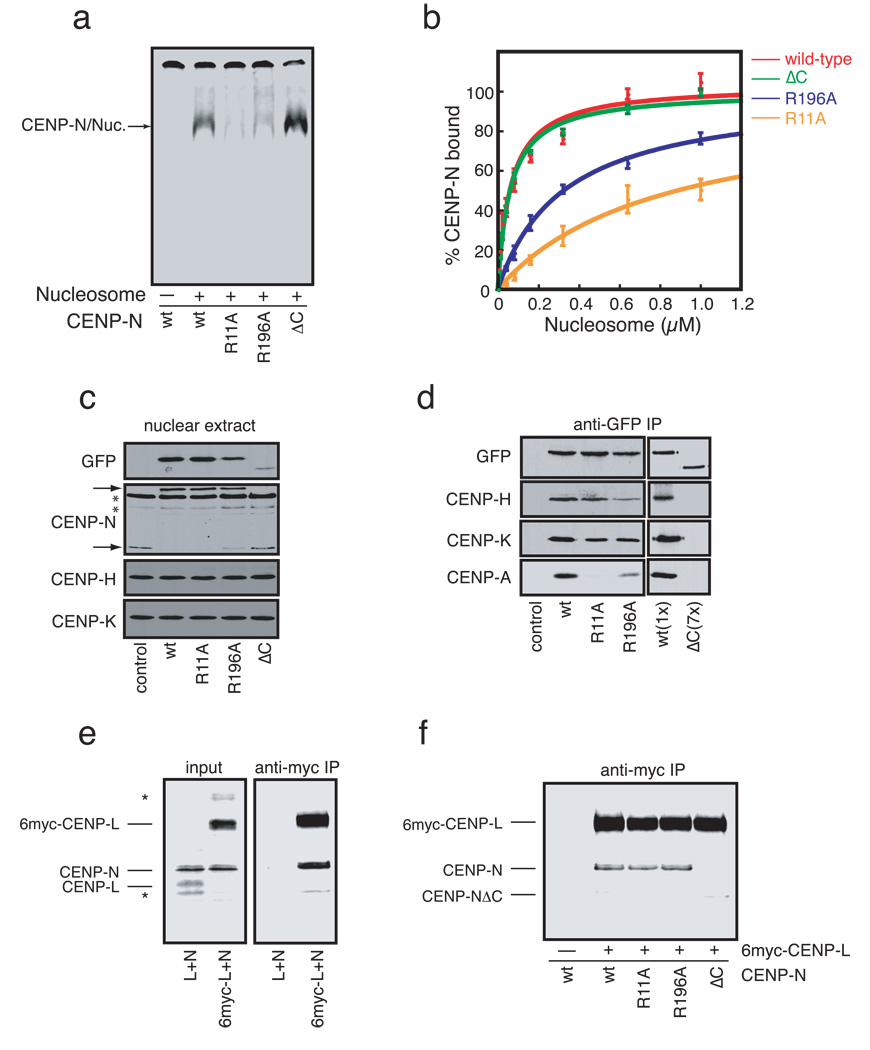
In order to directly determine the role of CENP-A nucleosome binding by CENP-N in centromere assembly we generated mutants in CENP-N that specifically reduced CENP-A nucleosome binding affinity. Conserved charged and polar amino acids within CENP-N were changed to alanine based on sequence alignments of CENP-N orthologs from several species (Figure S2a). Following an initial characterization of the mutants in vitro and in vivo (Figure S3 and Table S1), two of the point mutants, R11A and R196A, were selected for detailed analysis. Both the R11A and R196A mutants displayed reduced CENP-A nucleosome binding when compared to wildtype CENP-N (Figure 2a). Dose-response experiments indicated that the CENP-N R11A and R196A mutants had a 6-fold and a 2-fold reduction in CENP-A nucleosome binding, respectively (Figure 2b, Table S1), suggesting that both R11 and R196 in CENP-N contribute to the recognition of CENP-A nucleosomes. In addition, the C-terminus of CENP-N (amino acids 289–339 in human CENP-N) is more highly conserved among vertebrates than the rest of the protein (Figure S2b). We therefore constructed a truncation mutant of CENP-N lacking the C-terminus (called CENP-NΔC). The CENP-NΔC mutant did not affect nucleosome binding when compared to wildtype CENP-N (Figure 2a, 2b).
Figure 2. Identification and characterization CENP-N mutants defective in CENPA-nucleosome binding.
a) CENP-N mutants exhibit a range of affinities for CENP-A nucleosomes. CENP-N (wt) or the indicated CENP-N mutants were expressed and an equal concentration (~1nM) of each mutant was assayed for its ability to bind CENP-A nucleosomes (300nM). The control reaction (−) lacked CENP-A nucleosomes. b) Doseresponse experiments for each CENP-N mutant were performed as in (a) with increasing concentrations of CENP-A nucleosome added to each reaction (N=3, error bars represent SEM). c) Expression of CENP-N mutants in HEK293 cells. Nuclear extracts from stable HEK293 cell lines expressing GFP-CENP-N (wt) or the indicated GFP-CENP-N mutant were separated by SDS-PAGE and western blotted with the indicated antibodies. Two nonspecific bands (*) are present in the anti-CENP-N western blot. Upper and lower arrows indicate positions of GFP-CENP-N and endogenous CENP-N, respectively. Quantitation of each bands is presented in Figure S4a. d) Coimmunoprecipitation of CENP-H, K, and A from micrococcal-nuclease solubilized chromatin with CENP-N mutants. Anti-GFP immunoprecipitates from each cell line were probed with the indicated antibodies. 7X more nuclear extract from the CENP-NΔC cell line was required to achieve equal levels of CENP-N in the immunoprecipitations. Quantitation of each bands is presented in Figure S4b. e) CENP-N binds to CENP-L. 6xmyc-CENP-L, CENPL and CENP-N were expressed and labeled with 35S-methionine and the indicated proteins were mixed at equal stoichiometry. 20% of each mixture was resolved as input (left panel). The remaining 80% was immunoprecipitated with anti-myc antibodies (right panel). Two background bands (*) are present in the input reactions. f) CENP-L binding requires the C-terminus of CENP-N. Immunoprecipitations are identical to those in (e), except that equal amounts of wildtype CENP-N (wt) or the indicated CENP-N mutant was used in each reaction.
We generated stable HEK293 cell lines that expressed green-fluorescent protein (GFP) fusions to either wildtype CENP-N or the CENP-N mutants to determine how changing the affinity of CENP-N for the CENP-A nucleosome affected CENP-N localization and centromere assembly in vivo. Western blotting indicated that each cell line expressed comparable levels of the respective GFP-CENP-N protein, with the exception of the CENP-NΔC mutant, which was reduced ~7-fold compared to wildtype GFP-CENP-N (Figure 2c and S4a). The HEK293 cell lines express CENP-N from a single genomic locus under the same promoter (Methods) suggesting that the decreased protein level is a general property of the CENP-NΔC mutant. Interestingly, while the levels of CENP-H and CENP-K were not affected in any of the stable cells lines, the expression of wildtype GFP-CENP-N or either of the two GFP-CENP-N point mutants caused a significant reduction in endogenous CENP-N protein (Figure 2c). A reduction of endogenous CENP-N protein has also been described in cells depleted of CENP-H suggesting that CENP-N is unstable when not associated with other CCAN components14. We conclude that wildtype GFP-CENP-N and the GFP-CENP-N R11A and R196A mutant proteins effectively replaced endogenous CENP-N in these cell lines.
We directly determined the ability of the CENP-N point mutants to interact with other CCAN proteins and with CENP-A nucleosomes in vivo. Immunoprecipitation of wildtype GFP-CENP-N showed that GFP-CENP-N bound to CENP-H, CENP-K and CENP-A nucleosomes (Figure 2d, S4b). The GFP-CENP-N R11A and R196A mutants were also associated with CENP-H and CENP-K, though the R196A mutant bound CENP-H less well when compared to wildtype CENP-N. Importantly, the R11A and R196A mutants both displayed defects in CENP-A nucleosome binding, the severity of which was consistent with the binding of each mutant to reconstituted CENP-A nucleosomes. Thus, residues R11 and R196 within CENP-N contribute to CENP-A nucleosome binding in vitro and in vivo. The comparatively low levels of the GFP-CENP-NΔC present in stable cells and the inability of this mutant to down-regulate endogenous CENP-N protein levels suggested that the GFP-CENP-NΔC mutant might not be stably associated with other CCAN proteins. Indeed, GFP-CENP-NΔC did not bind to CENP-H, CENP-K or CENP-A nucleosomes (Figure 2d).
In order to understand the mechanism by which the C-terminus of CENP-N mediates CENP-N association with CCAN proteins we expressed epitope-tagged centromere proteins and untagged CENP-N in reticulocyte extracts and performed pairwise binding experiments by anti-myc immunoprecipitation. Myc-tagged CENP-L, but not untagged CENP-L, efficiently co-precipitated with CENP-N indicating a direct interaction between CENP-N and CENP-L (Figure 2e). Myc-tagged CENP-L bound the CENP-N R11A and R196A mutants as efficiently as wildtype CENP-N (Figure 2f), consistent with the efficient association of these CENP-N mutants with CENP-H and CENP-K in vivo. However, the GFP-CENP-NΔC mutant did not bind to CENP-L. These data suggest that CENP-N associates with other CCAN components via a direct interaction with CENP-L that requires the highly conserved C-terminus of CENP-N.
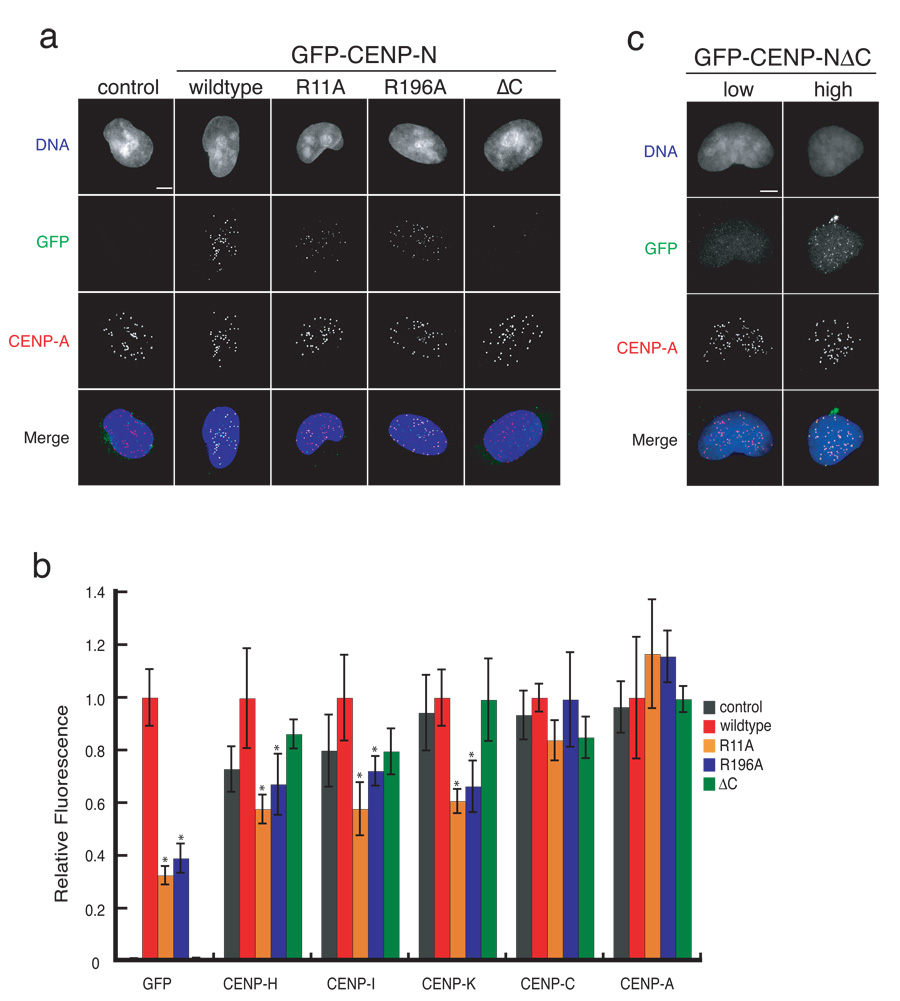
We compared the localization of the CENP-N R11A, R196A and ΔC mutants to wildtype CENP-N. Wildtype GFP-CENP-N localized exclusively to centromeres as indicated by colocalization with endogenous CENP-A (Figure 3a). Both the R11A and R196A mutants also localized to centromeres but did so inefficiently. Quantification showed that the levels of the CENP-N R11A and R196A mutants at centromeres was reduced to 32 ± 4% and 39 ± 6%, respectively, of the levels of wildtype CENP-N (Figure 3b). The difference in localization efficiency between the GFP-CENP-N R11A and R196A mutants was not as great as would be predicted from their relative CENP-A nucleosome binding affinities (Figure 2b, 2d), suggesting that the other CCAN proteins likely contribute to CENP-N localization efficiency in vivo. Nevertheless, these data show that mutations that reduce CENP-A nucleosome binding by CENP-N result in quantitative defects in the centromere-specific localization of CENP-N.
Figure 3. CENP-N mutants exhibit centromere assembly defects.
a) Representative images of control HEK293 cells and stable cell lines expressing wildtype or the indicated CENP-N mutant. Scale bar represents 5 µm. b) CENP-N mutants cause centromere assembly defects. The centromere fluorescence intensity of the indicated centromere protein at individual centromeres in stable cell lines was measured (see Methods). Error bars represent SEM for three independent experiments including >300 centromeres from >20 cells for each cell line. Asterisks indicate p<0.05 significance compared to wildtype. c) The C-terminus of CENP-N is not required for centromere localization. Representative images from transiently transfected HeLa cells show the localization of GFP-CENP-NΔC in low and high expressing cells. Scale bar represents 5 µm.
We did not detect the CENP-NΔC mutant at centromeres in our stable cell line suggesting that association with other CCAN subunits is an important step in the recruitment of CENP-N to centromeres (Figure 3a,b). However, transiently transfected cells expressing the GFP-CENP-NΔC mutant from a strong promoter occasionally contained detectable levels of the mutant protein at centromeres (Figure 3c). Thus, the C-terminus of CENP-N is not absolutely required for CENP-N centromere localization. Instead, our data indicate that association with the CCAN stabilizes CENP-N and likely increases the efficiency of CENP-N centromere localization.
Mutations in CENP-N that affected CENP-A nucleosome binding caused dominant defects in centromere assembly in our stable cell lines. Quantification of CENP-H, CENP-I and CENP-K in cells expressing the CENP-N R11A and R196A mutants indicated that the levels of each protein at centromeres in interphase cells was significantly reduced when compared to cells expressing wildtype CENP-N (Figure 3b). Thus, reducing the level of CENP-N at centromeres led to a reduction in the levels of a subset of other CCAN subunits. The defect in CENP-H, CENP-I and CENP-K localization in the mutant cells was not as severe as that of CENP-N itself suggesting that a small amount of remaining endogenous CENP-N may also contribute to centromere assembly in these cell lines (Figure 3b). The CENP-N R11A and R196A mutants did not alter the levels of CENP-A or CENP-C at centromeres.
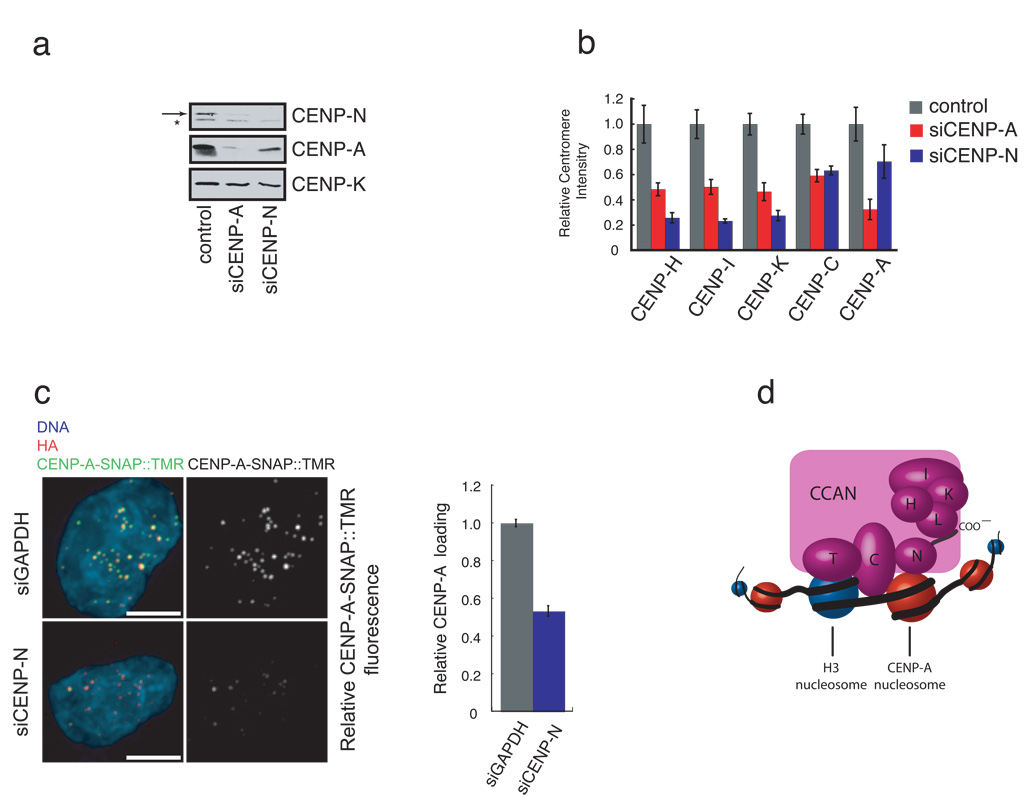
We next examined the localization dependence of CENP-H, CENP-I, and CENPK, on CENP-N by depleting CENP-N with siRNA in HeLa cells (Figure 4a, S5a). CENP-N depletion led to a substantial reduction in the levels of CENP-H, CENP-I and CENP-K at centromeres (Figure 4b) consistent with previous studies demonstrating CENP-H reduction in cells depleted of CENP-N 14. The localization defects of CENP-H, CENP-I and CENP-K in CENP-N depleted cells were more severe than in cells depleted of CENP-A, indicating that the observed reduction in CENP-H, CENP-I and CENP-K levels at centromeres is not an indirect consequence of the reduced CENP-A levels in the CENP-N depeleted cells (Figure 4b, see below). CENP-C centromere localization was also reduced in CENP-N depleted cells but not to the extent that other CCAN proteins were (Figure 4b). Thus, the localization dependence of CCAN proteins on CENP-N function was similar between CENP-N-depleted cells and the stable cell lines expressing CENP-N mutants that have defects in CENP-A nucleosome binding.
Figure 4. Depletion of CENP-N affects centromere assembly.
a) CENP-N depletion leads to a reduction in CENP-A levels. HeLa cells were treated with the indicated siRNA for 56 hours and the abundance of the indicated centromere protein was determined by western blotting. Arrow (top panel) indicates the position of CENP-N. A background band (*) was present in the anti-CENP-N western blot. CENP-K is included as a loading control. b) CENP-N depletion causes centromere assembly defects. HeLa cells were treated as in (a), except that the centromere fluorescence intensity of the indicated protein was measured. Error bars represent SEM for three independent experiments including >150 centromeres from >10 cells for each experiment. c) Reduced CENP-A assembly after CENP-N RNAi. CENP-A-SNAP cells were synchronized, transfected with siRNA’s against CENP-N or GAPDH and assayed for CENP-A loading by specifically labeling nascent CENP-A-SNAP using TMR-Star as outlined in Figure S5c. Representative images are shown (left panel). HA labels steady state pool of CENP-A-SNAP and is used as centromere marker. Scale bar is 5 µm. Fluorescence intensity of 200 centromeres from 20 cells was quantified in 3 experiments and normalized to the GAPDH siRNA control. Error bars represent SEM. d) A model depicting the multiple roles of CENP-N in CENP-A nucleosome recognition, centromere assembly through the recruitment of CCAN proteins via the CENP-N carboxy-terminal region, and propagation of centromeric chromatin through the CCAN dependent regulation of CENP-A nucleosome assembly.
Depletion of CENP-N caused a reduction in total CENP-A protein (Figure 4a) and CENP-A levels at centromeres (Figure 4b, S5b). CENP-N is therefore required for the maintenance of centromeric chromatin. Similar defects have been described in S. pombe cells with mutations in the CENP-N ortholog Mis15, suggesting that CENP-N function is evolutionarily conserved16. CENP-H, CENP-I and CENP-K are required for the deposition of newly synthesized CENP-A at centromeres4. We therefore asked whether the reduction in CENP-A levels within centromeric chromatin in CENP-N depleted cells was due to a failure to load new CENP-A at centromeres. To address this we employed the pulse labeling method based on the SNAP-tag to determine the fate of newly synthesized protein. Using this strategy, the assembly of nascent CENP-A was previously shown to be restricted to early G1 phase 17. siRNA mediated reduction of CENP-N levels resulted in a significant reduction of newly synthesized CENP-A-SNAP recruitment to centromeres (Figure 4c, S5c) indicating that the loss of steady state levels of centromeric CENP-A is caused, at least in part, by a defect in CENP-A assembly.
We have identified CENP-N as the first protein to bind specifically to CENP-A nucleosomes and shown that the direct binding of CENP-A nucleosomes by CENP-N is required for centromere assembly. The DNA sequence-independent binding of CENP-A nucleosomes by CENP-N suggests that CENP-N recognizes the epigenetic mark in chromatin that specifies centromere identity. Importantly, while CENP-N targets directly to CENP-A nucleosomes, we find that CENP-N itself is required for recruiting new CENP-A to the centromere, suggesting that CENP-N is part of a feedback loop responsible for propagating centromeric chromatin in dividing cells (Figure 4d).
CENP-N mutants with a two-fold or six-fold reduction in apparent binding affinity for CENP-A nucleosomes localized equally efficiently to centromeres in vivo, suggesting that CCAN proteins other than CENP-N may also provide direct interactions with chromatin that are important for centromere assembly. The interdependence of several CCAN proteins, including CENP-N and CENP-T, for centromere localization is consistent with this possibility5. A complex of CENP-T and CENP-W was recently shown to bind directly to DNA in vitro and the CENP-T/W complex associates with histone H3-containing nucleosomes18. CENP-N and the CENP-T/W complex are therefore likely to cooperate in providing multiple distinct chromatin contacts that are required for the localization of a subset of other CCAN proteins, including CENP-H, CENP-I and CENP-K, to centromeres.
CENP-C assembly at centromeres is less sensitive to CENP-N depletion than the other CCAN proteins we examined, consistent with previous results suggesting that CENP-I and CENP-C are independently recruited to centromeric chromatin in human cells19. Nevertheless, CENP-A is required for both CENP-N and CENP-C centromere localization indicating that multiple centromere localization pathways downstream of CENP-A exist5, 20. CENP-C has previously been shown to bind directly to DNA in vitro but it is unclear how such an activity could translate into the centromere-specific localization observed for CENP-C in vivo 21, 22. Identifying the molecular mechanisms by which CENP-C is recruited to CENP-A chromatin in the absence of CENP-N and understanding how these distinct centromere recognition pathways are integrated at the level of chromatin will provide important insights into centromere assembly and structure.
Material and Methods
cDNA isolation and expression
cDNA’s encoding CENP-C, N, K, T, and I were amplified from HeLa cell mRNA with gene-specific oligonucleotides using the Super-Script III kit (Invitrogen, Carlsbad, CA) and cloned by blunt end ligation into pCR4 Blunt-TOPO using the Zero-Blunt cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturers instructions. cDNA’s encoding CENP-B, CENP-H, CENP-L, CENPM, CENP-O, CENP-P, CENP-Q, CENP-R, CENP-S, Mis18α, Mis18β and Mis18BP1/KNL2 were produced by gene synthesis (DNA 2.0, Palo Alto, CA). All cDNA’s were sequenced and subcloned into the AscI and PacI sites of a modified pCS2+-based plasmid for coupled transcription and translation in reticulocyte lysates (Promega, Madison, WI). 34S-methionine (Amersham, Uppsala, Sweden) was added to specifically label each protein. The abundance of the centromere proteins was estimated by western blotting.
Histone expression
Histone H2A, H2B, H3 and H4 were expressed, purified, and refolded as described23, except that soluble H3/H4 tetramer and H2A/H2B dimer, rather than histone octamer, were assembled and frozen on liquid nitrogen. CENP-A was expressed as a soluble tetramer with histone H4 essentially as described11. Briefly, 6 liters of BL21 bacterial cells transformed with a bi-cistronic vector encoding human CENP-A and Xenopus laevis histone H4 were grown to OD600 0.2 at 37°C. The culture was then switched to 23°C and grown until reaching OD600 0.6, at which point IPTG was added to 0.3 mM to induce protein expression for 6 hrs. The cells were spun down and frozen directly on liquid nitrogen. Cell pellet was resuspended in 100 ml of lysis buffer (10 mM KPO4, 0.9 M NaCl, 10 mM _beta_-mercaptoethanol (BME) and 1 mM PMSF, pH 6.8), sonicated, and centrifuged at 100,000xg to clarify the extract. The high-speed supernatant was applied to a hydroxyapatite column, which was washed with 5 column volumes of lysis buffer and eluted with a 0.9 M to 3.5 M NaCl gradient in 10 mM KPO4, pH 6.8. Soluble CENP-A/H4 typically elutes in a broad peak between 1.5 and 3.5 M NaCl. Fractions containing CENP-A/H4 tetramers were pooled and dialyzed 2X against SP-loading buffer (10 mM Tris-HCl, 0.75 M NaCl, 10 mM BME, 0.5 mM EDTA, pH 7.4). The dialysate was loaded onto a 1 ml Hi-Trap SP column (Amershan, Uppsala, Sweden) and eluted with a 0.75 to 2M NaCl gradient in 10 mM Tris-HCl, 0.5 mM EDTA, pH 7.4. Fractions containing CENP-A/H4 were pooled, concentrated, and applied to a sephadex 200 gel-filtration column equilibrated with 10mM Tris-HCl, 2M NaCl, 0.5mM EDTA, pH 7.4. Peak fractions were pooled, concentrated, and frozen down for nucleosome reconstitution. ~5 mgs of CENP-A/H4 tetramer can be expected from 6 liters of cells.
Nucleosome reconstitution
186-bp DNA containing the centromere repeat or the 601 nucleosome-positioning sequence were generated by polymerase chain reaction (PCR). CENP-A/H4 tetramer, H2A/H2B dimer and DNA were mixed at a stoichiometry of 1.1:2.2:1 in high-salt buffer (10 mM Tris-HCl, 2M NaCl, 0.5 mM EDTA, pH7.4) and the salt concentration was slowly lowered to 2.5 mM NaCl over ~60 hours by gradient dialysis. Mononucleosomes were then purified on a 5 ml 5–30% glycerol gradient as described23. Fractions containing pure mononucleosomes were concentrated and stored at 4°C.
Gel shift assays
Centromere proteins were run out alone or in the presence of the indicated nucleosome on 5% acrylamide gels in 0.5x TBE for 70 minutes at 10 mAmp. Typical binding reactions were carried out for 30 minutes at room temperature in 10 µl and contained the indicated concentration of nucleosome and 0.5–1 µl of ivt mix in 10 mM Tris-HCl, 20% glycerol, pH 7.4. Binding reactions were loaded directly on the gel. After electrophoresis, gels were first stained with ethidium bromide or SYBR-gold (Invitrogen, Carlsbad, CA) to visualize and/or quantify the nucleosomes followed by staining with coomassie blue to visualize proteins. Gels were then dried and the 35S-methionine labeled centromere proteins were visualized and quantified with a phosphorimager.
CENP-N mutagenesis and stable cell lines
Point mutants in CENP-N were made with a site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) as described by the manufacturer. Stable cell lines were created by co-transfecting HEK293 flp-in target cells (Invitrogen, Carlsbad, CA) with pOG44 and a modified version of pcDNA5/FRT engineered to express CENP-N with an N-terminal GFP-tag according to the manufacturers instructions. For western blotting and co-immunoprecipitation experiments, nuclei were isolated from ~5×107 cells and chromatin was solubilized with micrococcal nuclease as described5, 8. After micrococcal nuclease treatment, extracts were centrifuged at 14K for 10 minutes in a microfuge and the concentration of each supernatant was determined using Bradford reagent (Biorad, Hercules, CA). 10 µg of supernatant was loaded on a 12.5% poly-acylamide gel to determine the levels of the centromere proteins present (panel 2c) and the remaining extract (~2 mg) was used in anti-GFP immunoprecipitations. For CENP-A immunofluorescence, cells were fixed with methanol at −20°C for 2 minutes without prior extraction and processed using standard techniques. For staining CCAN proteins, HEK293 cells were pre-extracted with 0.1% Triton X-100 in PBS for 1 minute and then fixed for 10 minutes in PBS + 4% formaldehyde and processed using standard techniques. Similar methods were used for staining CCAN proteins in HeLa cells except that cells were pre-extracted for 3 minutes in 0.3% Triton X-100 before fixation.
siRNA
CENP-N (GUAAAUUUCCGACAGAGAAUU, CUACCUACGUGGUGUACUAUU, GAUAUUACCGAAAUGAAGAUU, CCAGAAAGUUUGGGAUGUUUU) and CENP-A (5’-AACACAGUCGGCGGAGACAAG) siRNAs were obtained from Dharmacon (Lafayette, CO) and used according to the manufacturer’s instructions. Buffer alone or GAPDH siRNA was used in control experiments. Cells were processed for immunofluorescence as described above. For western blots, cell extracts were prepared by resuspending cells in 20 mM HEPES, 0.5 M NaCl, 1% Triton X-100, 1 mM DTT, 10 mM EDTA, 10 mM EGTA, 1 mM PMSF followed by centrifugation for 10 minutes at 14,000 rpm in a microfuge. Protein concentration in each extract was determined using Bradford reagent (Biorad, Hercules, CA) and identical amounts of total protein were loaded in each well of the gel.
SNAP pulse labeling
HeLa cells stably expressing near endogenous levels of CENP-A-SNAP- 3xHA17 were treated with 2mM thymidine for 17 hours to arrest cells in S phase. Following release in 24 µM deoxycytidine for 3 hours cells were transfected with CENP-N and GAPDH siRNA pools (Dharmacon (Lafayette, CO)) according to manufacturer’s instructions. 9 hours after release from thymidine fresh thymidine was added to synchronize cells at the next G1/S boundary. SNAP-tag was quenched with non-fluorescent BTP (Covalys, Witterswil, Switzerland) after which cells were released into S phase. Newly synthesized CENP-A-SNAP is labeled 7.5 hours after release (in G2 phase) with TMR-Star (Covalys, Witterswil, Switzerland) and cells are allowed to proceed through the cell cycle for CENP-A assembly and are collected at the next G1/S boundary by thymidine addition. Cells were fixed (without pre extraction) and processed for microscopy (see Figure S5c for schematic).
Microscopy
Stacks of fixed cell immunofluorescence images encompassing the entire cell (or cells) were captured at 0.2 µm axial steps using a motorized stage mounted on an Olympus IX70 microsope. Immuno fluorescent images were acquired using a 60X 1.4NA PlanApo objective and a CoolSnap-HQ CCD camera (Photometrics, Tucson, AZ) on a DeltaVision Spectris system (Applied Precision, Seattle, WA). TMR-Star labeled CENP-A-SNAP images were acquired using a 100X 1.4NA UPlanSApo objective and a Cascade2 EMCCD camera (Photometrics, Tucson, AZ). Centromere fluorescence intensity in each channel was quantified as described except that intensity measurements were performed on non-deconvolved maximal intensity projections of each z-series24. Images presented are maximum intensity projections of deconvolved images.
Supplementary Material
1
2
3
4
5
6
7
Acknowledgements
The authors would like to thank members of the Straight Lab for helpful comments and support, Jeremy Minshull and DNA2.0 (Palo Alto, CA) for gene synthesis, Silva H. Hanissian for the CENP-U(50) cDNA, Janet Yang, Geeta Narlikar, Mike Resch, Karolin Luger and Jeff Hansen for reagents and help with nucleosome reconstitution, Kazuo Todokoro for CENP-H antibody, Song-Tao Liu for CENP-H and CENP-I antibodies and Dan Herschlag for advice. CWC was generously supported by a postdoctoral fellowship from the Helen Hay Whitney Foundation. KG was supported by a predoctoral fellowship from the National Science Foundation and by NIH grant T32GM007276. AFS is the Gordon Family Scholar supported by the Damon Runyon Cancer Research Foundation, and this work was supported by National Institutes of Health grant R01GM074728. MCCS is supported by the Fundação para a Ciência e a Tecnologia (FCT) (SFRH/BD33219/2007). LETJ is supported by FCT, Fundação Calouste Gulbenkian and the EU Seventh Framework Programme (FP7).
References
- 1.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 2.Choo KH. Domain Organization at the Centromere and Neocentromere. Dev Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 3.Carroll CW, Straight AF. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 5.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 6.Izuta H, et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 7.Saitoh H, et al. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 8.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita Y, et al. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 12.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClelland SE, et al. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. Embo J. 2007;26:5033–5047. doi: 10.1038/sj.emboj.7601927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K Complex Coordinately Direct Kinetochore Assembly in Vertebrates. Mol Biol Cell. 2007 doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hori T, et al. CCAN Makes Multiple Contacts with Centromeric DNA to Provide Distinct Pathways to the Outer Kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang CH, Tomkiel J, Saitoh H, Johnson DH, Earnshaw WC. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trazzi S, et al. In vivo functional dissection of human inner kinetochore protein CENP-C. J Structural Biology. 2002;140:39–48. doi: 10.1016/s1047-8477(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 23.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7