Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs (original) (raw)
Abstract
Fragile X mental retardation is caused by loss-of-function of a single gene encoding FMRP, an RNA-binding protein that harbors three canonical RNA-binding domains, two KH-type and one RGG box. Two autosomal paralogs of FMRP, FXR1P and FXR2P, are similar to FMRP in their overall structure, including the presence of putative RNA-binding domains, but to what extent they provide functional redundancy with FMRP is unclear. Although FMRP has been characterized as a polyribosome-associated regulator of translation, less is known about the functions of FXR1P and FXR2P. For example, FMRP binds intramolecular G-quadruplex and kissing complex RNA (kcRNA) ligands via the RGG box and KH2 domain, respectively, although the RNA ligands of FXR1P and FXR2P are unknown. Here we demonstrate that FXR1P and FXR2P KH2 domains bind kcRNA ligands with the same affinity as the FMRP KH2 domain although other KH domains do not. RNA ligand recognition by this family is highly conserved, as the KH2 domain of the single Drosophila ortholog, dFMRP, also binds kcRNA. kcRNA was able to displace FXR1P and FXR2P from polyribosomes as it does for FMRP, and this displacement was FMRP-independent. This suggests that all three family members recognize the same binding site on RNA mediating their polyribosome association, and that they may be functionally redundant with regard to this aspect of translational control. In contrast, FMRP is unique in its ability to recognize G-quadruplexes, suggesting the FMRP RGG domain may play a non-redundant role in the pathophysiology of the disease.
INTRODUCTION
Gene duplication during evolution has given rise to both increased functionality through diversification of homologous genes and increased potential for rescuing the effects of deleterious gene mutation through conservation of cellular function. Although the impact of redundancy of function between paralogous genes is difficult to assess in human disease, studies of loss-of-function in mouse models suggest that many human diseases may be ameliorated to some extent by the existence of functional paralogs. Understanding the potential for functional overlap within a disease caused by loss-of-function of a single family member may uncover specific functions of the affected protein, as well as increase the potential for therapeutic intervention.
Fragile X syndrome, the leading cause of inherited mental retardation and a common genetic cause of autism, is caused by loss-of-function of the FMRP RNA-binding protein (reviewed in 1). This most frequently results from CGG repeat expansion in the 5′-UTR of the FMR1 gene, leading to abnormal methylation, cessation of transcription and complete loss-of-function. FMRP has three canonical RNA-binding domains, two of the KH type and an RGG box (2–4). Interestingly, one patient has been described with a CGG repeat copy number in the normal range but with a single-point mutation in the second KH-type RNA-binding domain (KH2) (5). This isoleucine-to-asparagine mutation (I304N) lies within the hydrophobic platform of the RNA-binding pocket of all KH domains studied to date (6,7) and is predicted to disrupt sequence-specific RNA binding by this domain (8), suggesting that the RNA-binding properties of FMRP are central to its cellular function and role in disease pathogenesis.
FMRP has two autosomal paralogs, FXR1P and FXR2P (9,10), which likely arose from gene duplication of a common ancestral gene (11) and have been identified in all mammals studied as well as in zebrafish. Though yeast and Caenorhabditis elegans lack FXR proteins, a single FXR family member, dfmr1, exists in Drosophila (12). At the sequence level, FMRP, FXR1P and FXR2P are highly homologous through the first 13 exons (of FMRP) and diverge significantly thereafter (11). The presence of conserved domains including a nuclear localization signal, two KH domains and a nuclear export signal suggests that all three FXR proteins may share some cellular functions. In support of this, all three have been shown to bind RNA (3,4,9,13,14), to associate with free ribosomes (15–18) and polyribosomes (14,17,19–22). Treatment of transfected cells with leptomycin B to block exportin1-dependent nuclear export resulted in the nuclear accumulation of all three FXRPs (23), suggesting that they use the same mechanism for nucleocytoplasmic shuttling though they have different distributions between the nucleus and nucleolus. All three homo- and heterodimerize through a conserved domain encoded by their respective seventh exons (18), though evidence suggests that homodimerization predominates in vivo (24).
The presence of divergent sequences also implies the potential for specialized functions, including two exons (exons 11 and 12) present in the KH2 domain of FMRP that are not present in FXR1P and FXR2P (11,25), nor in dFMRP (12). These exons were likely acquired during the mammalian radiation as they are absent from chicken (26) and Xenopus FMRP (27). Exon 12 is alternatively spliced, whereas exon 11 is constitutively included (28,29). The C-termini following the nuclear export signal have diverged considerably, including the acquisition of two nucleolar localization signals, NoS1 and NoS2, in FXR1P/2P, that are lacking in FMRP (30).
The tissue distribution of the three FXR proteins, as well as their subcellular localization, also suggests both conserved and divergent functions. They have largely overlapping expression in human tissues, especially in the nervous system (31), though some marked differences exist outside of the nervous system (32–34). In neurons, at a subcellular level, all three proteins were found to be mainly cytoplasmic with minor staining in the nucleus and nucleolus (33,35), and within the cytoplasm, all three could be found in association with ribosomes by immuno-EM, again hinting at similar function (33). Outside the nervous system, FXR1P is highly expressed in muscle cells with little or no expression of FMRP (19,33,34,36,37). FXR1P has at least four alternatively spliced isoforms of 70, 78, 81 and 84 kDa, the latter two being muscle specific (19,38).
In many cases, the levels of functionally redundant paralogs increase to compensate for the loss of one. This does not appear to be the case for the FXR family. In the absence of FMRP, the levels of FXR1P and FXR2P are unchanged in the null mouse model (33,39), in affected human fetuses (31) or in lymphoblastoid cells from Fragile X patients (19,22). In the FXR2 null mouse, levels of FMRP and FXR1P are unchanged in the somatodendritic compartment (39). Moreover, in a mouse model of the human I304N mutation, the levels of FXR1P and FXR2P are not altered in the brain (Julie B. Zang, Elena D. Nosyreva, Corinne M. Spencer, Lenora J. Volk, Kiran Musunuru, Ru Zhong, Elizabeth F. Stone, Lisa A. Yuva-Paylor, Kimberly M. Huber, Richard Paylor, Jennifer C. Darnell and Robert B. Darnell, submitted for publication). Taken together, there is no evidence for compensatory increases in homolog levels that would suggest functional redundancy, despite co-expression of all three in neurons.
In order to determine whether the three Fragile X family members may be functionally redundant with respect to RNA binding and polysome-associated functions, we have compared their RNA-binding specificities and find that the RNA-binding properties of FMRP, FXR1P and FXR2P KH2 domains are very similar. Moreover, this specificity has been conserved through evolution, as dFMRP-KH2 binds an _in vitro_-selected RNA ligand of the human KH2 domain (40) (kissing complex RNA, or kcRNA) with the same affinity as the mammalian KH2 domains. kcRNA binding is specific to the FMRP family of proteins, as KH domains from other related proteins fail to bind this RNA ligand. FMRP, FXR1P or FXR2P can all be specifically displaced from polyribosomes by kcRNA, suggesting that all three family members may be functionally redundant with regard to the regulation of translation of specific mRNAs on polysomes. In contrast, we find that only FMRP recognizes an _in vitro_-selected ligand of the human FMRP RGG box (41) (G-quadruplex RNA) with high affinity. Understanding the role of RGG box RNA binding in FMRP function may therefore lead to greater understanding of the pathogenesis of Fragile X syndrome, as FXR1P and FXR2P may compensate for other functions of FMRP mediated by the KH domains.
RESULTS
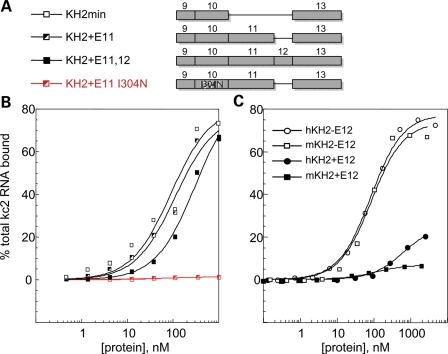
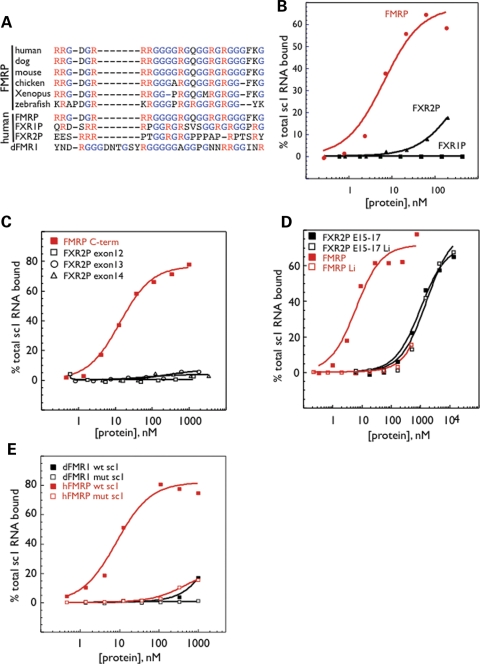
KH-type RNA-binding domains are frequently present in two or more copies in proteins that mediate splicing, translational control or mRNA transport. In some cases, the avidity of proteins for RNA targets may be achieved by the presence of multiple KH domains of similar specificity that binds to repeats or clusters of similar RNA motifs (reviewed in 7). FMRP appears to use a different mechanism, as the two KH domains of FMRP are among the most divergent from each other in sequence, and a crystal structure of the tandem domains suggests that they may function independently (42). The structures of KH domains are typified by a characteristic array of beta sheets and alpha helices (S1-H1-H2-S2-S3-H3) and two loops, the GXXG invariant loop between H1 and H2 and a variable loop between S2 and S3. NMR solution structures and KH domain-nucleic acid co-crystals have revealed that KH domains pinion nucleic acids between the conserved GXXG loop and the nonconserved variable loop allowing specific recognition of 4–5 nucleotides (8). FMRP KH2 harbors the longest variable loop of any KH domain yet described (43,44), encoded by exons 11 (E11) and 12 (E12) of the FMR1 gene. Since these two exons are present in mammalian FMR1 mRNA, but are lacking in chicken, Xenopus and zebrafish, the FXR1 and FXR2 family members and dfmr1, we examined whether they participate in KH2:RNA ligand recognition. Previous studies used in vitro RNA selection to identify a high-affinity RNA ligand for the KH2 domain of human FMRP which folds into a loop–loop pseudoknot or kissing complex motif (referred to here as kcRNA; kc2 RNA is one of many individual clones fitting this consensus) (40). Human FMRP KH2 domains were expressed as T7- and His-tagged fusion proteins, with or without the 45 amino acids from exon 11 and the 21 amino acids from exon 12 (Fig. 1A), and kcRNA binding was assessed by filter binding assay (Fig. 1B). A KH2 construct lacking both exons 11 and 12 (KH2-min, open squares) binds similarly (_K_d = 90 nm) to KH2 constructs lacking the alternatively spliced exon 12 (KH2 + E11, _K_d = 110 nm) and better than the full-length KH2 domain (KH2 + E11,12, _K_d = 310 nm). Thus, the variable loop of KH2 is dispensable for kcRNA binding.
Figure 1.
The KH2 domain variable loop encoded by exons 11 and 12 (E11 and E12), absent from FXR1P and FXR2P, does not contribute to kcRNA binding. (A) Diagram depicting the sequences present in the recombinant human KH2 domains that were produced in bacteria and purified. Domains differed in the presence or absence of exons 11 and 12 and in the presence of the I304N mutation in the ‘parent’ construct as a negative control for kcRNA binding. (B) Filter binding assays were performed to determine the affinity of the indicated domains in (A) for binding to the kc2 RNA. Domains were tested for RNA binding immediately after purification. (C) Mouse (m, squares) and human (h, circles) KH2 domains with and without the alternatively spliced exon 12 (filled symbols, +E12; open symbols, -E12) were assayed for binding to kcRNA by filter binding assay. Although kcRNA binding is conserved between mouse and human, the presence of exon 12 sequences inhibits binding. In this experiment, domains were tested for RNA binding following storage at 4°C (see text for further explanation of the observed differences in binding for the constructs containing E12 in B and C).
Approximately 80% of mature FMR1 transcripts in the brain lack exon 12 due to alternative splicing (29). In that context, it is interesting that KH2+E11,12 binds kcRNA with lower affinity than the form with that exon spliced out (KH2 + E11). We have repeated this experiment several times and find that the KH2 domains from mouse or human FMRP containing the 21 amino acids of exon 12 bind kcRNA if assayed immediately after purification (as in Fig. 1B). If stored at 4°C for more than a day, binding rapidly decreases (to levels shown in Fig. 1C), although exon 12(−) forms retain the same binding affinity for up to 6 months under the same storage conditions. These results suggest that the presence of the 21 amino acids encoded by exon 12 may destabilize the KH2 domain in vitro.
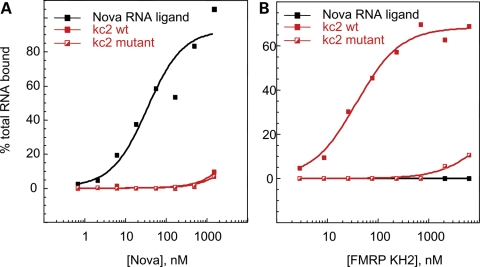
Without its lengthy variable loop, the FMRP KH2 domain is very similar to its paralogs FXR1P and FXR2P (Fig. 2) (42,44). The first KH:RNA co-crystal structure, that of a Nova KH domain bound to a high-affinity _in vitro_-selected RNA ligand, revealed a novel method of single-stranded RNA binding for KH domains. The RNA ligand lies on a hydrophobic α/β platform, where it is gripped by the conserved GXXG loop and the variable loop (8). Among the FXRP KH2 domains, the amino acids involved in forming this aliphatic α/β platform are largely conserved (Fig. 2, asterisks). Nova recognizes its RNA target through a combination of van der Waals contacts, hydrogen bonds and stacking interactions, and several of these important contacts between KH domain amino acid side chains and the RNA ligand are absolutely conserved between Nova-1 and Nova-2 (Fig. 2, red dots). This is likely to explain how these related domains recognize almost exactly the same RNA ligands (8,45) and suggests the possibility that FMRP might bind the same ligands as the Nova proteins. Filter binding assays were performed comparing human full-length Nova protein and FMRP KH2 domain (Fig. 3). Although Nova-1 bound with an affinity of 37 nm to one of its in vivo targets, the GABA-Aγ2 intron 9-binding site (46), it failed to bind kc2 RNA or mutant kc2 RNA with high affinity (Fig. 3A). In contrast, FMRP KH2 bound kc2 RNA with an affinity of 38 nm but failed to bind the GABA-Aγ2 RNA ligand or a single-point mutation in kc2 (C50G) that disrupts one of the loop–loop interactions in the pseudoknot (40) (Fig. 3B) demonstrating specificity in the binding of these KH domains to their respective RNA ligands. We also tested FMRP KH1 and found that it fails to bind kcRNA (data not shown). In addition, filter binding assays done with WT full-length hFMRP or hFMRP containing point mutations which disrupt either the KH1 or KH2 domains (I241N and I304N) demonstrate that the I304N mutation in KH2 disrupted kcRNA binding, while the I241N mutation in KH1 did not affect kcRNA binding by full-length FMRP (40).
Figure 2.
KH2 domains from human (h), mouse (m) and Drosophila (d) FXR family members are conserved in key positions. Signature alpha helices (red boxes), beta sheets (blue boxes) and loops (green boxes) are shown. The asterisks denote hydrophobic amino acids whose side chains make up the aliphatic α/β platform of the domain. Red dots denote amino acids important for interactions of human Nova-2 KH3 with its RNA ligand. An open triangle indicates the isoleucine mutated to asparagine in the I304N patient, and the dashed gray line denotes the exon–exon junction between exons 10 and 13 of FMRP, with exons 11 and 12 removed, or exons 10 and 11 of FXR1P and FXR2P. In the dFMRP sequence, number 3 represents nonaligned amino acids ‘AIA’, and number 2, ‘NI’. RNA contacts from the Nova2 KH3:RNA co-crystal structures are shown below the sequences (8,44).
Figure 3.
The KH domains of Nova-1 and FMRP bind different RNA ligands. Human Nova-1 protein was expressed and HisTag-purified from a bacterial lysate. Filter binding assays were performed using Nova-1 and the human FMRP KH2 domain at the indicated concentrations with one of Nova's in vivo targets, the GABA-Aγ2 intron 9-binding site (46) (black squares, Nova ligand), or the 96 nucleotide _in vitro-_selected RNAs, kc2 (filled red squares, kc2wt) or the C50G point mutant in kc2 (half-filled red squares, kc2 mutant). (A) Nova bound its cognate ligand with an affinity of 37.1 nm (±16.0 nm). Binding to kc2wt or kc2 mutant was too low to determine a _K_d value. (B) FMRP KH2 bound its kcRNA ligand with _K_d = 37.8 ± 5.6 nm. Binding to kc2 mutant or the Nova ligand was not quantifiable.
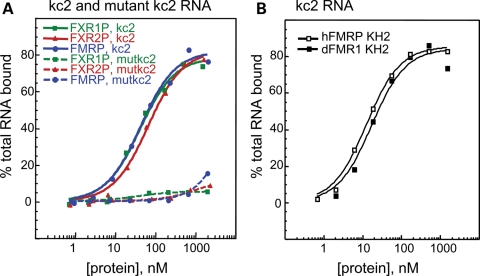
The high conservation between FMRP and FXR1/2P KH domains also suggested the possibility that FXR1P and FXR2P might bind the same RNA ligands as does FMRP. Filter binding assays with kc2 RNA demonstrated that the KH2 domains of human FXR1P and FXR2P bind kcRNA indistinguishably from FMRP KH2 (Fig. 4A). Specific interactions with this motif are indicated by the failure of all three to bind the kc2 RNA with a single-point mutation. The KH2 domain of the Drosophila homolog, dFMRP, also binds to kcRNA with the same affinity as human FMRP KH2 (Fig. 4B). Although the observation that the RNA-binding properties of the KH2 domain of the Fragile X paralogs have been conserved during evolution is important, these data additionally suggest that the use of _dfmr1_-null Drosophila as a disease model for Fragile X syndrome is likely to be fruitful, as RNA binding by this family may have been largely conserved through evolution.
Figure 4.
KH2 domains from human FMRP, FXR1P, FXR2P and dFMRP bind kcRNA specifically and indistinguishably. (A) FMRP KH2 (blue circles), FXR1P KH2 (green squares) or FXR2P KH2 (red triangles) were produced in bacteria and purified. Equilibrium filter binding assays with 32P-labeled kcRNA were used to determine the affinity of the KH2 domains for kc2 RNA ligand (_K_d value for FMRP = 43 nm, FXR1P = 42 nm and FXR2P = 60 nm, solid lines). Assays were repeated with the C50G mutant kcRNA (dashed lines) indicating the specificity of the FXRP interaction with this RNA. (B) The KH2 domains from human (open squares) and Drosophila (closed squares) were expressed and purified side-by-side and kcRNA binding assessed by filter binding assay as in (A).
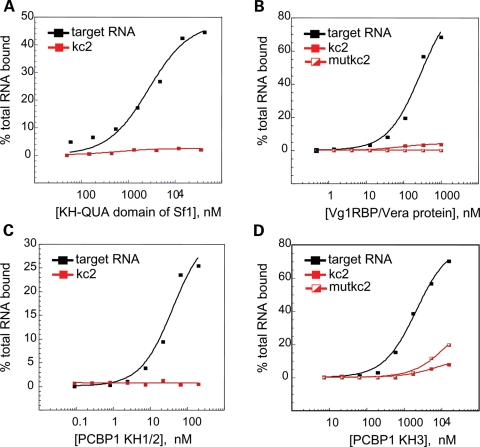
KH domains can be identified in at least 300 eukaryotic proteins of varying cellular functions. To more generally assess the ability of KH domains to recognize the kcRNA motif, we generated fusion protein constructs for KH domains of human poly-C-binding protein 1 (PCBP1, αCP1 or hnRNPE1) which is involved in translational control (47 and references therein), the extended KH-quaking domain of human Sf1 which binds the branch point motif (BPS) during splicing (48) and the full-length Xenopus Vg1RBP/Vera protein (four KH domains) which binds Vg1 mRNA and localizes it to the vegetal pole of the frog oocyte (49). Each KH domain was assayed for kcRNA binding by filter binding assay (Fig. 5). We found that none of these KH domains bound their reported RNA ligands in SBB buffer (data not shown), which contains 200 mm salt and so is a moderately stringent buffer for protein–RNA association. Therefore, we assayed the KH-QUA2 domain of Sf1 (Fig. 5A), all four KH domains of Xenopus Vg1RBP/Vera (Fig. 5B), the paired KH1 and two domains of mouse PCBP1 (Fig. 5C) and the third KH domain of mouse PCBP1 (Fig. 5D) for binding to kcRNA in the buffers used in their respective published studies (47,50–52). Under these conditions, each KH domain bound to their reported ligands but none recognized the kcRNA ligand bound by the FXRP family KH2 domain. Taken together, these results demonstrate that FMRP, FXR1P and FXR2P KH2 domains bind the same kcRNA motif, this binding has been conserved through evolution from fly to man and kcRNA binding is specific to the FMRP family, as all other KH domains tested fail to bind it.
Figure 5.
The KH domains of Sf1, Vg1RBP/Vera or PCBP1 bind their previously identified RNA ligands, but do not bind with significant affinity to kcRNA or mutant kcRNA. (A) The isolated KH-QUA domain of human Sf1 bound the intronic branch point sequence with _K_d = 2.83 ± 0.71 µm (black squares). (B) All four KH domains of Xenopus Vera bound to the Vg1 RNA (black squares) with _K_d = 292 ± 80 nm. (C) Binding of the mouse PCBP1 tandem KH1 and KH2 domains to the R7a1 ligand (black squares) had a _K_d value of 42.3 ± 12 nm. (D) Binding of the KH3 domain of PCBP1 to the R7a1 ligand (black squares) had a _K_d value of 2.17 ± 0.25 µm. In all cases, binding to kc2 RNA (red squares) or C50G mutant kc2RNA (half-filled red squares) was too low to accurately determine a _K_d value.
Redundant role for FXRP:kcRNA binding in neurons
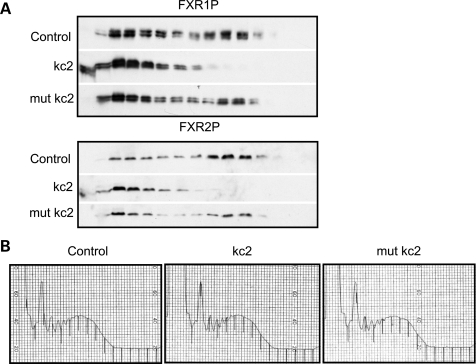
We have previously demonstrated that kcRNA added to a mouse brain lysate can compete FMRP off brain polyribosomes, presumably by mimicking the in vivo RNA ligand through which FMRP associates with polysomes (40). As all three FXRP family members bind the same ligand in vitro, we hypothesized that kcRNA might compete FXR1P and FXR2P off polysomes if all three also bind the same RNA ligand in vivo. We assayed the ability of kcRNA to shift FXR1P and FXR2P off polyribosomes in IMR32 human neuroblastoma cell lysate and found that they were effectively shifted to the lighter mRNP-containing sucrose fractions by kcRNA but not by G-quadruplex RNA (sc1) (41) or mutant kcRNA (data not shown). It is possible, however, that FXR1P and FXR2P are not directly binding RNA but are associated with polysomes due to protein–protein interactions with FMRP, since they have been reported to heterodimerize with FMRP (10,24). To assess the ability of kcRNA to compete FXR1/2P off polysomes in the absence of FMRP, we repeated the assay in cortical lysates generated from Fmr1 null mice (Fig. 6). A significant proportion of FXR1P and FXR2P were associated with heavy polysomes in these lysates, and kcRNA (kc2) significantly shifted both FXRPs to lighter fractions corresponding to small mRNPs or free protein. A single-point mutation in kcRNA (mut kc2) abolished this effect in both cases. Importantly, the global polysome profiles as assessed by A254 trace were not changed by the addition of 500 nm kcRNA or point mutant kcRNA (Fig. 6B). These data demonstrate that the association of both FXR1P and FXR2P with polysomes is independent of FMRP, and that they associate with polysomes using the same mechanism used by FMRP—binding an RNA ligand that can be competed by a loop–loop pseudoknot motif RNA. These data suggest that there is likely to be functional redundancy in translational control by the FXR family members in the brain.
Figure 6.
FMRP, FXR1P and FXR2P use the same mechanism to associate with polysomes. (A) Postmitochondrial supernatants prepared from cortex and cerebellum of Fmr1 null mice were treated with buffer (control), 500 nm kc2 (kc2) or 500 nm mutant kc2 (mut kc2) containing a point mutation that disrupts the loop–loop interaction in the kissing motif. Lysates were then fractionated over linear 20–50% sucrose gradients, proteins recovered from each fraction by TCA precipitation and resolved by SDS–PAGE. FXR1P or FXR2P was visualized by western blot with ML13 or 1G2 antibodies respectively. (B) kcRNA treatment has no effect on overall ribosome distribution on transcripts as evidenced by A254 traces.
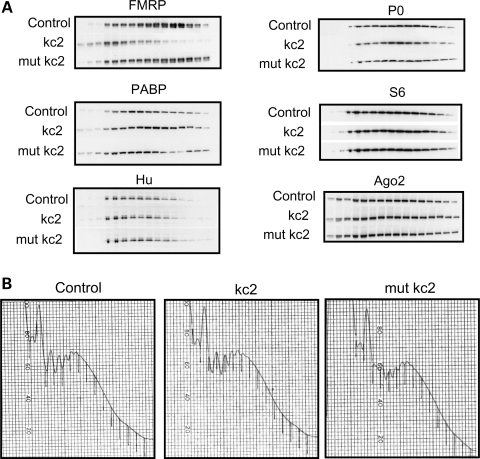
We assessed whether other polysome-associated proteins, many of which may have a role in translational regulation, can also be competed off polysomes by kcRNA. The distribution of ribosomal proteins S6 and P0, the mRNA-binding proteins PABP, Hu B, C, D and Ago2 was not significantly altered after incubation with kcRNA (Fig. 7A), underscoring the specificity of the binding of kcRNA to the FXR family of translation factors. Again, the overall polysome profile was not affected by kcRNA or mutant kcRNA addition (Fig. 7B). Eukaryotic elongation factor eEF1a and hnRNPA1 also showed no change in polysome association after kcRNA or mutant kcRNA treatment (data not shown).
Figure 7.
Several polysome-associated proteins are not competed off polysomes by kcRNA treatment. (A) Postmitochondrial supernatants prepared from the cortex of WT P21 mice were treated with buffer (control), with 500 nm kc2 (kc2), or 500 nm mutant kc2 (mut kc2). Lysates were then fractionated over linear 20–50% sucrose gradients, proteins recovered from each fraction by TCA precipitation and resolved by SDS–PAGE. Half as much lysate was precipitated for lanes 1–3 as for 4–16 to permit full solubilization of large TCA pellets. Proteins were visualized by western blot with antibodies as detailed in Materials and Methods. (B) kcRNA and mutant kcRNA treatment have no effect on the polysome profile as evidenced by A254 traces.
Lack of functional redundancy for G-quadruplex binding
Members of the FXR family have been reported to harbor a second type of RNA-binding domain, the RGG box. In vitro RNA selection experiments with the FMRP C-terminus identified a G-quadruplex RNA ligand as the protein's highest affinity RNA ligand, an interaction that was mapped to the FMRP RGG box (41). The RGG box in human FMRP that binds the G-quadruplex ligand is shown aligned with the FMRP RGG boxes from five other species, demonstrating significant conservation of the RGG domain (Fig. 8A). Alignment of human FMRP, FXR1P, FXR2P and dFMRP sequences (from 12) shows little conservation of R/G motifs in these homologs, consistent with prior observations made of the FXR homologs in zebrafish (53). The lack of conservation in the Drosophila homolog is particularly evident. To compare RGG box–G-quadruplex binding by these different proteins, we expressed the longest C-terminus present in brain isoforms (starting immediately after the KH2 domain) of each family member. Each was assayed for binding to sc1 (the ‘winning’ _in vitro_-selected G-quadruplex RNA) (Fig. 8B) (41). As expected, the FMRP C-terminus bound sc1 RNA with high affinity (_K_d = 6.7 ± 2.1 nm). However, the FXR1P C-terminus showed no detectable binding to the G-quadruplex RNA ligand. FXR2P demonstrated potential G-quadruplex RNA binding with estimated _K_d value of 306.5 ± 99 nm but RNA binding was not saturated in this pilot experiment. To further investigate this, the FXR2P C-terminus was expressed in four fragments corresponding to exons 12, 13 (containing the putative RGG box (10,11), 14 and 15–17. Compared with wt FMRP C-terminus that bound sc1 with a _K_d value of 13.8 ± 1.5 nm, FXR2P exons 12, 13 and 14 showed no binding to sc1 (Fig. 8C). Instead, the observed G-quadruplex RNA-binding activity of the FXR2P C-terminus was localized to protein encoded by exons 15–17 (Fig. 8D, black squares). To test whether this binding by FXR2P was specific for G-quadruplexes, we repeated binding assays in the presence of Li+ versus K+, as G-quadruplex structures are unstable in Li+ (41) (Fig. 8D, open symbols). Although sc1 binding by the C-terminus of FMRP decreased >150-fold in the presence of Li+, RNA binding by FXR2P exons 15–17 was unaffected by the nature of the monovalent cation, demonstrating that the observed RNA-binding activity by the C-terminus of FXR2P is not dependent on G-quadruplex formation. It is likely that exons 15–17 of FXR2P, which contain two arginine rich motifs (30,38), bind sc1 RNA nonspecifically through those positive charges.
Figure 8.
G-quadruplex binding by the RGG box is specific to FMRP. (A) Alignment of FXR family RGG box sequences from the indicated species by ClustalW analysis. (B) The C-termini of human FMRP (red circles), FXR1P (black squares) and FXR2P (black triangles) were assayed for binding to sc1 by filter binding assay. (C) Fusion proteins corresponding to sequences encoded by exons 12 (open squares), 13 (circles) or 14 (triangles) of the FXR2P C-terminus were assayed for binding to sc1 RNA compared with the full-length C-terminus of FMRP (closed red squares). (D) Filter binding assay of sc1 binding by fusion protein corresponding to exons 15–17 of FXR2P (black, _K_d = 1.05 ± 0.18 µm in K+) and the C-terminus of FMRP (red, _K_d = 6.5 ± 1.9 nm in K+) was conducted in buffer containing 200 mm K+ (closed squares) or 200 mm Li+ (open squares). (E) The C-terminus of dFMRP (black) was assayed for sc1 (closed) or mutant sc1 (open) binding compared with the C-terminus of human FMRP (red) by filter binding assay.
Finally, we assayed whether the Drosophila dFMRP C-terminus could bind sc1 or a point mutant in sc1 RNA by filter binding assay. Although human FMRP C-terminus bound sc1 RNA with a _K_d value of 8.5 ± 1.8 nm (Fig. 8E, red filled squares), and a point mutation in sc1 severely abrogated binding (red open squares), the fly C-terminus failed to bind sc1 or mutant sc1 with high affinity (Fig. 8E, black symbols). Taken together, these results demonstrate that G-quadruplex RNA binding is a specific function of FMRP that is absent in its autosomal paralogs FXR1P and FXR2P. Furthermore, our data suggest that this activity of FMRP has been acquired after the gene duplication events that generated FXR1P and FXR2P, as it is not present in the Drosophila dFMRP ancestral homolog. Understanding the functional significance of G-quadruplex binding in vivo may therefore be of substantial importance in elucidating the molecular pathogenesis of Fragile X syndrome.
DISCUSSION
In humans, the loss of FMRP activity leads to Fragile X syndrome despite the presence of FXR1P and FXR2P. Since all three proteins have a similar expression pattern in the brain, there may be critical activities of FMRP that are not redundantly provided by FXR1/2P in neurons. Recent genetic evidence in mouse models has led to uncertainty regarding the degree of functional overlap between FMRP autosomal paralogs. For example, Fmr1 and Fxr2 null mice have some behavioral phenotypes in common (54), and doubly mutant mice lacking both FMRP and FXR2P (Fmr1/Fxr2 DKO mice) show increased exploratory behavior in open-field tests, decreased prepulse inhibition and decreased freezing in a conditioned fear test relative to either Fmr1 null or Fxr2 null littermates (55). Strikingly, both single-knockouts display only subtle defects in circadian rhythm, whereas Fmr1/Fxr2 DKO mice are completely arrhythmic when maintained in normal light:dark cycles (56). Likewise, Fmr1/Fxr2 DKO mice have significant defects in basal synaptic transmission which are not detectable in either single null (57). Loss of FMRP expression leads to alterations in long-term synaptic plasticity including exaggerated mGluR-dependent long-term depression (LTD) in hippocampal CA1 cells as well as loss of protein synthesis dependence for its maintenance (58–60). Surprisingly, Fxr2 null mice have decreased mGluR-LTD that remains protein synthesis dependent, whereas Fmr1/Fxr2 DKO mice have a dramatically exaggerated LTD, which led to the suggestion that there is some level of compensation by the homologs (57). However, since these are opposite phenotypes, it may be that the proteins are functioning in different pathways in controlling local translation at the synapse. Little is known about functional redundancy with FXR1P because loss of expression leads to neonatal lethality (37). We now show evidence for functional redundancy between FMRP and both FXR1P and FXR2P in KH domain-mediated RNA binding and polyribosome association on a biochemical level, as well as a unique function for FMRP in RGG box RNA recognition. In light of these findings, the RGG box interaction with G quadruplex RNA, specific to FMRP, may play an important function in the regulation of translational control at the synapse, and understanding this function may shed light on the striking differences in long-term synaptic plasticity between loss of FMRP and FXR2P.
G-quadruplexes are tertiary structures formed from stacks of planar G-quartets and stabilized by certain monovalent cations (Na+ and K+, but not Li+). In vitro selection experiments identified the G-quadruplex as the highest affinity ligand of the FMRP C-terminal RGG box, and indeed, of the full-length FMRP protein (41). Although an impressive amount of recent work has documented the role of these structures in DNA, including their destabilization by pharmaceutical agents, the potential importance of G-quadruplexes in RNA is just beginning to be appreciated. Bioinformatic studies predict many G-quadruplexes in RNA in the transcriptome (61,62) and have catalogued over 50 000 predicted G-quadruplexes near splice sites and alternative polyadenylation sites (63). There are also 2922 bioinformatically predicted G-quadruplexes in 5′-UTRs, including 13 in protooncogene 5′-UTRs, and this motif is thought to modulate their translation (64). In addition to FMRP, other RNA-binding proteins, such as the G4R1/RHAU helicase, recognize G-quadruplexes; G4R1/RHAU binds to G-quadruplexes with an affinity in the pM range, unwinding this ‘knot’ and permitting proper RNA metabolism (65). Several in vivo mRNA targets of FMRP, including PSD-95 (66), MAP1B (41) and amyloid precursor protein (67) as well as several mRNAs that co-IP with FMRP in mouse brain (41,68), contain G-quadruplex motifs, although direct binding to any of these motifs in vivo has not yet been demonstrated. In vitro interactions of FMRP with the G-quadruplex motif from MAP1B, semaphorin 3F and the SELEX ligand sc1 have been confirmed by structural studies (69–72).
RGG boxes have been reported to be present in numerous nucleic acid-binding proteins, yet a strict definition of an RGG box and its potential for specific RNA recognition remain unclear. The RGG box was originally described in hnRNP U on the basis of molecular studies that identified the domain responsible for RNA binding (73,74). Alignment of this domain in hnRNP U with other RNA-binding proteins including fibrillarin, nucleolin and hnRNP A1 suggested a consensus consisting of three to four closely spaced repeats of the RGG tripeptide. Examination of the human FMRP sequence identified an arginine- and glycine-rich motif fitting this consensus (2,4). Subsequently, FXR1P and FXR2P (9,10) and dFMRP (12) have also been considered to harbor C-terminal arginine-rich RNA-binding domains, although their sequence homologies and alignments are not well conserved (Fig. 8). Therefore, it has been unclear whether these domains share the same RNA-binding properties as the RGG box of FMRP.
A single-point mutation present in a severely affected Fragile X patient (5) has directed attention to the KH2 domain as a critical RNA-binding domain in FMRP. A mouse model of this mutation is sufficient to cause the Fragile X phenotype (Julie B. Zang, Elena D. Nosyreva, Corinne M. Spencer, Lenora J. Volk, Kiran Musunuru, Ru Zhong, Elizabeth F. Stone, Lisa A. Yuva-Paylor, Kimberly M. Huber, Richard Paylor, Jennifer C. Darnell and Robert B. Darnell, submitted for publication). This mutation disrupts KH2-RNA binding and polysome association in the mouse brain (Julie B. Zang, Elena D. Nosyreva, Corinne M. Spencer, Lenora J. Volk, Kiran Musunuru, Ru Zhong, Elizabeth F. Stone, Lisa A. Yuva-Paylor, Kimberly M. Huber, Richard Paylor, Jennifer C. Darnell and Robert B. Darnell, submitted for publication), in lymphoblastoid cells derived from the human patient (22) and in tissue culture cells transfected with FMRP reporter constructs (75,76). In contrast, deletion of the RGG box has little-to-no effect on polysome association (75,76). Moreover, FMRP can be completely competed off polysomes by the RNA ligand of the KH2 domain, kcRNA, but not by the G-quadruplex ligand of the RGG box (40). Here, we find that FMRP, FXR1P and FXR2P all share the properties of kcRNA binding and kcRNA-dependent polysome association, suggesting that the FXR1/2 proteins are likely to provide functional redundancy for this activity of FMRP in the brain of Fragile X patients (or in vertebrate models), whereas the RNA binding of the RGG box of FMRP may confer a specific function.
Very little is known about the in vivo function of the RGG box. When overexpressed, an RGG deletion mutant behaves much like wild-type FMRP in its ability to form dendritic puncta (77), to cause synaptic overgrowth in mouse neurons (77), and in its ability to associate with polysomes (75,76). A clue to the role of the RGG box in FMRP function may relate to FMRP's ability to nucleate stress granules when overexpressed. Stress granules are physiologic correlates of the translational inhibition that results from exposure of cells to a variety of stressors including heat shock, oxidative stress, viral infection and arsenite poisoning (reviewed in 78 and 79). Cells react by inhibiting translation of housekeeping (and likely other) transcripts (80) to preserve resources for the synthesis of stress response proteins. mRNAs from disassembled polysomes are sorted into pools destined for degradation or storage as repressed mRNPs. When stress abates, stress granules dissolve and polyribosomes form again on these ‘stored’ mRNAs. FMRP has been reported to shuttle between polysomes and stress granules induced to form by heat shock (81), hippuristanol or pateamine (inhibitors of initiation) (82), arsenite treatment (oxidative stress) (83) or neuronal trauma in vivo (83). FMRP, like several other proteins including TIA-1, TTP, G3BP, RCK, CPEB, caprin-1, FAST, Ago2 and SMN (78), is believed to nucleate stress granules when overexpressed. Khandjian and colleagues have reported that the deletion of the FMRP RGG box specifically abrogates its ability to nucleate stress granules when overexpressed (75,81). Interestingly, FXR1P is found in stress granules as well, and its presence in granules after heat shock is inhibited by the deletion of the RGG box in FMRP, suggesting that FMRP may recruit FXR1P to granules (81). FXR1P has also been reported to bind G-quadruplex RNA (84); however, this activity was mapped to sequences in the longest form of FXR1P, which is muscle specific and therefore not present in neurons. The C-terminus of FXR1P tested here is the longest form expressed in brain, which nonetheless lacks the G-quadruplex-binding domain present in muscle isoforms. Since skeletal muscle expresses little, if any, FMRP (33), it is intriguing to speculate that muscle-specific isoforms of FXR1P may have gained this function, in the absence of FMRP expression, in order to participate in stress granule formation.
Taken together, our observations suggest a model whereby the KH domains of FMRP are necessary for an action on polyribosomes that can be compensated for by FXR1/2P, but that the FMRP RGG box is uniquely necessary for stress-related transition from polyribosomes to stress granules. The role of FMRP in regulating the translation of specific mRNAs in cells is likely to be complex and to depend on levels of FMRP as well as the ‘state’ of the cell, including stage of the cell cycle, influence of ‘stress’, or in the case of neurons, synaptic activity. Our findings may help elucidate how the loss of FMRP leads to aberrant mRNA translation and synaptic dysfunction present in Fragile X syndrome.
MATERIALS AND METHODS
Cloning and purification of KH domains and C-termini
RNA-binding domains were amplified by PCR, using either human or mouse cDNA (Clontech), dfmr1 plasmid (a gift from Dr Tom Jongens) or previously cloned FMRP or I304N FMRP plasmids (40) as template, using the indicated primers and cloned into the multiple cloning site of pET21a or b (Novagen) to introduce an N-terminal T7 tag and a C-terminal His-Tag for purification. Mouse FMRP KH2 domain was cloned using the same primers as were used to amplify human FMRP KH2. The minimal KH2 domain lacking exons 11 and 12 was cloned by deleting exon 11 from the isoform lacking exon 12 (ISO7) as follows: Two PCR reactions were assembled using hKH2F (CCGGATCCTGCTGAAGATGTAATACAAGTTCC) and e11delR (GCTGTCCTTTGTTCCCACAAAAACAAATGGTACCATA CCCTCTTCTTGTGGAACATTTTTCTCATTTTCAGC) for one, and hKH2R (AAGCGGCCGCTAAATAGTTCAGGTGATAATCC) and e11delF (GGTATGGTACCATTTGTTTTTGTGGGAACAAAGGACAGCATCGCT) for the other, with human pet21-FMRP as a template and amplified with Pfu DNA polymerase. Expected products were gel-purified, denatured together in equimolar amounts and allowed to anneal. Following fill-in with Klenow and Pfu, this template was used in a new PCR reaction with hKH2F and hKH2R as terminal primers. The PCR product of the correct size was gel-purified, cloned into pET21b and confirmed by sequencing. Full-length Nova-1 was previously described (85). Human Sf1 KH-QUA2 domain was expressed from the Sf1-KH-STAR-pET-24d plasmid, a gift from Dr Michael Sattler and Dr Angela Kramer (50). After sequence confirmation, fusion proteins were expressed in BL21DE3(Star), induced for 3–4 h with IPTG, and proteins purified by His-Tag affinity chromatography as described previously (40). Purity of fusion protein preparations was assessed by Coomassie stain and western blots with anti-T7 antibody. Proteins were quantified by Bradford assay (BioRad) and stored in His-Tag elution buffer (20 mm HEPES, pH 8.4, 1 M KCl, 200 mm imidazole) until use.
- hFXR1P KH2 F: CCGGATCCGTTTGTGGAGGATTTTATTCAGG;
- hFXR1P KH2 R: CCGCGGCCGCTAGATAGGCAATATGATACTCTAGAAGAACC;
- hFXR1P C-terminus F: CCGGATCCGCAGCTAAGAATGGAACGCCTACAGATTGATGAACAGCTGC;
- hFXR1P C-terminus R: CCGCGGCCGCTGAAACACCATTCAGGACTGCTGC;
- hFXR2P KH2 F: CCGGATCCGTTTTCTGAGGACTCAGTGCAAGTGC;
- hFXR2P KH2 R: CCGCGGCCGCCAGGTAGGAGAGGTGATACTCC;
- hFXR2P C-terminus F: CCGAATTCTCCTACCTGCAGGAGGTAGAGCAGC;
- hFXR2P C-terminus R: CCCTCGAGTGAAACCCCATTCACCATACTACCCAACTCCAAGGGGGCG;
- exon 12 F: GGGAATTCGAGGTAGAGCAGCTTCGCTTGGAGAGGC;
- exon 12 R: GGCTCGAGATAGGCAGGACCGCCTGTCCTCCGGCC;
- exon 13 F: GGGAATTCGGCCCCAGCTCAGATGTGTCTACAGC;
- exon 13 R: GGCTCGAGTGAGCTAATAGATGAAGAATTGTATCTCG;
- exon 14 F: GGGAATTCGTGCTGAAGGATCCAGACAGTAATCCC;
- exon 14 R: GGCTCGAGCAGGCCATTCTCTGTCATGTTGGGCCC;
- exon 15–17 F: GGGAATTCGAAGATGAATCAAGACCTCAACG;
- exon 15–17 R: GGCTCGAGTGAAACCCCATTCACCATACTACC;
- Xenopus Vg1RBP/Vera KH1-4 F: GCGAATTCGGAGGTTCCGCTGAGAATGCTGGTTCCC;
- Vg1RBP/Vera KH1-4 R: CCGCTCGAGCAATATTTCCTGAATTTTCCTTTGTGCAAGC;
- Mouse PCBP1 KH1/2 F: GCGAATTCGACTCTCACCATTCGGCTGCTGATGCACGG;
- PCBP1 KH1/2 R: CCGCTCGAGGAGCGTCTCCAGCATGACCAGGCAGATCTGCTTCACACACTCGG;
- PCBP1 KH3 F: GCGAATTCGCAAACCACCCATGAACTCACCATTCCAAATAACTTAATCGGC;
- PCBP1 KH3 R: CCGCTCGAGAAGCCTGGCATTGATTAGATACTGGGC;
- dFMRP KH2 F: GCGAATTCGTACGCCGAGGAGTTCTTCCAGGTGCCCAGGG;
- dFMRP KH2 R: CCGCTCGAGCAGGTGCGACAGATGATACTCCAACAGCACTTTGGCATTTGC;
- dFMRP C-terminus F: CGGGATCCGAAGGAAGTAGAACAGTTGCGT CAGGAGAAGATGGAGATTGATCAGC;
- dFMRP C-terminus R: ATAGTTTAGCGGCCGCGGACGTGCCATTGA CCAGGCCCTCCTTTTTGACATTCTCCGC.
In vitro transcription of 32P-labeled RNA ligands
Transcription templates were prepared by PCR and purified over G-25 columns as described in what follows. In vitro transcription reactions included 13 µl of DNA template, 5 µl transcription buffer (Stratagene), 4 µl of 10 mm NTP mix (Amersham), 1 µl of 32P-alpha-UTP (Amersham), 1 µl of RNAsin (Promega) and 1 µl of T7 RNA polymerase (Stratagene). Labeled RNA was treated with three units of RQ1 DNAse and gel-purified on 8% urea-polyacrylamide gels.
Transcription templates for RNA ligands
sc1, mutant sc1, kc2 and mutant kc2 were prepared as described previously (40,41). For PCBP1-binding curves, we used the R7a1 stem-loop SELEX ligand (47) (the template sequence was AGTAATACGACTCACTATAGGAGTGACCTTCTCAA CTTTATATTCCCTTTACCCCTTCCCCCAAGGCACT, and reverse complementary primer was AGTGCCTTGGGGGAAGGGG). These oligos were annealed, filled in with Pfu and amplified by PCR using the reverse primer and a T7 promoter primer (AGTAATACGACTCACTATAG). PCR products were purified over G-25 columns and used as templates for in vitro transcription. For Vg1RBP/Vera binding, we used the pVLE plasmid, a gift from Dr Nancy Standart, linearized with MscI (51). The Sf1 ligand, the BPS RNA UAUACUAACAA (50), was synthesized by Dharmacon, end-labeled with 32P-gamma-ATP using T4 PNK and gel-purified.
Filter binding assay
Filter binding assays were performed as described previously (40,41). Briefly, 120 000 cpm of _in vitro_-transcribed 32P-labeled RNA was diluted to 120 µl in 1× SBB buffer (200 mm potassium acetate, 50 mm Tris–acetate, pH 7.4, 5 mm magnesium acetate), heated to 75°C for 10 min and bench-cooled for 5 min to renature the RNA. Ten microliters of RNA (10 000 cpm, 1–5 fmol) was added to tubes containing serial 3-fold dilutions of RNA-binding protein in SBB buffer in 40 µl aliquots. After 10 min of equilibration at room temperature, samples were captured on MF nitrocellulose membranes (Millipore HAWP-02500) by filtration on a Millipore 12-well vacuum manifold and washed with 5 ml of 1× SBB. Bound RNA was quantified by scintillation counting in 5 ml of Readi-Safe scintillant and background (RNA added to 40 µl SBB with no protein and filtered) was subtracted from each value. Total counts per sample were determined by spotting 10 µl of RNA on a dry filter. Data are expressed as percentage of total RNA and plotted against log of the protein concentration using Kaleidograph software (Synergy Software). _K_d values were determined by the Kaleidograph binding curve algorithm. SBB was substituted with 1× binding buffer (10 mm Tris–Cl, pH 7.4, 150 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT) for PCBP1 curves (47), (25 mm NaCl, 25 mm Tris–HCl, pH, 1 mm EDTA, 0.5 mg/ml tRNA) for Sf1 curves (48) and FBA buffer (100 mm NaCl, 50 mm Tris, pH 8, 1 mm MgCl2) for Vg1RBP/Vera curves (51).
Polysome gradient analysis of mouse brain
Two-to-three-week-old mice were sacrificed by isoflurane anesthesia and decapitation. Postmitochondrial supernatants of cerebral cortex and cerebellum, dissected free from white matter, were prepared and separated by 20–50% sucrose gradients as described previously (40). Then, 720 µl fractions (16 per gradient) were collected with continuous monitoring at 254 nm using an ISCO UA-6 UV detector. Where indicated, in vitro_-transcribed RNAs (unlabeled) were added to the S1 lysate (after heating to 75°C for 10 min and bench-cooling for 5 min in 1× SBB buffer) for 15 min at room temperature prior to the 20 000_g spin to generate the S2 supernatant which was loaded on the sucrose gradient.
SDS–PAGE and western blot
The proteins contained in each fraction of the sucrose gradients were TCA-precipitated and analyzed by western blot using the indicated antibody and the appropriate anti-HRP secondary antibody (Jackson Immunochemicals). Chemiluminescence was quantitated with a Versadoc Imaging System (BioRad).
Antibodies used for western blot
Anti-FMRP polyclonal antibody ab17722 (Abcam) was used at 1:1000, anti-FXR1P rabbit polyclonal antibody ML13 (gift from Dr E. Khandjian) was used at 1:25 000, anti-FXR2P mouse monoclonal antibody 1G2 (obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA 52242, USA) was used at 1:2000, anti-ribosomal protein S6 rabbit monoclonal 5G10 (Cell Signalling) was used at 1:1000, anti-PABP rabbit polyclonal antibody ab21060 (Abcam) was used at 1:2000, anti-Ago2 (eIF2C2) rabbit polyclonal antibody ab32381 (Abcam) was used at 1:1000, anti-P0 human antibody (USBiological R2031-25) was used at 1:20 000 and anti-Hu was a human Hu syndrome patient serum used at 1:10 000.
FUNDING
This work was supported by National Institutes of Health (R01s NS34389, NS40955 to R.B.D. and 5R01 HD40647 to J.C.D.), and MSTP-GM07749 to C.E.F. R.B.D. is a Howard Hughes Medical Institute Investigator. Funding to pay the Open Access publication charges for this article was provided by R01 HD40647.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Eduoard Khandjian for the anti-FXR1P antibody, Dr Tom Jongens for cDNA-encoding dfmr1, Dr Angela Kramer and Dr Michael Sattler for the Sf1 KH-QUA2 expression construct and Dr Nancy Standart for the pVLE plasmid. We thank members of the Darnell Laboratory including Sarah van Driesche, Tina Marney and Dr Maria Frias for helpful discussions and critical review of the manuscript.
Conflict of Interest Statement. None declared.
REFERENCES
- 1.Bassell G.J., Warren S.T. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson T.J., Rice P.M., Thompson J.D., Heringa J. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem. Sci. 1993;18:331–333. doi: 10.1016/0968-0004(93)90068-x. [DOI] [PubMed] [Google Scholar]
- 3.Ashley C.T., Wilkinson K.D., Reines D., Warren S.T. FMR-1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 4.Siomi H., Siomi M.C., Nussbaum R.L., Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 5.DeBoulle K., Verkerk A.J., Reyniers E., Vits L., Hendrickx J., Van Roy B., Van Den Bos F., de Graaff E., Oostra B.A., Willems P.J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 6.Ramos A., Hollingworth D., Pastore A. The role of a clinically important mutation in the fold and RNA-binding properties of KH motifs. RNA. 2003;9:293–298. doi: 10.1261/rna.2168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valverde R., Edwards L., Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 8.Lewis H.A., Musunuru K., Jensen K.B., Edo C., Chen H., Darnell R.B., Burley S.K. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 9.Siomi M.C., Siomi H., Sauer W.H., Srinivasan S., Nussbaum R.L., Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., O'Conner J.P., Siomi M.C., Srinivasan S., Dutra A., Nussbaum R.L., Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkpatrick L.L., McIlwain K.A., Nelson D.L. Comparative genomic sequence analysis of the FXR gene family: FMR1, FXR1 and FXR2. Genomics. 2001;78:169–177. doi: 10.1006/geno.2001.6667. [DOI] [PubMed] [Google Scholar]
- 12.Wan L., Dockendorff T.C., Jongens T.A., Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verheij C., de Graaff E., Bakker C.E., Willemsen R., Willems P.J., Meijer N., Galjaard H., Reuser A.J., Oostra B.A., Hoogeveen A.T. Characterization of FMR1 proteins isolated from different tissues. Hum. Mol. Genet. 1995;4:895–901. doi: 10.1093/hmg/4.5.895. [DOI] [PubMed] [Google Scholar]
- 14.Corbin F., Bouillon M., Fortin A., Morin S., Rousseau F., Khandjian E.W. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 15.Tamanini F., Meijer N., Verheij C., Willems P.J., Galjaard H., Oostra B.A., Hoogeveen A.T. FMRP is associated to the ribosomes via RNA. Hum. Mol. Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- 16.Khandjian E.W., Corbin F., Woerly S., Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y., Gutekunst C.A., Eberhart D.E., Yi H., Warren S.T., Hersch S.M. Fragile X mental retardation protein–nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siomi M.C., Zhang Y., Siomi H., Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khandjian E.W., Bardoni B., Corbin F., Sittler A., Giroux S., Heitz D., Tremblay S., Pinset C., Montarras D., Rousseau F., Mandel J. Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum. Mol. Genet. 1998;7:2121–2128. doi: 10.1093/hmg/7.13.2121. [DOI] [PubMed] [Google Scholar]
- 20.Khandjian E.W., Huot M.E., Tremblay S., Davidovic L., Mazroui R., Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl Acad. Sci. USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefani G., Fraser C.E., Darnell J.C., Darnell R.B. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., Warren S.T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 23.Tamanini F., Bontekoe C., Bakker C.E., van Unen L., Anar B., Willemsen R., Yoshida M., Galjaard H., Oostra B.A., Hoogeveen A.T. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum. Mol. Genet. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- 24.Tamanini F., Van Unen L., Bakker C., Sacchi N., Galjaard H., Oostra B.A., Hoogeveen A.T. Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P. Biochem. J. 1999;343:517–523. [PMC free article] [PubMed] [Google Scholar]
- 25.Eichler E.E., Richards S., Gibbs R.A., Nelson D.L. Fine structure of the human FMR1 gene. Hum. Mol. Genet. 1994;3:684–685. [PubMed] [Google Scholar]
- 26.Price D.K., Zhang F., Ashley C.T.J., Warren S.T. The chicken FMR1 gene is highly conserved with a CCT 5′-untranslated repeat and encodes an RNA-binding protein. Genomics. 1996;31:3–12. doi: 10.1006/geno.1996.0002. [DOI] [PubMed] [Google Scholar]
- 27.Blonden L., van ‘t Padje S., Severijnen L.A., Destree O., Oostra B.A., Willemsen R. Two members of the Fxr gene family, Fmr1 and Fxr1, are differentially expressed in Xenopus tropicalis. Int. J. Dev. Biol. 2005;49:437–441. doi: 10.1387/ijdb.051974lb. [DOI] [PubMed] [Google Scholar]
- 28.Ashley C.T., Sutcliffe J.S., Kunst C.B., Leiner H.A., Eichler E.E., Nelson D.L., Warren S.T. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat. Genet. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 29.Verkerk A., de Graaff E., De Boulle K., Eichler E.E., Konecki D.S., Reyniers E., Manca A., Poustka A., Willems P.J., Nelson D.L., Oostra B.A. Alternative splicing in the fragile X gene FMR1. Hum. Mol. Genet. 1993;2:399–404. doi: 10.1093/hmg/2.4.399. [DOI] [PubMed] [Google Scholar]
- 30.Tamanini F., Kirkpatrick L.L., Schonkeren J., van Unen L., Bontekoe C., Bakker C., Nelson D.L., Galjaard H., Oostra B.A., Hoogeveen A.T. The fragile X-related proteins FXR1P and FXR2P contain a functional nucleolar-targeting signal equivalent to the HIV-1 regulatory proteins. Hum. Mol. Genet. 2000;9:1487–1493. doi: 10.1093/hmg/9.10.1487. [DOI] [PubMed] [Google Scholar]
- 31.Agulhon C., Blanchet P., Kobetz A., Marchant D., Faucon N., Sarda P., Moraine C., Sittler A., Biancalana V., Malafosse A., Abitbol M. Expression of FMR1, FXR1 and FXR2 genes in human prenatal tissues. J. Neuropathol. Exp. Neurol. 1999;58:867–880. doi: 10.1097/00005072-199908000-00009. [DOI] [PubMed] [Google Scholar]
- 32.Tamanini F., Willemsen R., van Unen L., Bontekoe C., Galjaard H., Oostra B.A., Hoogeveen A.T. Differential expression of FMR1, FXR1 and FXR2 proteins in human brain and testis. Hum. Mol. Genet. 1997;6:1315–1322. doi: 10.1093/hmg/6.8.1315. [DOI] [PubMed] [Google Scholar]
- 33.Bakker C.E., de Diego Otero Y., Bontekoe C., Raghoe P., Luteijn T., Hoogeveen A.T., Oostra B.A., Willemsen R. Immunocytochemical and biochemical characterization of FMRP, FXR1P, and FXR2P in the mouse. Exp. Cell Res. 2000;258:162–170. doi: 10.1006/excr.2000.4932. [DOI] [PubMed] [Google Scholar]
- 34.Coy J.F., Sedlacek Z., Bachner D., Hameister H., Joos S., Lichter P., Delius H., Poustka A. Highly conserved 3′-UTR and expression pattern of FXR1 points to a divergent gene regulation of FXR1 and FMR1. Hum. Mol. Genet. 1995;4:2209–2218. doi: 10.1093/hmg/4.12.2209. [DOI] [PubMed] [Google Scholar]
- 35.Devys D., Lutz Y., Rouyer N., Bellocq J., Mandel J. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 36.Dube M., Huot M.E., Khandjian E.W. Muscle specific fragile X related protein 1 isoforms are sequestered in the nucleus of undifferentiated myoblast. BMC Genet. 2000;1:4. doi: 10.1186/1471-2156-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mientjes E.J., Willemsen R., Kirkpatrick L.L., Nieuwenhuizen I.M., Hoogeveen-Westerveld M., Verweij M., Reis S., Bardoni B., Hoogeveen A.T., Oostra B.A., Nelson D.L. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum. Mol. Genet. 2004;13:1291–1302. doi: 10.1093/hmg/ddh150. [DOI] [PubMed] [Google Scholar]
- 38.Kirkpatrick L.L., McIlwain K.A., Nelson D.L. Alternative splicing in the murine and human FXR1 genes. Genomics. 1999;59:193–202. doi: 10.1006/geno.1999.5868. [DOI] [PubMed] [Google Scholar]
- 39.Christie S.B., Akins M.R., Schwob J.E., Fallon J.R. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J. Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darnell J.C., Fraser C.E., Mostovetsky O., Stefani G., Jones T.A., Eddy S.R., Darnell R.B. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G Quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 42.Valverde R., Pozdnyakova I., Kajander T., Venkatraman J., Regan L. Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure. 2007;15:1090–1098. doi: 10.1016/j.str.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 43.Musco G., Stier G., Joseph C., Morelli M.A.C., Nilges M., Gibson T., Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the Fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 44.Lewis H.A., Chen H., Edo C., Buckanovich R.J., Yang Y.Y., Musunuru K., Zhong R., Darnell R.B., Burley S.K. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure. 1999;7:191–203. doi: 10.1016/S0969-2126(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 45.Jensen K.B., Musunuru K., Lewis H.A., Burley S.K., Darnell R.B. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl Acad. Sci. USA. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dredge B.K., Darnell R.B. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol. Cell. Biol. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thisted T., Lyakhov D.L., Liebhaber S.A. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alpha CP-2KL, suggest distinct modes of RNA recognition. J. Biol. Chem. 2001;276:17484–17496. doi: 10.1074/jbc.M010594200. [DOI] [PubMed] [Google Scholar]
- 48.Berglund J.A., Abovich N., Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deshler J.O., Highett M.I., Schnapp B.J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 50.Liu Z., Luyten I., Bottomley M.J., Messias A.C., Houngninou-Molango S., Sprangers R., Zanier K., Kramer A., Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 51.Git A., Standart N. The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA. 2002;8:1319–1333. doi: 10.1017/s135583820202705x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deshler J.O., Highett M.I., Abramson T., Schnapp B.J. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 53.Tucker B., Richards R., Lardelli M. Expression of three zebrafish orthologs of human FMR1-related genes and their phylogenetic relationships. Dev. Genes Evol. 2004;214:567–574. doi: 10.1007/s00427-004-0438-9. [DOI] [PubMed] [Google Scholar]
- 54.Bontekoe C.J., McIlwain K.L., Nieuwenhuizen I.M., Yuva-Paylor L.A., Nellis A., Willemsen R., Fang Z., Kirkpatrick L., Bakker C.E., McAninch R., et al. Knockout mouse model for Fxr2: a model for mental retardation. Hum. Mol. Genet. 2002;11:487–498. doi: 10.1093/hmg/11.5.487. [DOI] [PubMed] [Google Scholar]
- 55.Spencer C.M., Serysheva E., Yuva-Paylor L.A., Oostra B.A., Nelson D.L., Paylor R. Exaggerated behavioral phenotypes in Fmr1/Fxr2 double knockout mice reveal a functional genetic interaction between Fragile X-related proteins. Hum. Mol. Genet. 2006;15:1984–1994. doi: 10.1093/hmg/ddl121. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J., Fang Z., Jud C., Vansteensel M.J., Kaasik K., Lee C.C., Albrecht U., Tamanini F., Meijer J.H., Oostra B.A., Nelson D.L. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am. J. Hum. Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J., Hou L., Klann E., Nelson D.L.N. Altered hippocampal synaptic plasticity in the Fmr1 gene family knockout mouse models. J. Neurophysiol. 2009;101:2572–2580. doi: 10.1152/jn.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huber K.M., Gallagher S.M., Warren S.T., Bear M.F. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl Acad. Sci. USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou L., Antion M.D., Hu D., Spencer C.M., Paylor R., Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 60.Nosyreva E.D., Huber K.M. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J. Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- 61.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kostadinov R., Malhotra N., Viotti M., Shine R., D'Antonio L., Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5′-UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Creacy S.D., Routh E.D., Iwamoto F., Nagamine Y., Akman S.A., Vaughn J.P. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Todd P.K., Mack K.J., Malter J.S. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc. Natl Acad. Sci. USA. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Westmark C.J., Malter J.S. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Darnell R.B., Warren S.T. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in Fragile X Syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 69.Ramos A., Hollingworth D., Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menon L., Mader S.A., Mihailescu M.R. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA. 2008;14:1644–1655. doi: 10.1261/rna.1100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Menon L., Mihailescu M.R. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zanotti K.J., Lackey P.E., Evans G.L., Mihailescu M.R. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 73.Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 75.Mazroui R., Huot M.E., Tremblay S., Boilard N., Labelle Y., Khandjian E.W. Fragile X mental retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet. 2003;12:3087–3096. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- 76.Darnell J.C., Mostovetsky O., Darnell R.B. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 77.Pfeiffer B.E., Huber K.M. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J. Neurosci. 2007;27:3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 79.DeGracia D.J., Hu B.R. Irreversible translation arrest in the reperfused brain. J. Cereb. Blood Flow Metab. 2007;27:875–893. doi: 10.1038/sj.jcbfm.9600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawai T., Fan J., Mazan-Mamczarz K., Gorospe M. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell. Biol. 2004;24:6773–6787. doi: 10.1128/MCB.24.15.6773-6787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mazroui R., Huot M.E., Tremblay S., Filion C., Labelle Y., Khandjian E.W. Trapping of messenger RNA by Fragile X mental retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 82.Mazroui R., Sukarieh R., Bordeleau M.E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim S.H., Dong W.K., Weiler I.J., Greenough W.T. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J. Neurosci. 2006;26:2413–2418. doi: 10.1523/JNEUROSCI.3680-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bechara E., Davidovic L., Melko M., Bensaid M., Tremblay S., Grosgeorge J., Khandjian E.W., Lalli E., Bardoni B. Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure. Nucleic Acids Res. 2007;35:299–306. doi: 10.1093/nar/gkl1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buckanovich R.J., Yang Y.Y., Darnell R.B. The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J. Neurosci. 1996;16:1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]