Variation in aggregation propensities among ALS-associated variants of SOD1: Correlation to human disease (original) (raw)
Abstract
To date, 146 different mutations in superoxide dismutase 1 (SOD1) have been identified in patients with familial amyotrophic lateral sclerosis (ALS). The mean age of disease onset in patients inheriting mutations in SOD1 is 45–47 years of age. However, although the length of disease duration is highly variable, there are examples of consistent disease durations associated with specific mutations (e. g. A4V, less than 2 years). In the present study, we have used a large set of data from SOD1-associated ALS pedigrees to identify correlations between disease features and biochemical/biophysical properties of more than 30 different variants of mutant SOD1. Using a reliable cell culture assay, we show that all ALS-associated mutations in SOD1 increase the inherent aggregation propensity of the protein. However, the relative propensity to do so varied considerably among mutants. We were not able to explain the variation in aggregation rates by differences in known protein properties such as enzyme activity, protein thermostability, mutation position or degree of change in protein charge. Similarly, we were not able to explain variability in the duration of disease in SOD1-associated ALS pedigrees by these properties. However, we find that the majority of pedigrees in which patients exhibit reproducibly short disease durations are associated with mutations that show a high inherent propensity to induce aggregation of SOD1.
INTRODUCTION
Familial amyotrophic lateral sclerosis (ALS) is an invariably fatal neurodegenerative disease that principally affects upper and lower motor neurons. A subset of familial ALS cases (12–20%) are caused by mutations in superoxide dismutase 1 (SOD1), with 146 different mutations described in families or more rarely in an individual apparently sporadic case (www.alsod.org). Eleven different mutations have been expressed in transgenic mice and rat models (1–15), which develop ALS-like phenotypes that include hindlimb weakness, progressive generalized paralysis and muscle atrophy. Recently, recessive inheritance of a SOD1 mutation has been described in dogs that develop ALS-like disease (16). In all these models, there is evidence that the levels of expression are critical to induce disease. For example, there are mice that express the G37R or D90A SOD1 that do not develop disease unless bred to homozygosity (8,12). Similarly, disease is absent in mice expressing low levels of A4V SOD1 but evident when the levels of total SOD1 are raised by co-expression with wild-type (WT) SOD1 (1). Additionally, very aggressive phenotypes are found in the rare consanguineous human cases (G27ΔGP, L84F, N86S, L126S) (17–19). Thus, the levels of expression of SOD1 mutant proteins seem to play an important role in disease.
In all mouse models, the manifestation of disease symptoms is accompanied by the accumulation of detergent-insoluble aggregated forms of mutant SOD1 (1,12,13,15,20–22). In human SOD1-associated ALS, there is similar evidence that mutant SOD1 aggregation is a pathological feature (20). Thus, there seems to be a clear correlation between the presence of detergent-insoluble aggregated forms of mutant SOD1 in spinal cords and disease (23). Importantly, aggregated forms of mutant SOD1 that display similar properties of detergent insolubility can be produced in cultured cells (22–25), representing an efficient system to screen and study aggregation of ALS mutants. SOD1-associated ALS mutations are spread throughout the 153 amino acid protein sequence with the vast majority of point mutations occurring at highly conserved amino acids (24). Eighty codons in SOD1 are known to be targets of mutation that give rise to the ALS phenotype; in some cases, multiple amino acid substitutions occur at one site (up to six for G93). It is well established that specific mutations are associated with disease of short- or long-clinical course (26). Examples of short disease course include the A4V mutation (less than 2 years) (27), whereas mutations such as H46R are associated with a long-disease course (more than 10 years) (28). A recent study used a variety of biophysical data to calculate aggregation rates for different ALS mutants, suggesting that aggregation of mutant protein could be a key factor in disease progression (29). Here, we have used our cell culture model to analyze a total of 33 SOD1-associated ALS mutations in regards to their ability to form detergent-insoluble aggregates, including different mutation substitutions at the same codon. By this approach, we assess how measured aggregation potentials relate to known biophysical/biochemical characteristics and examine whether aggregation propensities correlate to disease features in human ALS patients.
RESULTS
Large variability in aggregation among SOD1-associated ALS mutants
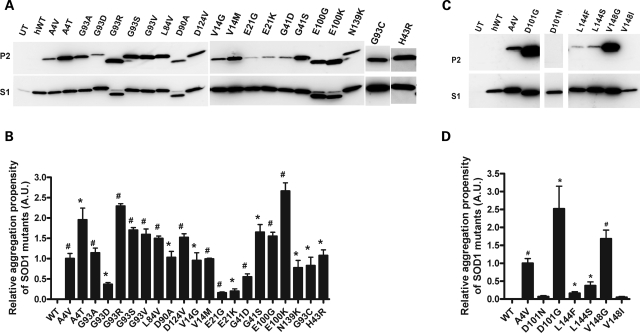
Our first analysis of 21 mutant SOD1 proteins demonstrated that all are capable of forming detergent-insoluble SOD1 aggregates in cells within 24 h (Fig. 1A). Quantification of the aggregation propensity, which is calculated from the ratio of insoluble to soluble forms of mutant SOD1 in cell lysates, of different mutations showed significant differences from WT SOD1 (Fig. 1B). To normalize data from different experiments, we chose to use the aggregation propensity of A4V SOD1 as the reference mutant [assigning one to the mean aggregation propensity of A4V, as previously described (22,23)]. Most mutations analyzed in Fig. 1A are of similar, or higher, aggregation propensity to A4V. Several mutants possessed aggregation propensities lower than that of A4V (G93D, E21G, E21K and G41D). In the cases in which more than one mutation occurred at a particular codon, we often observed significantly different levels of aggregated protein for each individual mutation at one position (Fig. 1B); examples include A4V versus T (P = 0.0168); G93A versus D (P = 0.0078), R (P = 0.0056) or S (P = 0.0222); G41D versus S (P = 0.0128) and E100G versus K (P = 0.0160). We also observed examples in which different amino acid changes at the same position did not differentially affect the aggregation propensity: G93A versus C (P = 0.4934), V (P = 0.111); V14G versus M (P = 0.8766) and E21G versus K (P = 0.6213). Further studies on additional SOD1 mutants involving different amino acid substitutions at the same site demonstrated very high variability in aggregation propensities. Mutation of D101 or V148 to G induced the formation of very high levels of detergent-insoluble proteins in 24 h (Fig. 1C). D101G together with E100K SOD1 represent the most aggregation prone proteins analyzed so far. However, D101N and V148I showed dramatically lower aggregation levels at 24 h, not different from that of WT SOD1 (Fig. 1D).
Figure 1.
Large variability in aggregation among SOD1-associated ALS mutants. (A and C) Immunoblots of detergent insoluble (P2) and soluble fractions (S1) of HEK293FT cells transfected with WT or mutant SOD1 for 24 h. UT: untransfected cells. (B and D) Quantification of the relative aggregation propensity of the WT and mutant SOD1 as described in Materials and Methods. Bars represent mean ± SEM of three or more independent transfection experiments. *P < 0.05, #P < 0.001.
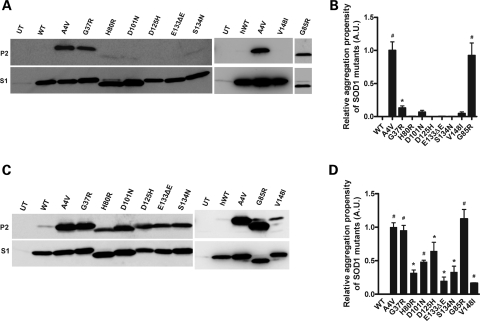
In addition to the slow aggregating D101N and V148I mutants, we found other mutations in which aggregation levels resemble those of WT SOD1 protein at 24 h (Fig. 2A). When compared with G37R, which to date had been representative of mutants with low potentials to aggregate (23), the H80R, D101N, D125H, E133ΔE, S134 N and V148I SOD1 mutants showed even lower potentials to aggregate (Fig. 2A, upper panels). In all cases, high levels of soluble SOD1 protein were detected for each of the mutants indicating robust protein expression (Fig. 2A, lower panels). Quantification and statistical analysis indicated that the aggregation propensity of these mutants in 24 h was not different from WT SOD1 (Fig. 2B). Thus, we identified, for the first time, SOD1 mutants that do not readily form detergent-insoluble aggregates in 24 h, showing a similar behavior to WT SOD1. These findings were initially viewed as an indication that aggregation, a priori, may not be necessarily linked to disease development. In order to more rigorously determine whether these mutants remain completely soluble, we extended the interval between transfection and harvest of the cells from 24 to 48 h. With longer incubation times all mutants formed detectable levels of detergent-insoluble protein, whereas WT SOD1 still remained completely soluble (Fig. 2C). At 48 h, the aggregation propensities of these mutants were significantly different from WT SOD1 (Fig. 2D).
Figure 2.
Some SOD1-associated ALS mutant proteins aggregate slowly. (A and C) Immunoblots of detergent insoluble (P2) and soluble fractions (S1) of HEK293FT cells transfected with WT or mutant SOD1 for 24 h (A) or 48 h (C). UT: untransfected cells. (B and D) Quantification of the relative aggregation propensity of SOD1 proteins at 24 h (B) and 48 h (D). Bars represent mean ± SEM of three or more independent transfection experiments. *P < 0.05, #P < 0.001. Note that D101N and V148I SOD1 mutants have been included in this figure for comparison.
Do specific biophysical or biochemical properties of SOD1 predict aggregation propensity?
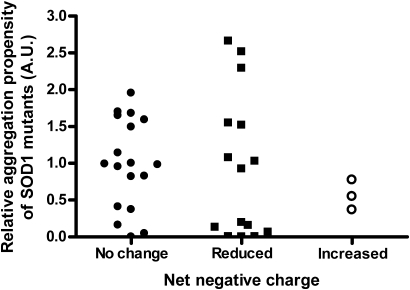
It has been suggested that mutations in SOD1 that decrease the net negative charge of the protein (eliminating negatively charged or introducing positively charged amino acids) causes misfolding and aggregation of mutant SOD1 (30,31). Many of the ALS-associated point mutations in SOD1 reduce the net negative charge of SOD1; however, three ALS mutants would be expected to possess a protein charge more negative than WT SOD1 (Fig. 3, Table 1). Thus, we asked whether differences in protein charge among the ALS mutants may explain the variability in the relative rates of mutant SOD1 aggregation. Some of these changes in protein charge can be observed in mutations occurring at amino acid G93. Although some mutations in G93 do not alter the overall protein charge (Table 1), there is a reduction in the negative charge of SOD1 when mutating G93 to R, increasing the aggregation propensity significantly compared with those mutants that do not produce a change in protein charge (G93R versus A, P = 0.0056; G93R versus C, P = 0.0262; G93R versus S, P = 0.0012; G93R versus V, P = 0.0325). Additionally, when G93 is mutated to D, which increases the negative charge of SOD1, then the levels of aggregated protein are significantly reduced compared with neutral change mutants (e.g. G93D versus A, P = 0.0078). Another example of similar findings is that of mutations at G41, where G41S presented an aggregation propensity higher than that of the more negatively charged G41D (P = 0.0128). Additionally, when mutations occur in E100 (E100G, E100K), the resulting mutant proteins present less negative charge than WT SOD1; the E100K mutant shows a more substantial decrease in negative charge and significantly higher aggregation potential when compared with E100G (P = 0.016). Overall, these findings support the hypothesis that changes in negative charge may play an important role in modulating aggregation of SOD1. However, this apparent correlation does not apply to all mutants that alter the negative charge of SOD1. A good example is the case of mutations in E21, which can be mutated to either G or K, similar to mutations in E100. Although E21K SOD1 has a higher decrease in negative charge, the aggregation propensity levels of E21G and E21K are not statistically different from each other (P = 0.6213), with both being very low (Fig. 1). Another example is that of changes in D101. Decreasing the negative charge of SOD1 by substituting amino acid D101 by either G or N produces mutant proteins in which aggregation levels are either very high (D101G) or very low (D101N). In this case, the same magnitude of decrease in negative charge produces a very large variability in aggregation rates (D101G versus D101N, P = 0.0296). Additional examples, in which a reduction in the negative charge of SOD1 does not produce dramatic increases in aggregation, are the cases of H80R, D125H and E133ΔE (Table 1). To further probe as to whether the location of the mutation within the protein may interact with the change in net charge, we graphed measured aggregation propensity as a function of mutation charge (Fig. 3) and location (Supplementary Material, Fig. S1). No obvious pattern emerges to link change in net charge to inherent aggregation propensities. Further, there were no obvious correlations between changes in net charge and disease onset or duration (Supplementary Material, Fig. S2). Thus, our data suggest that the relative aggregation propensity of mutant SOD1 is not inextricably linked to changes in net protein charge. However, we do not discard the possibility that changes in protein charge, along with other protein characteristics, could explain the different aggregation propensities of ALS mutants.
Figure 3.
Changes in the net negative charge of SOD1 do not predict aggregation propensity. Mutations in SOD1 that reduce, increase or do not modify the negative charge of SOD1 present aggregation propensity values that range from very low to very high; with no particular group representing mutants of high or low aggregation propensities. Unpaired Student _t_-tests: no change in charge mutants versus mutants with reduced charge (P = 0.8711), no change in charge mutants versus mutants with increased charge (P = 0.2556), mutants with reduced charge versus mutants with increased protein charge (P = 0.5213).
Table 1.
Changes in protein charge do not explain aggregation propensity
| Aggregation propensity at 24 h | No change in net negative charge | Reduce net negative charge | Increase net negative charge |
|---|---|---|---|
| Low | S134N, V148I | H80R, D101N, D125H, E133ΔE | |
| Moderate | I113T, L144F, L144S, C111Y | E21G, E21K, G37R | G41D, G93D, N139K |
| High | C6G, G93C, V14G, C6F, V14M, A4V, G93A, L84V, G93V, G41S, V148G, G93S | H43R, G85R, D90A, E100G, D124V | |
| Extreme | A4T | G93R, E100K, D101G |
Aggregation versus disease
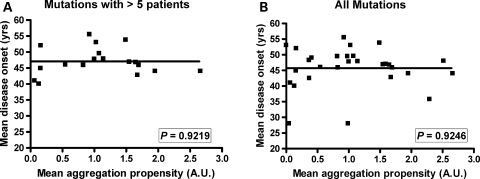
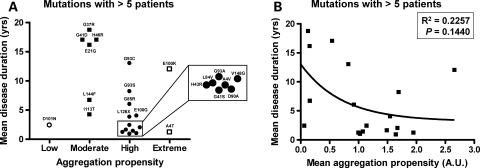
A recent study, using mathematical models, suggested that the predicted aggregation propensity of SOD1 mutants correlates with disease duration in humans (29). Here we used data from our cell culture model to analyze whether our measured aggregation propensity correlates with patient data on disease onset and/or duration. We imposed a filter on the patient data, focusing on cases in which the number of affected individuals with a particular mutation met or exceeded five individuals. We chose five individuals as the lower limit, because it emerged as a natural breakpoint in our data and because this number of patients allows for a more rigorous estimation of reproducibility of observed phenotypes. After filtering our patient data set, we were able to identify 21 different mutations for which we possessed data on an adequate number of patients. The data on aggregation propensities of the different mutants were stratified into four categories as explained in Figure 4. For the 21 mutations in the patient data sets we analyzed, two mutations were categorized as exhibiting extreme aggregation propensity, 12 of the mutations fit criteria for high-aggregation propensity, six fit criteria as moderate and one fits criteria for low aggregation propensity (see Supplementary Material, Table S1). No obvious correlation between aggregation propensity and age of onset was noted (Fig. 4). This outcome was expected because the mean age of onset is 45–47 years of age for all SOD1 mutants (Supplementary Material, Table S1). When we stratified the mutants by the four criteria described above and graphed these groups as a function of disease duration, we noted that mutants exhibiting aggregation propensities equivalent to or greater than the A4V mutation largely predicted shorter disease duration in patients (Fig. 5A). Statistical analysis demonstrated significant differences between the groups of high and moderate aggregation propensities (unpaired _t_-test, P = 0.0008). However, inverse correlations do not exist between aggregation propensity and disease duration (P = 0.1440, Fig. 5B), possibly because disease duration in patients with mutations of moderate or low-aggregation propensity is less predictable. Still, when we focus on the mutants that show high-aggregation propensities, we find that the majority of patients with these mutations exhibit a rapid disease course.
Figure 4.
Disease onset is not driven by changes in aggregation propensity. SOD1 mutations with a significant number of patients (more than five) (A), or all available compiled data (B) grouped in terms of aggregation propensity: Extreme (produces a level of insoluble mutant protein in 24 h that is equal to or greater than twice that of A4V SOD1—always set at 1), high (aggregate load is similar to A4V in 24 h; aggregation propensities range between 0.7 and 1.7), moderate (aggregate load is detectable but less than 0.5 in 24 h), slow (no aggregates detected in 24 h—only visible at 48 h). No correlation was found between aggregation and age of onset (P > 0.05). Note that the mean onset for ALS patients with a SOD1 mutation occurs between 45–47 years of age.
Figure 5.
Mutants possessing a higher aggregation propensity correlate with shorter disease duration. (A) Mutations with a significant number of patients (more than five) grouped in terms or aggregation propensity categories, as explained in Figure 4. Mutations associated with shorter disease durations belong to a group that present high or extreme aggregation propensities. (B) Non-linear regression of aggregation propensity and disease duration. A statistically significant correlation between aggregation and disease duration was not found (P > 0.05).
DISCUSSION
Studies to date, which have examined a relatively small percentage of all ALS mutants, have demonstrated that ALS mutations in SOD1 increase the propensity of the protein to form detergent-insoluble aggregates. Data from our present study raises the total number of SOD1-associated mutants for which we have measured aggregation propensities to 30% of all known mutants, providing definitive evidence that increased aggregation propensity is highly likely to be a universal feature of mutant SOD1. However, the inherent propensity of different mutants to produce aggregated protein varies, even in cases in which multiple mutations target a single amino acid position. We could not identify a specific biochemical or biophysical property of the SOD1-associated ALS mutants that adequately explains the variability in the propensity of these mutant proteins to aggregate. Although causality for variability is unknown, we find that the inherent aggregation propensity of ALS mutants is related to the duration of disease in ALS patients such that mutants that show high-aggregation propensities are associated with a greater risk for short disease duration.
One aspect of our study sought to determine whether variability in mutant SOD1 aggregation propensity could be explained by the nature of the amino acid mutation. Mutations in SOD1 that bring the protein charge to neutrality, or decrease the negative charge, have been suggested to make it more prone to aggregate (32,33). More than half of the SOD1 proteins studied here represent mutations affecting a charged residue or introduce a charged amino acid in place of a non-charged (Fig. 3). Within this group of mutations that modify the negative charge of SOD1, we can find SOD1 mutants with all defined levels of aggregation propensity (low, moderate, high or extreme), not following a clear correlation between a decrease or increase in protein charge and aggregation rates. For example, eliminating aspartate in amino acid 101 produces a low (D101N) or very high (D101G) aggregating protein, with both proteins producing a decrease in negative charge. However, when mutations occur at E100, the mutant with a larger increase in net negative charge has the highest aggregation propensity (E100K versus G). The E100 and D101 amino acids are in such close proximity that it seems unlikely that the different changes in charged amino acids at these two residues could have very different structural effects on the protein. Additionally, we have found no obvious correlation between aggregation propensity and change in charge in amino acids that concentrate to a particular structure (beta strand versus non-beta strand regions, surface versus interior; see Supplementary Material, Fig. S1). Finally, we found no obvious correlation between the magnitude of change in charge by mutation and disease onset or duration (see Supplementary Material, Fig. S2). Collectively, these data indicate that changes in protein charge cannot be the only determining factor that drives aggregation of mutant SOD1 and that there is no obvious correlation between charge changes and a disease feature.
Thermostability is viewed as a measure of the inherent stability of protein conformation. The mutants we identify here as slow to aggregate (H80R, D101N, D125H, E133ΔE, S134N and V148I) exhibit some of the characteristics of WT SOD1 (see Supplementary Material, Fig. S3). Biophysically, these mutants show similar H/D exchange kinetics (which assesses the exposure of residues in the folded protein to solvent) to WT SOD1 as apo-proteins (34), as well as high levels of activity (only when metallated) and high thermostability (in both fully metallated or demetallated states) (34) (see Supplementary Material, Table S2). In contrast, the mutants that show higher aggregation propensities generally show reduced thermostability (see Supplementary Material, Table S2). However, in our tabulation of data on aggregation levels at 24 h in comparison to thermostability, we noted no obvious association between low thermostability of the apo-protein (lacking Cu) and aggregation propensity (see Supplementary Material, Table S2).
Sorting out the role of metal binding in aggregation propensity is somewhat complicated in that different approaches of assessing metal binding have yielded different outcomes depending upon whether the protein was produced and isolated from yeast or Sf21 insect cell expression systems. From the available data, we find little evidence to relate poor metal-binding capacity to the formation of detergent-insoluble SOD1 aggregates. Three of the low aggregating SOD1 mutants identified here appear to bind metals weakly (see Supplementary Material, Table S2). Moreover, experimental SOD1 mutants in which copper binding ligands have been abolished do not present a higher propensity to form detergent-insoluble aggregates than other SOD1-associated ALS mutations (23). Thus, it does not appear that low metal-binding capacity correlates to high-aggregation propensity.
To date, the specific role or impact that aggregates of mutant SOD1 have on disease pathogenesis remains unclear. In some settings, aggregation of mutant protein has been suggested to reduce toxicity by concentrating an otherwise toxic protein to a specific subcellular compartment (35,36); however, evidence linking aggregates to toxicity is still largely correlative. All of the SOD1 ALS murine models that have been analyzed accumulate significant amounts of detergent-insoluble aggregates in tissues most affected by the disease process (12,15,23,24,37–40). In certain cases, in which the expression of mutant protein was low, disease development and/or aggregation was not observed (1,12). The two interpretations of these findings are that: (i) aggregation of mutant SOD1 is critical to the development of disease or (ii) some other process initiates disease onset and that cells damaged by the disease process are prone to aggregate mutant SOD1. Our cell model, however, suggests that a high propensity to aggregate is an inherent characteristic of ALS mutant SOD1. Our studies have shown that SOD1-associated ALS mutants display a wide variety of aggregation propensities that are dependent upon the type and location of the mutation. Thus, if aggregates have an important role in disease pathogenesis, then some characteristic of human disease should correlate to the aggregation propensity of the individual mutant.
As noted in Results, the mean age of disease onset in SOD1-associated ALS is between 45–47 years of age, and we find no obvious correlation between aggregation propensity and age of onset (Fig. 4). However, in our analysis of clinical data regarding mutations with reliable patient information (see Supplementary Material, Table S1), we have observed that in general there is an inverse relationship between high-aggregation propensity and disease duration (Fig. 5). Mutants with higher aggregation propensities compose a group of mutations in which the clinical data available is more abundant. In this set of mutants, survival intervals tend to be shorter; all exhibit a survival of less than 12 years, although most of them are characterized by survival times of 4 years or less. In contrast, SOD1 mutants with moderate aggregation propensities present survival times ranging 10–18 years. However, these relationships were not absolute as we found examples of mutants with moderate aggregation propensity in which disease duration is shorter than 10 years: I113T and L144F SOD1 (see Supplementary Material, Table S1, Fig. 5). The mutant I113T is known to have incomplete penetrance, thus it might be possible that additional factors regulate disease appearance in certain generations, which would be responsible for the large variability in disease duration among patients. Patients harboring the L144F mutation present complete penetrance, but exhibit a wide range of survival times (see Supplementary Material, Table S1). It is possible that as more data becomes available, the number of exceptions (low aggregation potential linked to rapid progression) may rise. However, when we include in our analysis of patient data, all available data including cases with only one or two patients (see Supplementary Material, Fig. S4), we continue to observe that high-aggregation propensity shows a strong association with disease of short duration (less than 5 years). Mutants that show low or moderate aggregation potentials are essentially unpredictable. Overall, we regard the high propensity of a particular mutant to aggregate as a risk factor for a more rapidly progressing disease.
All of our data in the present study, as well as data derived from prior studies (23,25) have used the HEK293FT cell as the model system. We note that we were able to find one report in the literature in which inducible vectors were used in mouse NSC-34 cells to express a limited number of human mutant proteins (including A4V, C6F, H46R, G93A, C146R) (41). These investigators used a very similar approach of determining the levels of mutant SOD1 in detergent soluble and insoluble fractions. After 48 h of induced expression, they reported that A4V and G93A mutants showed ratios of insoluble to soluble SOD1 that were similar to what we report in our current study. We have previously examined the aggregation of the C6F and C146R mutants in HEK293FT cells and found high inherent rates of aggregation (25). Cozzolino reported a similar high rate of aggregation of these mutants in mouse NSC-34 cells. In contrast, the H46R mutant showed much lower levels of aggregated mutant SOD1 in NSC-34 cells; which again is similar to what we have previously reported for this mutant in our HEK293FT cell system (23). We also examined aggregation propensities of human SOD1 by transient transfection of mouse neuroblastoma N2a cells (Supplementary Material, Fig. S5). One of the more interesting pairs of mutants, D101G and D101N, showed similar aggregation propensities in the N2a cells as was observed in the HEK293FT cells. Thus, we think that our findings in HEK293FT cells reflect inherent aggregation propensities of these mutants that are manifest in other cell types. It is possible that there are factors unique to motor neurons that modulate aggregation and that such factors could significantly impact mutant SOD1 folding when present at physiological levels. However, we argue that the HEK293FT cell model, which is dependent upon high levels of expression, overwhelms most of the cellular systems that might otherwise modulate mutant SOD1 aggregation (proteasome degradation, chaperone levels, etc.) providing insight in the inherent propensity of these proteins to self associate. Even in the face of other modulating factors, the inherent propensity to aggregate would be the basic force behind aggregation. At this basic level, we find that mutants that exhibit a high-aggregation propensity are often associated with disease of short duration.
One other factor to consider is that most of the human cases for which we have significant data are examples of disease of relatively short duration. Because the longer duration cases are not equally represented, the apparent association between high-aggregation rates and short disease duration could be due to bias in data set. However, statistical analyses of the data indicate a non-random distribution of disease duration among the classes of mutants, with high-aggregation propensity more often being found in patients with short duration. Moreover, within the group of patients that show short duration, 9 of 12 mutants that exhibited high-aggregation potential are associated with disease durations of less than 5 years. However, we clearly identify mutants that in cell culture show aggregation propensities that do not fit with expectations for disease duration. Whether these exceptions are truly examples of mutants that aggregate slowly and yet are associated with rapidly progressing disease (e.g. D101N mutant) or mutants that aggregate rapidly and yet are associated with slowly progressing disease (e.g. E100K) is uncertain. These may be examples in which the cell culture system is not accurately predicting the in vivo situation or these may be examples in which other modifying factors overshadow the role of aggregation in disease duration.
The association between aggregation of mutant SOD1 and symptomatic disease duration, rather than onset, is a particularly intriguing finding. In comparison to other neurodegenerative diseases that have been associated with protein misfolding, SOD1-associated ALS appears unique. For example, in neurodegenerative disorders with expansions of glutamine repeats regions, there is an association between aggregation and age of disease onset rather than progression. Individuals with polyglutamine expansions in the huntingtin gene develop Huntington's disease (HD). In HD, the length of the polyglutamine tract strongly correlates with disease onset, the larger the expansion the earlier onset (42,43); and longer repeat lengths correlate with higher aggregation propensities of mutant huntingtin (44). In HD, disease duration is not noted to be variable and the length of the polyglutamine repeat does not correlate with disease duration (43). Another example is Alzheimer's disease (AD), which is defined by pathological accumulation of aggregated of β-amyloid peptides in the brain. Familial forms of AD linked to mutations in amyloid precursor protein or presenilin 1, lead to a net increase in the amount of the highly aggregating β-amyloid peptide 1–42 (45,46). These inherited forms of AD generally show much earlier disease onsets, but disease durations are similar to the sporadic disease (47,48). Thus, in other examples of neurodegenerative disease associated with protein misfolding and aggregation, the aggregation rates of the causative protein appears to best correlate with disease onset.
We provide evidence that mutations in SOD1 that are associated with a high-aggregation propensity generally predict a more rapidly progressing disease. However, several exceptions are noted, and it becomes less predictable for mutants with lower aggregation propensities. Thus, it appears that at least two factors regulate disease progression; one of which is high inherent aggregation propensity of SOD1 mutant protein. In this view, we would categorize high-aggregation propensity as a risk factor for rapidly progressing disease. Studies from our laboratory, and others, have demonstrated that, in mouse models of SOD1-associated ALS, the most significant accumulations of large mutant SOD1 aggregates occurs late in disease (12,13,15,23,24,37–40). Importantly, significant accumulation of mutant SOD1 aggregates occurs well after the appearance of multiple pathological abnormalities in these mouse models (40). Thus, we concur with others that have suggested that there must be a toxic form of mutant SOD1 that is distinct from larger protein aggregates with these entities initiating disease. However, aggregation of the mutant protein may be one of the forces capable of modulating the rate of disease progression. The mechanisms by which aggregation of mutant SOD1 promotes disease progression are unclear at present. One hypothesis that could apply is the idea that the accumulation of SOD1 aggregates impairs the ability of the cell to maintain protein homeostasis (49). The idea is that as chaperones become occupied in unproductive attempts to dissolve protein aggregates, these activities are not available for productive functions in protein folding. There is also evidence that cells accumulating protein aggregates show reduced proteasome function (50). Together, the disruption of these critical protein homeostatic processes could cause a feed-forward cascade of impairment in protein folding and metabolism that could underlie the rapid progression of disease that is seen in many of the mouse models. If these mechanisms apply, then compounds that modulate mutant SOD1 aggregation could be useful therapeutics in slowing the progression of this disease in humans.
MATERIALS AND METHODS
SOD1 cDNA constructs
The majority of the methods in this paper have been previously described (22,25). All mutant SOD1 proteins were expressed in HEK293FT cells using the pEF-BOS expression vector. A few of the SOD1 mutants cDNAs used have been previously created into the pEF-BOS system (WT, A4V, G37R, G41D, G85R, G93A and G93C SOD1) (23,51); however, many other mutants were kindly provided, in the YEp351 yeast vector, by Dr Stephen Holloway (University of Texas Health Sciences Center, San Antonio, TX, USA), which we subcloned into our pEF-BOS mammalian vector. The sequences of all mutants in the pEF-BOS vector were verified by automated sequence analysis.
Transfections and detergent extraction and centrifugation assay
All mutants were transfected, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), into HEK293FT cells for 24 or 48 h, as indicated in figure legends. Then cells were collected, washed in 1× PBS and lysed by sonication in non-ionic detergent (10 mm Tris, pH 7.5; 1 mm EDTA, pH 8.0; 100 mm NaCl; 0.5% NP-40, 1:100 v/v protease inhibitor cocktail from Sigma, St Louise, MO, USA). Detergent soluble (S1) and insoluble (P2) fractions were separated by our previously described detergent extraction and centrifugation technique (22,25). Detergent extractions for each mutant were repeated a minimum of three times, up to 24 times for WT and A4V SOD1 controls.
SDS–PAGE and immunoblotting
In each case, S1 (5 µg) and P2 (20 µg) fractions for each mutant are boiled in Laemmli sample buffer that contains 5% β-mercaptoethanol (βME) and electrophoresed in 18% SDS–PAGE gels. Transfer onto nitrocellulose membranes lasted only 2 h and immunoblotting by human SOD1 at 1:2500 was done overnight. PBS-T (1× PBS, 0.1% Tween 20) was used for membrane washes between antibody incubations. As secondary antibody we used a goat anti-rabbit antibody at 1:5000 dilution (KPL, Gaithersburg, MD, USA) for 1 h before developing with ECL reagents (Thermo Scientific Inc., Rockford, IL, USA).
Statistical analysis
The aggregation propensity of SOD1 mutants is defined as the ratio of band intensity of detergent-insoluble versus soluble fractions (P2/S1) (22,23,25). As means to compare SOD1 mutants in different gels, the mean aggregation propensities were normalized by assigning a value of one for our A4V SOD1 positive control (22,23,53). Differences between WT and mutant SOD1 proteins were assessed by paired Student _t_-tests. Correlations and _t_-tests were performed using GraphPad Prism 5.0 software (San Diego, CA, USA).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
[Supplementary Data]
FUNDING
This work was supported by a grant from the National Institutes of Neurological Disorders and Stroke [P01 NS049134-01 to P.J.H. and D.R.B.; and by R01 NS39112 to P.J.H.]; the Judith and Jean Pape Adams Charitable Foundation (P.J.H.). This project has also been generously supported by the Swedish Brain Research Foundation, the Hållstens Research Foundation, the Swedish Medical Society and the Swedish association for the neurologically disabled. Funding to pay the Open Access publication charges for this article was provided by NINDS P01 NS049134.
ACKNOWLEDGEMENTS
We thank Stephen Holloway (University of Texas Health Science Center at San Antonio) and Hilda Slunt-Brown (McKnight Brain Institute at University of Florida, Gainesville) for their efforts in generating some of the cDNA mutants of SOD1 used in this study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Deng H.X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G.H., Khan M.S., Hung W.Y., Bigio E.H., et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong P.C., Pardo C.A., Borchelt D.R., Lee M.K., Copeland N.G., Jenkins N.A., Sisodia S.S., Cleveland D.W., Price D.L. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki S., Nagai M., Aoki M., Komori T., Itoyama Y., Iwata M. Motor neuron disease in transgenic mice with an H46R mutant SOD1 gene. J. Neuropathol. Exp. Neurol. 2007;66:517–524. doi: 10.1097/01.jnen.0000263868.84188.3b. [DOI] [PubMed] [Google Scholar]
- 4.Nagai M., Aoki M., Miyoshi I., Kato M., Pasinelli P., Kasai N., Brown R.H., Jr, Itoyama Y. Rats expressing human cytosolic copper–zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J. Neurosci. 2001;21:9246–9254. doi: 10.1523/JNEUROSCI.21-23-09246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tobisawa S., Hozumi Y., Arawaka S., Koyama S., Wada M., Nagai M., Aoki M., Itoyama Y., Goto K., Kato T. Mutant SOD1 linked to familial amyotrophic lateral sclerosis, but not wild-type SOD1, induces ER stress in COS7 cells and transgenic mice. Biochem. Biophys. Res. Commun. 2003;303:496–503. doi: 10.1016/s0006-291x(03)00353-x. [DOI] [PubMed] [Google Scholar]
- 6.Bruijn L.I., Becher M.W., Lee M.K., Anderson K.L., Jenkins N.A., Copeland N.G., Sisodia S.S., Rothstein J.D., Borchelt D.R., Price D.L., Cleveland D.W. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 7.Bruijn L.I., Beal M.F., Becher M.W., Schulz J.B., Wong P.C., Price D.L., Cleveland D.W. Elevated free nitrotyrosine levels, but not protein-bound nitrotyrosine or hydroxyl radicals, throughout amyotrophic lateral sclerosis (ALS)-like disease implicate tyrosine nitration as an aberrant in vivo property of one familial ALS-linked superoxide dismutase 1 mutant. Proc. Natl Acad. Sci. USA. 1997;94:7606–7611. doi: 10.1073/pnas.94.14.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonsson P.A., Graffmo K.S., Brannstrom T., Nilsson P., Andersen P.M., Marklund S.L. Motor neuron disease in mice expressing the wild type-like D90A mutant superoxide dismutase-1. J. Neuropathol. Exp. Neurol. 2006;65:1126–1136. doi: 10.1097/01.jnen.0000248545.36046.3c. [DOI] [PubMed] [Google Scholar]
- 9.Gurney M.E. Transgenic-mouse model of amyotrophic lateral sclerosis. N. Engl. J. Med. 1994;331:1721–1722. doi: 10.1056/NEJM199412223312516. [DOI] [PubMed] [Google Scholar]
- 10.Howland D.S., Liu J., She Y., Goad B., Maragakis N.J., Kim B., Erickson J., Kulik J., DeVito L., Psaltis G., et al. Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS) Proc. Natl Acad. Sci. USA. 2002;99:1604–1609. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedlander R.M., Brown R.H., Gagliardini V., Wang J., Yuan J. Inhibition of ICE slows ALS in mice. Nature. 1997;388:31. doi: 10.1038/40299. [DOI] [PubMed] [Google Scholar]
- 12.Wang J., Xu G., Slunt H.H., Gonzales V., Coonfield M., Fromholt D., Copeland N.G., Jenkins N.A., Borchelt D.R. Coincident thresholds of mutant protein for paralytic disease and protein aggregation caused by restrictively expressed superoxide dismutase cDNA. Neurobiol. Dis. 2005;20:943–952. doi: 10.1016/j.nbd.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Xu G., Li H., Gonzales V., Fromholt D., Karch C., Copeland N.G., Jenkins N.A., Borchelt D.R. Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: alphaB-crystallin modulates aggregation. Hum. Mol. Genet. 2005;14:2335–2347. doi: 10.1093/hmg/ddi236. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe Y., Yasui K., Nakano T., Doi K., Fukada Y., Kitayama M., Ishimoto M., Kurihara S., Kawashima M., Fukuda H., et al. Mouse motor neuron disease caused by truncated SOD1 with or without C-terminal modification. Brain Res. Mol. Brain Res. 2005;135:12–20. doi: 10.1016/j.molbrainres.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson P.A., Ernhill K., Andersen P.M., Bergemalm D., Brannstrom T., Gredal O., Nilsson P., Marklund S.L. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 16.Awano T., Johnson G.S., Wade C.M., Katz M.L., Johnson G.C., Taylor J.F., Perloski M., Biagi T., Baranowska I., Long S., et al. Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2009;106:2794–2799. doi: 10.1073/pnas.0812297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boukaftane Y., Khoris J., Moulard B., Salachas F., Meininger V., Malafosse A., Camu W., Rouleau G.A. Identification of six novel SOD1 gene mutations in familial amyotrophic lateral sclerosis. Can. J. Neurol. Sci. 1998;25:192–196. doi: 10.1017/s0317167100034004. [DOI] [PubMed] [Google Scholar]
- 18.Hayward C., Brock D.J., Minns R.A., Swingler R.J. Homozygosity for Asn86Ser mutation in the CuZn-superoxide dismutase gene produces a severe clinical phenotype in a juvenile onset case of familial amyotrophic lateral sclerosis. J. Med. Genet. 1998;35:174. doi: 10.1136/jmg.35.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato M., Aoki M., Ohta M., Nagai M., Ishizaki F., Nakamura S., Itoyama Y. Marked reduction of the Cu/Zn superoxide dismutase polypeptide in a case of familial amyotrophic lateral sclerosis with the homozygous mutation. Neurosci. Lett. 2001;312:165–168. doi: 10.1016/s0304-3940(01)02212-1. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe M., Dykes-Hoberg M., Culotta V.C., Price D.L., Wong P.C., Rothstein J.D. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 21.Jonsson P.A., Graffmo K.S., Andersen P.M., Brannstrom T., Lindberg M., Oliveberg M., Marklund S.L. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 22.Prudencio M., Durazo A., Whitelegge J.P., Borchelt D.R. Modulation of mutant superoxide dismutase 1 aggregation by co-expression of wild-type enzyme. J. Neurochem. 2009;108:1009–1018. doi: 10.1111/j.1471-4159.2008.05839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N.G., Jenkins N.A., Borchelt D.R. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Xu G., Borchelt D.R. Mapping superoxide dismutase 1 domains of non-native interaction: roles of intra- and intermolecular disulfide bonding in aggregation. J. Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karch C.M., Borchelt D.R. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cudkowicz M.E., kenna-Yasek D., Sapp P.E., Chin W., Geller B., Hayden D.L., Schoenfeld D.A., Hosler B.A., Horvitz H.R., Brown R.H. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann. Neurol. 1997;41:210–221. doi: 10.1002/ana.410410212. [DOI] [PubMed] [Google Scholar]
- 27.Rosen D.R., Bowling A.C., Patterson D., Usdin T.B., Sapp P., Mezey E., kenna-Yasek D., O'Regan J., Rahmani Z., Ferrante R.J. A frequent ala 4 to val superoxide dismutase-1 mutation is associated with a rapidly progressive familial amyotrophic lateral sclerosis. Hum. Mol. Genet. 1994;3:981–987. doi: 10.1093/hmg/3.6.981. [DOI] [PubMed] [Google Scholar]
- 28.Arisato T., Okubo R., Arata H., Abe K., Fukada K., Sakoda S., Shimizu A., Qin X.H., Izumo S., Osame M., Nakagawa M. Clinical and pathological studies of familial amyotrophic lateral sclerosis (FALS) with SOD1 H46R mutation in large Japanese families. Acta Neuropathol. (Berl) 2003;106:561–568. doi: 10.1007/s00401-003-0763-5. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q., Johnson J.L., Agar N.Y., Agar J.N. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS. Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw B.F., Valentine J.S. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem. Sci. 2007;32:78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Sandelin E., Nordlund A., Andersen P.M., Marklund S.S., Oliveberg M. Amyotrophic lateral sclerosis-associated copper/zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins. J. Biol. Chem. 2007;282:21230–21236. doi: 10.1074/jbc.M700765200. [DOI] [PubMed] [Google Scholar]
- 32.Calamai M., Taddei N., Stefani M., Ramponi G., Chiti F. Relative influence of hydrophobicity and net charge in the aggregation of two homologous proteins. Biochemistry. 2003;42:15078–15083. doi: 10.1021/bi030135s. [DOI] [PubMed] [Google Scholar]
- 33.Chiti F., Calamai M., Taddei N., Stefani M., Ramponi G., Dobson C.M. Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. Proc. Natl Acad. Sci. USA. 2002;99(Suppl. 4):16419–16426. doi: 10.1073/pnas.212527999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez J.A., Shaw B.F., Durazo A., Sohn S.H., Doucette P.A., Nersissian A.M., Faull K.F., Eggers D.K., Tiwari A., Hayward L.J., Valentine J.S. Destabilization of apoprotein is insufficient to explain Cu,Zn-superoxide dismutase-linked ALS pathogenesis. Proc. Natl Acad. Sci. USA. 2005;102:10516–10521. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 36.Gong B., Lim M.C., Wanderer J., Wyttenbach A., Morton A.J. Time-lapse analysis of aggregate formation in an inducible PC12 cell model of Huntington's disease reveals time-dependent aggregate formation that transiently delays cell death. Brain Res. Bull. 2008;75:146–157. doi: 10.1016/j.brainresbull.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Xu G., Gonzales V., Coonfield M., Fromholt D., Copeland N.G., Jenkins N.A., Borchelt D.R. Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol. Dis. 2002;10:128–138. doi: 10.1006/nbdi.2002.0498. [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Xu G., Borchelt D.R. High molecular weight complexes of mutant superoxide dismutase 1: age-dependent and tissue-specific accumulation. Neurobiol. Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- 39.Johnston J.A., Dalton M.J., Gurney M.E., Kopito R.R. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karch C.M., Prudencio M., Winkler D.D., Hart P.J., Borchelt D.R. Role of mutant SOD1 disulfide oxidation and aggregation in the pathogenesis of familial ALS. Proc. Natl Acad. Sci. USA. 2009;12:7774–7779. doi: 10.1073/pnas.0902505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cozzolino M., Amori I., Pesaresi M.G., Ferri A., Nencini M., Carri M.T. Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:866–874. doi: 10.1074/jbc.M705657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Persichetti F., Srinidhi J., Kanaley L., Ge P., Myers R.H., D'Arrigo K., Barnes G.T., MacDonald M.E., Vonsattel J.P., Gusella J.F. Huntington's disease CAG trinucleotide repeats in pathologically confirmed post-mortem brains. Neurobiol. Dis. 1994;1:159–166. doi: 10.1006/nbdi.1994.0019. [DOI] [PubMed] [Google Scholar]
- 43.Gusella J.F., MacDonald M.E. Huntington's disease: seeing the pathogenic process through a genetic lens. Trends Biochem. Sci. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G.P., Lehrach H., Wanker E.E. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc. Natl Acad. Sci. USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duff K., Eckman C., Zehr C., Yu X., Prada C.M., Perez-tur J., Hutton M., Buee L., Harigaya Y., Yager D., et al. Increased amyloid-beta42(43) in brains of mice expressing mutant presenilin 1. Nature. 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 46.Borchelt D.R. Metabolism of presenilin 1: influence of presenilin 1 on amyloid precursor protein processing. Neurobiol. Aging. 1998;19:S15–S18. doi: 10.1016/s0197-4580(98)00026-8. [DOI] [PubMed] [Google Scholar]
- 47.Bertram L., Tanzi R.E. 30 years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat. Rev. Neurosci. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- 48.Bird T.D. Genetic aspects of Alzheimer disease. Genet. Med. 2008;10:231–239. doi: 10.1097/GIM.0b013e31816b64dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gidalevitz T., Krupinski T., Garcia S., Morimoto R.I. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS. Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 51.Borchelt D.R., Lee M.K., Slunt H.S., Guarnieri M., Xu Z.S., Wong P.C., Brown R.H., Jr, Price D.L., Sisodia S.S., Cleveland D.W. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl Acad. Sci. USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zetterstrom P., Stewart H.G., Bergemalm D., Jonsson P.A., Graffmo K.S., Andersen P.M., Brannstrom T., Oliveberg M., Marklund S.L. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc. Natl Acad. Sci. USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borchelt D.R., Wong P.C., Becher M.W., Pardo C.A., Lee M.K., Xu Z.S., Thinakaran G., Jenkins N.A., Copeland N.G., Sisodia S.S., et al. Axonal transport of mutant superoxide dismutase 1 and focal axonal abnormalities in the proximal axons of transgenic mice. Neurobiol. Dis. 1998;5:27–35. doi: 10.1006/nbdi.1998.0178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]