The genomic basis of trophic strategy in marine bacteria (original) (raw)
Abstract
Many marine bacteria have evolved to grow optimally at either high (copiotrophic) or low (oligotrophic) nutrient concentrations, enabling different species to colonize distinct trophic habitats in the oceans. Here, we compare the genome sequences of two bacteria, Photobacterium angustum S14 and Sphingopyxis alaskensis RB2256, that serve as useful model organisms for copiotrophic and oligotrophic modes of life and specifically relate the genomic features to trophic strategy for these organisms and define their molecular mechanisms of adaptation. We developed a model for predicting trophic lifestyle from genome sequence data and tested >400,000 proteins representing >500 million nucleotides of sequence data from 126 genome sequences with metagenome data of whole environmental samples. When applied to available oceanic metagenome data (e.g., the Global Ocean Survey data) the model demonstrated that oligotrophs, and not the more readily isolatable copiotrophs, dominate the ocean's free-living microbial populations. Using our model, it is now possible to define the types of bacteria that specific ocean niches are capable of sustaining.
Keywords: microbial adaptation and ecology, microbial genomics and metagenomics, monitoring environmental health, trophic adaptation
The marine environment is the largest habitat on Earth, accounting for >90% of the biosphere by volume and harboring microorganisms responsible for ≈50% of total global primary production. Within this environment, marine bacteria (and archaea) play a pivotal role in biogeochemical cycles while constantly assimilating, storing, transforming, exporting, and remineralizing the largest pool of organic carbon on the planet (1).
Nutrient levels in pelagic waters are not uniform. Large expanses of water are relatively nutrient depleted (e.g., oligotrophic open ocean water), whereas other zones are relatively nutrient rich (e.g., copiotrophic coastal and estuarine waters). Local variations in nutrient content can occur because of physical processes, including upwelling of nutrient rich deep waters or aeolian and riverine deposition, or biological processes such as phytoplankton blooms or aggregation of particulate organic matter. In addition, heterogeneity in ocean waters is not limited to gross differences in nutrient concentrations, but extends to microscale patchiness that occurs throughout the continuum of ocean nutrient concentrations (2).
In ecological terms, bacteria are generally defined as r-strategists, having a small body, short generation time, and highly dispersible offspring. Although this strategy is broadly true compared with macroorganisms, bacteria have evolved a wide range of growth and survival strategies to maximize reproductive success. In particular, nutrient type and availability have provided strong selective pressure for defining lifestyle strategies among marine bacteria. However, although a large number of copiotrophic marine organisms (and fewer oligotrophs) have been cultured, the study of trophic strategy has been impaired by a lack of understanding of the molecular basis of adaptation. Here, we show that trophic strategy is strongly reflected in genomic content and genomic signatures can be used as a proxy for determining the ecological characteristics of uncultured microorganisms, thereby allowing the assessment of trophic life strategies from bacterial genome sequences.
Results and Discussion
Defining Genomic Signatures of Copiotrophs and Oligotrophs.
We sequenced and annotated the genomes of two marine bacteria, the copiotroph Photobacterium angustum S14 and the oligotroph Sphingopyxis alaskensis RB2256, which are model representatives of the two major classes of heterotrophic marine bacteria and for which extensive physiological data are available (Table 1). P. angustum S14 was isolated from surface, coastal waters in Botany Bay (Sydney), New South Wales, Australia (3). It is characterized by a relatively large cell size (growing cells >1 μm3), and displays a “feast and famine” strategy with rapid rates of growth (>1 h_−1_) in rich media and pronounced size reduction and other adaptive traits in response to nutrient limitation induced stasis. S. alaskensis RB2256 was isolated by extinction dilution as a numerically abundant member (>10_5_ cells mL_−1_) of surface waters (10 m depth) in Resurrection Bay, Alaska and the North Sea, and a closely related strain, AF01, was isolated from 350 m deep, oligotrophic ocean waters near Japan (4–6). Oligotrophic traits of S. alaskensis include the ability to grow slowly with a constant maximum specific growth rate (<0.2 h_−1_) on low concentrations (nanomolar) of substrates and maintain a relatively small cell volume (<0.1 μm_3_) and constant cell size in the shift between starvation and growth conditions (6). The small size provides a mechanism for avoidance of predation, and the high surface area-to-volume ratio allows for efficient nutrient acquisition through high-affinity, broad-specificity uptake systems that are predicted to enable S. alaskensis to be able to achieve doubling times typically observed for bacteria in oligotrophic waters (6, 7).
Table 1.
Physiological characteristics of the model copiotroph P. angustum S14 and oligotroph S. alaskensis RB2256
| Physiological characteristic | P. angustum S14 | S. alaskensis RB2256 |
|---|---|---|
| Trophic strategy | Copiotroph | Oligotroph |
| Growth strategy | Feast and famine | Equilibrium |
| Cell size | Large (>1 μm3) | Small (<0.1 μm3) |
| Maximum growth rate | >1 h−1 | <0.2 h−1 |
| Growth rate dependence on media richness | Yes | No |
| Starvation cross-protection to high levels of other stress inducing agents | Yes | No |
| Growing cells inherently resistant to stress inducing agents | No | Yes |
| Lag phase after starvation | Yes | No |
| _rpoS_-dependent reductive cell division | Yes | No |
| Consistent cell yield during nutrient limited growth | No | Yes |
Genomic markers derived from the comparison of the S. alaskensis and P. angustum genomes were expanded and validated by searching 32 additional genome sequences, representing (i) related species of Vibrionaceae and Sphingomonadaceae, (ii) other known aquatic copiotrophs and oligotrophs, and (iii) marine bacteria with high or low rRNA operon copy numbers. A total of 43 genetic markers were identified (Fig. S1 and Table 2) that accurately predict trophic strategy and are consistent with physiological adaptations for enhancing microbial reproduction under specific nutrient regimes. The findings accord well with the understanding of the physiology of P. angustum S14 and S. alaskensis RB2256. Furthermore, they validate many of the characteristics for nutrient uptake and utilization of oligotrophic organisms that were predicted by a comprehensive group report at the Dahlem Conference on life under conditions of low nutrient concentrations ≈30 years ago (8) and greatly extend insight into the lifestyles of copiotrophic and oligotrophic bacteria.
Table 2.
Genomic features defining a lifestyle
| Marker | Copiotroph | Oligotroph | Median |
|---|---|---|---|
| Genome size | Large (4,798,216 bp) | Small (3,850,272 bp) | 4,367,642 bp |
| rRNA operon number | Many (9) | Few (1) | 3 |
| Multiple localizations* | Low (3.431%) | High (4.11%) | 3.681% |
| Cytoplasmic* | Low (58.91%) | High (60.735%) | 60.111% |
| Cytoplasmic membrane* | High (28.654%) | Low (26.778%) | 28.577% |
| Periplasmic* | High (3.656%) | Low (2.624%) | 3.348% |
| Outer membrane* | High (5.039%) | Low (3.43%) | 4.330% |
| Extracellular* | High (1.232%) | Low (0.589%) | 0.873% |
| Prophages | Many (0–10)† | Few (0–5)† | 1 |
| Repeats within CRISPRs | Many (0–59)† | Few (0–3)† | 0 |
| COG category N (cell motility) | High (3.311%) | Low (1.006%) | 1.553% |
| COG category T (signal transduction mechanisms) | High (7.071%) | Low (3.632%) | 5.405% |
| COG category V (defense mechanisms) | High (1.474%) | Low (1.236%) | 1.352% |
| COG category K (transcription) | High (7.527%) | Low (6.621%) | 7.082% |
| COG category Q (secondary metabolites biosynthesis, transport, and catabolism) | Low (2.173%) | High (3.544%) | 2.929% |
| COG category I (Lipid transport and metabolism) | Low (2.958%) | High (4.408%) | 4.164% |
| COG0110 (acetyltransferase: isoleucine patch superfamily) | High (0.166%) | Low (0.068%) | 0.082% |
| COG0183 (acetyl-CoA acetyltransferase) | Low (0.114%) | High (1.171%) | 0.124% |
| COG0243 (anaerobic dehydrogenases, typically selenocysteine-containing) | High (0.179%) | Low (0.0675%) | 0.085% |
| COG0318 (Acyl-CoA synthetases [AMP-forming/AMP-acid ligases II) | Low (0.149%) | High (0.235%) | 0.152% |
| COG0483 (archaeal fructose-1,6-bisphosphatase and related enzymes of inositol monophosphatase family) | Low (0.033%) | High (0.115%) | 0.064% |
| COG0583 (transcriptional regulator) | High (1.489%) | Low (0.371%) | 1.016% |
| COG0596 (predicted hydrolases or acyltransferases: α /β hydrolase superfamily) | Low (0.327%) | High (0.514%) | 0.382% |
| COG0625 (GST) | Low (0.298%) | High (0.342%) | 0.298% |
| COG0737 (5′-nucleotidase/2′,3′-cyclic phosphodiesterase and related esterases) | High (0.098%) | Low (0.035%) | 0.085% |
| COG0814 (amino acid permeases) | High (0.114%) | Low (0%) | 0% |
| COG1024 (enoyl-CoA hydratase/carnithine racemase) | Low (0.149%) | High (0.308%) | 0.194% |
| COG1028 (dehydrogenases with different specificities: related to short-chain alcohol dehydrogenases) | Low (0.476%) | High (1.135%) | 0.932% |
| COG1228 (imidazolonepropionase and related amidohydrolases) | Low (0.064%) | High (0.089%) | 0.073% |
| COG1263 (phosphotransferase system IIC components, glucose/maltose/_N_-acetylglucosamine-specific) | High (0.036%) | Low (0%) | 0% |
| COG1680 (β -lactamase class C and other penicillin binding proteins) | Low (0.059%) | High (0.115%) | 0.061% |
| COG1804 (predicted acyl-CoA transferases/carnitine dehydratase) | Low (0%) | High (0.072%) | 0% |
| COG1960 (acyl-CoA dehydrogenases) | Low (0.186%) | High (0.417%) | 0.267% |
| COG2124 (cytochrome P450) | Low (0%) | High (0.078%) | 0.018% |
| COG2200 (FOG:EAL domain) | High (0.372%) | Low (0.048%) | 0.157% |
| COG2207 (AraC-type DNA-binding domain-containing proteins) | High (0.365%) | Low (0.245%) | 0.338% |
| COG2852 (very-short-patch-repair endonuclease) | Low (0%) | High (0.024%) | 0% |
| COG3293 (transposase and inactivated derivatives) | Low (0%) | High (0.073%) | 0% |
| COG3325 (chitinase) | High (0.06%) | Low (0%) | 0% |
| COG3386 (gluconolactonase) | Low (0%) | High (0.065%) | 0.032% |
| COG3710 (DNA-binding winged-HTH domains) | High (0.034%) | Low (0%) | 0% |
| COG3773 (Cell wall hydrolases involved in spore germination) | Low (0%) | High (0.032%) | 0% |
| COG3920 (Signal transduction histidine kinase) | Low (0%) | High (0.024%) | 0% |
Specific clusters of orthologous groups (COGs) that were consistently overrepresented in copiotrophs included components of phosphotransferase systems (PTS) that are central for regulation and transport of sugars (COG1263, COG1299), Na_+_ transporters (COG0733), and a wide array of other highly specific transporters (COG0697, COG1292, COG2116, COG2704) and permeases (COG0697, COG0814, COG1114, COG1275, COG1972, COG2271, COG3104). Oligotrophs may have evolved to minimize the number of energy-intensive transporters and instead rely on a relatively smaller number of broad-specificity and sufficiently high-affinity ATP-binding cassette (ABC) transporters. This genomic signature is concordant with the observation that oligotrophs, including S. alaskensis (7), use broad-specificity, multifunctional high-affinity uptake systems (9).
Our analysis predicts that copiotrophs have a larger diversity of proteins localized in the outer membrane, which is the case even after normalization to the total number of ORFs (P < 0.05). The high number of outer membrane localized proteins might explain the larger number of predicted prophages found in copiotrophic genomes (also see last four paragraphs of this section). Surface receptors are needed for phage attachment, and a larger number and variety of targets are likely to increase the opportunity for establishing phage infection.
The proportion of extracytoplasmic proteins (cytoplasmic membrane, periplasmic, outer membrane, and extracellular) is also higher in copiotrophs. These results do not infer that oligotrophs have fewer transporters or periplasmic proteins present in the cell (because higher levels of expression from fewer genes could compensate for this). In fact, the metaproteome of the Sargasso Sea is overrepresented in SAR11 peptides involved in transport and periplasmic substrate binding (10). However, oligotrophs do have a reduced variety. Copiotrophs are therefore characterized by transporter diversification and specialization (e.g., PTS), and oligotrophs are characterized by transporter protein minimization.
The relatively higher number of secreted proteins in copiotrophs is consistent with the higher ectoenzymatic activity observed in particle attached (mainly copiotrophic) vs. free-living (mainly oligotrophic) bacteria (11, 12). The genomic analysis showed an overrepresentation across all copiotrophic datasets of enzymes such as chitinases (COG3325) and collagenases (COG0826). Chitin is derived from arthropod exoskeletons and is a major component of marine snow. Particle-associated bacteria are likely to derive more value from chitinases (and other types of hydrolases that attack polymers in marine snow), than free-living oligotrophic bacteria. Higher metabolic turnover rates and anaerobic conditions known to be generated in particles (13) and aggregated microenvironments may also select for marine bacteria that possess efficient rates of respiration using a variety of alternative electron acceptors. Consistent with this idea, genomes of copiotrophs are overrepresented in highly efficient dissimilatory selenocysteine-containing dehydrogenases (COG0243), and to a lesser extent, components of alternative electron transport chains including COG0716 (flavodoxins), COG2863 (cytochrome c553), and COG3005 (nitrate/trimethylamine _N_-oxide reductases, membrane-bound tetraheme cytochrome c subunit).
In general, copiotrophs are enriched in COGs involved in motility (N), defense mechanisms (V), transcription (K), and signal transduction (T) (Table 2 and Fig. S1). The capacity of copiotrophs to rapidly and tightly regulate metabolism and use energetically expensive transporters for nutrient acquisition is consistent with the presence of a high proportion of COGs related to transcription and signal transduction. A particularly strong bias is evident for COG0583 (transcriptional regulator), COG2207 (AraC-type DNA-binding domain-containing proteins), and COG3710 [DNA-binding winged-helix–turn–helix (HTH) domains]. Consistent with predictions by Poindexter (14) that copiotrophic bacteria are likely to display an extensive range of cellular responses to environmental stimuli, regulators with FOG:GGDEF domains (COG2199) and FOG:EAL domains (COG2200) are more abundant in copiotrophs. These domains are involved in the modulation of the synthesis and hydrolysis of the intracellular concentration of the effector molecule, cyclic diguanylate (c-diGMP, bis(3′,5′)-cyclic diguanylic acid), which in turn regulates expression of genes involved in a number of important growth and survival phenotypes (e.g., biofilms, virulence) (15, 16). Compared with oligotrophs, copiotrophs appear to integrate a larger number of signal inputs through this pool of secondary messengers to regulate a range of downstream genes.
The overrepresentation of COGs involved in cell motility attests to the dependence of copiotrophs on accessing nutrient-enriched patches in the open ocean. Copiotrophs have evolved a considerable number of genes for motility and sensory systems to locate and efficiently exploit transient microscale nutrient sources. Bacterial chemotactic-signal transducers (COG0835, COG0840) and CheY-like receiver domains (COG0745, COG2197, COG3437, COG3706) respond to changes in the concentration of attractants and repellents in the environment, and the higher number and diversity of receptor domains provide a greater capacity for copiotrophs to sense signals and are likely to be important for cells to scavenge nutrients and/or avoid toxins and/or predators. With regards to the latter, the small size of oligotrophs may lessen the selective pressure for high-speed swimming as a means of avoiding grazers.
Copiotrophic genomes also contain significantly more acetyltransferases of the isoleucine patch superfamily (COG0110). This superfamily comprises several members involved in acetylation of antibiotics (e.g., chloramphenicol). There is just one of these genes in S. alaskensis RB2256 in contrast to seven in P. angustum S14. Chemical modification of antibiotics would facilitate resistance against certain types of antibiotics and provide advantage for copiotrophs during allelopathic interactions in crowded and dense, growth-competitive environments.
In contrast to copiotrophs, oligotrophs are enriched in COGs for lipid transport and metabolism (I) and secondary metabolite biosynthesis, transport, and catabolism (Q). The overrepresentation of genes involved in lipid transport and metabolism (I) may relate to the improved ATP yield derived from fats compared with sugars (e.g., approximately three times higher per g from fatty acids vs. glucose). A large array of gene families involved in the degradation of fatty acids is consistently overrepresented in oligotroph genomes, including COG0183 (acetyl-CoA acetyltransferase), COG0318 (acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II), COG1024 (enoyl-CoA hydratase/carnithine racemase), COG1804 (acyl-CoA transferase/carnitine dehydratase), and COG1960 (acyl-CoA dehydrogenase).
In addition to catabolic genes, genes involved in fatty acids biosynthesis are overrepresented in oligotrophs; i.e., most of the genes belonging to COG1028 are dehydrogenases with different specificities for short-chain alcohols. Also, a few of the genes in these clusters are involved in polyhydroxyalkanoate synthesis, indicating that lipids function as storage material in many oligotrophs, with lipid compounds mobilized under starvation conditions. It is possible that oligotrophs preferentially use lipids as immediate and stored sources of carbon and energy.
S. alaskensis RB2256 has an overrepresentation of genes important for the degradation of aromatic compounds (dienelactone hydrolases, COG0412; imidazolonepropionases, COG1228). Similarly, the oligotroph genome dataset consistently has a high number of genes within COG0625 (GST) that are involved in detoxification reactions, the degradation of xenobiotic compounds, and the catabolism of recalcitrant compounds for carbon and energy, and other gene groups (COG0179, COG1680, COG3485) involved in the degradation of several aromatic substrates including β-lactamic compounds, tyrosine, and catechol. In addition, cytochrome P450 genes (COG2124) are present in S. alaskensis RB2256 (six copies) and absent in P. angustum S14, and the high frequency by which these genes occur is a conserved feature of oligotroph genomes. P450 is a diverse family of proteins that perform a variety of oxidative reactions with both exogenous and endogenous substrates, including fatty acid oxidation, bioconversion of recalcitrant organic compounds, and macromolecular synthesis. The general overabundance of genes involved in secondary metabolite transport and metabolism (COG category Q) and P450 genes may relate to a capacity of oligotrophs to use aromatic compounds for growth and/or reflect a requirement for the detoxification of compounds imported by the broad-specificity, high-affinity transporters that are prevalent in oligotrophs. The additional genetic load required to detoxify a wide range of compounds may be maintained at the expense of regulatory systems and by the reduction of intergenic regions, as has been suggested for the genome of the oligotroph “_Candidatus Pelagibacter ubique_” (17).
Phage are thought to be important in oceanic environments for facilitating the transfer of small gene cassettes between hosts (18). By using a gene clustering method, seven potential prophages were identified in the genome of P. angustum S14 and none in S. alaskensis RB2256. Expanding this analysis to potential prophage regions in genomes of a larger number of aquatic bacteria (29 genomes with at least seven rRNA and 31 with one rRNA operon) revealed an average of 2.1 vs. 0.81, respectively (significant at P < 0.05 even after normalization to total genome size).
Copiotroph genomes also contain more repeats within clustered regularly interspaced short palindromic repeats (CRISPRs). CRISPRs are clusters of repeats interspersed with short phage-derived sequences that have been proposed to confer hosts with resistance to phages (19), and their abundance is therefore expected to correlate with resistance to phage infection. Our analysis of all 80 completed aquatic genomes that have at least one predicted prophage or one predicted CRISPR shows that the number of prophages is inversely correlated with the total number of repeats (Spearman rank correlation P < 0.01). Interestingly, although genomes of oligotrophs do not contain more repeats than those of the copiotrophs, they contain fewer predicted prophages, suggesting that oligotrophs have alternative mechanisms for resisting phage infection and/or lysogenization.
It appears that coupling occurs between trophic strategy and host interaction with prophages. Infection by lytic phages and adoption of “kill-the-winner” population dynamics occurs with copiotroph r-strategist hosts, whereas oligotroph K-strategists have evolved means of avoidance. It has previously been predicted that highly virulent lytic phages will take advantage of r-strategist hosts when they become transiently numerically abundant, whereas slow-growing members of the community will be infected by less virulent and possibly nonlytic phages (20). Lysogeny has been shown to depend on host growth rate (21), and copiotrophic environments have been generally found to have a higher frequency of infected cells and phage causing larger lytic burst sizes compared with oligotrophic environments (22).
However, it should also be noted that lysogenic phages with an excretion release strategy (Inoviridae, Rudiviridae, Plasmaviridae, Fuselloviridae) may be more prevalent in oligotrophs. The overrepresentation in the public databases of sequences from copiotroph-infecting phages with a lysis release strategy (primarily belonging to the Caudovirales group; Fig. S2), makes it difficult to identify prophages belonging to the types of phages that might infect oligotrophs.
Predicting the Lifestyle of a Bacterium from Genome Sequence Data.
The comparison of oligotrophs and copiotrophs enabled us to define a set of discriminative genomic features of trophic lifestyle (Table 2). To detect the variations existing within genomes spanning the continuum of trophic strategies and predict the lifestyle of a bacterium just from genome sequence data, we applied the genomic markers to a neural gas clustering method based on self-organizing maps (SOMs). The original set of oligotrophic and copiotrophic marine bacteria could be easily distinguished with this approach (Fig. 1). By changing the cluster granularity for higher-resolution analysis and adding 92 additional genomes of aquatic bacteria to the SOMs, we were able to detect more subtle variations in trophic strategy across the full range of genomes used (Fig. 2). Moreover, the inclusion of metagenomic datasets (see Materials and Methods) allowed the trophic strategy of the dominant microorganisms in whole environmental samples to be inferred.
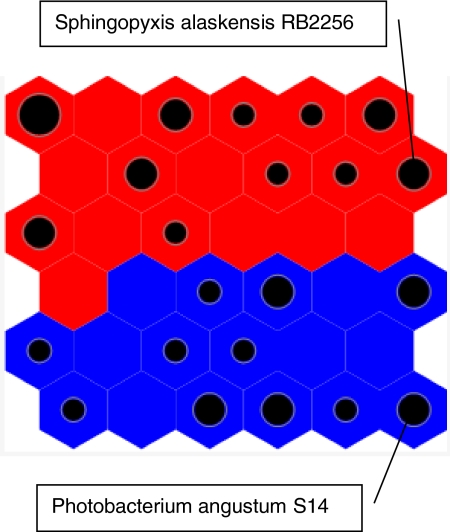
Fig. 1.
SOM showing the two clusters of organisms used to identify markers of trophic strategy. Maps were generated and visualized with Synapse as described in Materials and Methods. The clusters show a clear delimitation between copiotrophic (blue) and oligotrophic (red) genomes. The size of each black dot is proportional to the number of genomes occupying a cell in the map.
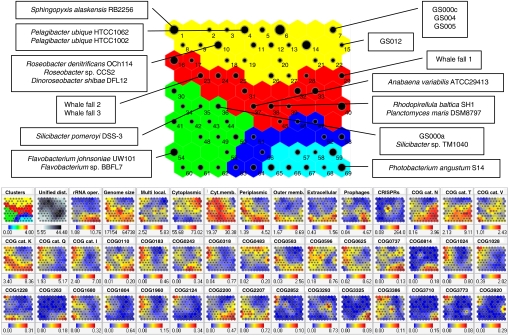
Fig. 2.
SOMs showing five clusters of organisms with different trophic strategies. Maps were generated and visualized with Synapse as described in Materials and Methods. (Upper) Enlargement of the clustering map with positions of relevant genomes marked. Yellow, extreme oligotrophs; red, moderate oligotrophs; green and blue, moderate copiotrophs; cyan, extreme copiotrophs. (Lower) The boxes labeled “Clusters” and “Unified dist.” show the clustering and distances, respectively, with all other boxes showing the component planes. The size of each black dot is proportional to the number of genomes occupying a cell in the map. The position on the map of all of the genomes and metagenomes used is provided in Table S2.
Well-known clades of marine copiotrophs, mostly belonging to the γ-proteobacterial orders Vibrionales and Alteromonadales were grouped within two clusters at one extreme of the spectrum (Fig. 2). At the other end of the spectrum, the genome sequences of two strains of the ubiquitous marine bacterium Candidatus Pelagibacter ubique clustered with S. alaskensis.
A number of observations were made with this approach. All cyanobacterial genomes clustered within the oligotrophs, with the open-ocean organisms (Prochlorococcus marinus) displaying more oligotrophic characteristics than those from freshwater (Nostoc and Anabaena). This distinction within the cyanobacteria is consistent with the inception of photosynthesis being associated with adaptation to environments deficient in carbon and energy (23); such conditions pervade in the bulk nutrient-depleted conditions of the open ocean where P. marinus resides (24). Planctomycetes, a group known to become dominant during phytoplankton blooms (25), also belonged to the cluster with Nostoc and Anabaena. The Planctomycetes are characterized by interesting genomic traits, having a single copy of the rRNA operon (a feature of oligotrophs), while also possessing large genomes (a feature of copiotrophs).
Away from the extremes, a distinct cluster containing members of the Cytophaga–Flavobacterium group, Silicibacter strains and some cold-adapted Alteromonadales (Pseudoalteromonas atlantica T6c, Colwellia psychrerythraea 34H) have trophic signatures that can be described as copiotrophic with some traits of oligotrophy. Silicibacter pomeroyi is proposed to have a physiology distinct from marine oligotrophs (26). The genomic clustering of S. pomeroyi and Silicibacter strain TM1040 was well separated from the closely related Roseobacter species (oligotroph), consistent with the lifestyle of moderate copiotrophs. The absence of chemotaxis genes suggests that S. pomeroyi may be limited in its ability to sense and exploit transient nutrient patches, and the lack of this important ability sets it apart from the extreme marine copiotrophs. However, the abundance of ABC transport genes in S. pomeroyi would facilitate an ability to capture a diverse range of low concentration substrates, including those that arise from phytoplankton blooms. The position of Silicibacter strains (α-proteobacteria) in the SOMs is one example of the strength of the model for identifying trophic differences that go beyond inferences of trophy based solely on phylogenetic affiliation; i.e., α-proteobacteria are generally considered oligotrophs compared with γ-proteobacteria as copiotrophs.
The SOMs analysis highlight that a broad range of trophic strategies have been selected throughout the evolution of marine species, perhaps even a continuum bridging oligotroph and copiotroph extremes. A benefit of our study is that by defining the genomic traits of the trophic extremes (i.e., S. alaskensis vs. P. angustum), our analytical approach can be used to rapidly assign the trophic strategy of a candidate bacterium from genomic data, and this fundamental understanding can form the basis for guiding ecological and physiological studies.
The metagenome data from the pelagic and coastal waters of the Global Ocean Survey (GOS) generally displayed characteristics of extreme oligotrophs, which is consistent with the sampling strategy (sequential size fractionation) that essentially excludes particle-attached bacteria (i.e., typically copiotrophs). Moreover the prevalence of genomic signatures of oligotrophs in the majority of the GOS data highlights the numerical dominance that free-living oligotrophs, rather than copiotrophs, have throughout ocean waters. The notable exception in the GOS data is GOS000a that has been shown to contain an atypically high number of reads from the copiotrophic genera Shewanella and Burkholderia (27). GOS000a clustered closely with Silicibacter strain TM1040 within the moderate copiotrophs. The whale-fall metagenomes clustered within the more moderate oligotrophs consistent with the higher nutrient availability of this environment when compared with the surrounding oligotrophic waters.
It was noteworthy that replacing S. alaskensis and P. angustum with genome sequences from other typical oligotrophs (e.g., Candidatus Pelagibacter ubique) and copiotrophs (e.g., Aeromonas hydrophila ATCC7966) produced a very similar trophic model with the same type of predictive outcomes. Furthermore, reducing by one the number of core characteristics (e.g., rRNA operon copy number) that define the model had little effect on the predicted outcomes, which illustrates that the model is robust and potentially useful for application to a wide range of datasets (e.g., low-coverage metagenome datasets and large datasets).
Conclusions and Perspectives
In this study we have shown that copiotrophs have higher genetic potential to sense, transduce, and integrate extracellular stimuli, characteristics that are likely to be crucial for their ability to fine-tune and rapidly respond to changing environmental conditions, such as sudden nutrient influx or depletion. Copiotrophs will also activate alternative catabolic pathways only after depletion of high-energy-yielding compounds have been exhausted. Oligotrophs, however, will need to detoxify and use a broader range of transported substances, whereas copiotrophs will avoid the problem by having a diverse array of transporters with much higher substrate specificity. Oligotrophs would be mainly oxybiontic, whereas copiotrophs would use a wide variety of electron acceptors.
With the distinct lack of ability to culture the majority of environmental microorganisms (particularly oligotrophs), metagenomics plays a critical role for inferring “who” is doing “what.” By defining trophy for metagenome datasets and applying functional metaanalyses (e.g., metatranscriptomics and metaproteomics), it will be possible to define the tempo and mode of microbial community processes on the basis of trophic adaptation. Continual development of the approach we describe here could include sequencing of additional genomes of individual microorganisms, particularly the inclusion of rare species, and marine archaea; genomic analysis of the latter would extend our conceptual understanding of the genomic basis of trophic adaptation beyond the domain, Bacteria, to the domain, Archaea, and enable trophic lifestyle to be inferred from metagenome data. Recent developments in single-cell manipulation and genome sequencing (28, 29) offer good avenues for generating the genomic signatures even of the unculturable majority of diverse marine microorganisms. Our method has the potential to be applied to 454 reads as long as reliable COG assignments and protein localization predictions can be made, which should be possible with the longer 454 reads (e.g., titanium).
Microbial observatories have been some of the last biological “sentinels” to be established for monitoring the impact of climate change and environmental health (30). We now provide the capacity to rapidly analyze the significant volume of DNA sequencing data that is being generated and effectively monitor the indigenous microbial populations for a defining characteristic of their growth, survival, and trophic lifestyle.
Materials and Methods
Strain Origin and Genome Sequencing.
P. angustum strain S14 (formerly Vibrio sp. S14) was isolated from a near-surface water sample (1 m depth) taken from Botany Bay, Sydney, Australia in June 1981 (3). Total genomic DNA of P. angustum S14 was extracted, and sequencing and annotation were performed at the J. Craig Venter Institute (JCVI). S. alaskensis strain RB2256 was isolated from Resurrection Bay, Seward, Alaska by extinction dilution (4). Total genomic DNA of S. alaskensis RB2256 was extracted, and sequencing and annotation were performed at the Department of Energy Joint Genome Institute (DOE JGI). Full details are provided in SI Text.
Comparison Genomes.
Completed microbial genomes were downloaded from the National Center for Biotechnology Information (NCBI) on December 19, 2007. The dataset was amended with genomes of marine bacteria individually downloaded from the DOE JGI or the JCVI web sites. A number of basic features of each genome were recorded: the total number of nucleotides, the number of rRNA operons, the genome size, and, if present, the number of extrachromosomal elements. For the draft genomes, the number of rRNA operons was estimated based on the 16S and 23S fragments present at the end of scaffolds. For the analysis of COG compositional differences, four datasets were compared: (i) the individual genomes of P. angustum S14 (copiotroph) vs. S. alaskensis RB2256 (oligotroph), (ii) a set of genomes within the Vibrionaceae (copiotrophs) vs. Sphingomonadaceae (oligotrophs), (iii) a selected set of genomes of known aquatic copiotrophs vs. known aquatic oligotrophs, and (iv) a set of selected genomes of aquatic bacteria with above- and below-average rRNA operons (Table S1). The fourth dataset was compiled based on the assumption that rRNA operon copy number correlates with the trophic lifestyle of a microorganism (31–33). The average rRNA copy number among the 549 completed bacterial genomes in the NCBI database (release of December 19, 2007) is 4.11 with a standard deviation of 2.86 and a skewed distribution. Therefore, comparison iv consisted of genomes of aquatic bacteria with seven or more rRNA operons (corresponding approximately to the 80th percentile of the distribution) that were considered copiotrophs vs. genomes with one rRNA operon (≈20th percentile) as oligotrophs (Table S1). We note that despite attempting to represent the full diversity of bacteria in each dataset the copiotroph dataset is overrepresented in γ-proteobacteria, whereas the oligotroph dataset is overrepresented in α-proteobacteria (Table S1). It is unclear whether this result reflects a correlation between trophic strategy and phylogeny or is caused by undersampling of marine microorganisms. For some purposes the four comparison sets were pooled into two groups and compared as described. Methods used for statistical analysis of COG composition, CRISPR and phage analysis, and protein localization are described in SI Text.
Generation of a Model for Trophic Strategy and Validation.
The values for each genome of the statistically significant features differentiating the trophic lifestyle (Table 2) were combined into a data matrix that was used to train 6 × 6 or 10 × 10 SOMs, depending on the size of the input data matrix, as implemented in Synapse (Peltarion). The construction of SOMs is a powerful technique for enabling the conversion of complex relationships between high-dimensional data into simple geometric relationships on a 2D display. The distance between any two elements on a grid is proportional to the dissimilarities between them. A clustering algorithm can be used to associate groups of elements on a SOM. In this study each cell contained one or more genomes whose positions on the map were defined by the values of 43 statistically relevant genomic markers. Different clustering granularities were set to infer intermediate trophic strategies. A total of eight ocean metagenomic samples, representing different oceanic environments, were also included in the data matrix. Five were selected from the Global Ocean Sampling expedition (34): GS000a (Sargasso station 13/11) and GS000c (Sargasso station 3) collected from the open ocean, GS004 collected from a coastal environment outside Halifax (Nova Scotia), GS005 collected from the embayment of Bedford basin (Nova Scotia), and GS012 collected from an estuary of the Chesapeake Bay. Three were representatives of the scavenging microbial communities surrounding a deep-sea whale carcass (whale fall) (35). The inclusion of the metagenomic samples into the matrix was accomplished by predicting ORF boundaries from all unassembled trimmed reads using MetaGene (36), conceptual translation of the ORFs and calculation of the overall percentages in COG composition, and protein localization distribution as described for individual bacterial genomes. Intrinsic genomic features that rely on the knowledge of genome borders were inferred as follows: the effective genome size restricted to bacteria was computed as described in Raes et al. (37); for rRNA operons, prophage numbers, and CRISPRs the median values from our training dataset were used (Table 2). These values will not skew the clustering of the metagenomic sample with either the copiotrophs or the oligotrophs.
Supplementary Material
Supporting Information
Acknowledgments.
The work of the Australian contingent was supported by the Australian Research Council. The work of JGI members was performed under the auspices of the DOE's Office of Science, Biological and Environmental Research Program and the University of California, Lawrence Berkeley National Laboratory. The work of JCVI members was supported by the Gordon and Betty Moore Foundation.
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AAOJ00000000 and CP000356–CP000357).
See Commentary on page 15519.
References
- 1.Whitman W, Coleman D, Wiebe W. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 3.Humphrey B, Kjelleberg S, Marshall KC. Responses of marine bacteria under starvation conditions at a solid-water interface. Appl Environ Microbiol. 1983;45:43–47. doi: 10.1128/aem.45.1.43-47.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schut F, et al. Isolation of typical marine bacteria by dilution culture: Growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eguchi M, et al. Sphingomonas alaskensis strain AF01, an abundant oligotrophic ultramicrobacterium from the north Pacific. Appl Environ Microbiol. 2001;67:4945–4954. doi: 10.1128/AEM.67.11.4945-4954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavicchioli R, Ostrowski M, Fegatella F, Goodchild A, Guixa-Boixereu N. Life under nutrient limitation in oligotrophic marine environments: An eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingomonas alaskensis) Microb Ecol. 2003;45:203–217. doi: 10.1007/s00248-002-3008-6. [DOI] [PubMed] [Google Scholar]
- 7.Schut F, et al. Substrate uptake and utilization by a marine ultramicrobacterium. Microbiology. 1995;141:351–361. doi: 10.1099/13500872-141-2-351. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch P, et al. Life under conditions of low nutrient concentrations. In: Shilo M, editor. Strategies of Microbial Life in Extreme Envrionments. Berlin: Dahlem Konferenzen; 1979. pp. 357–372. [Google Scholar]
- 9.Akagi Y, Taga N. Uptake of d-glucose and l- proline by oligotrophic and heterotrophic marine bacteria. Can J Microbiol. 1980;26:454–459. doi: 10.1139/m80-075. [DOI] [PubMed] [Google Scholar]
- 10.Sowell SM, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 11.Agis M, Unanue M, Iriberri J, Herndl G. Bacterial colonization and ectoenzymatic activity in artificial marine snow. Part II: Cleavage and uptake of carbohydrates. Microb Ecol. 1998;36:66–74. doi: 10.1007/s002489900094. [DOI] [PubMed] [Google Scholar]
- 12.DeLong EF, Franks DG, Alldredge AL. Phylogenetic diversity of aggregate-attached versus free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 13.Alldredge AL, Cohen Y. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science. 1987;235:689–691. doi: 10.1126/science.235.4789.689. [DOI] [PubMed] [Google Scholar]
- 14.Poindexter JS. Oligotrophy: Fast and famine existence. Adv Microb Ecol. 1981;5:63–89. [Google Scholar]
- 15.D'Argenio DA, Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- 16.Römling U, Gomelsky M, Galperin MY. C-di-GMP: The dawning of a novel bacterial signaling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 18.Lindell D, et al. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorek R, Kunin V, Hugenholtz P. CRISPR: A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 20.Suttle CA. Marine viruses: Major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 21.Long A, McDaniel LD, Mobberley J, Paul JH. Comparison of lysogeny (prophage induction) in heterotrophic bacterial and Synechococcus populations in the Gulf of Mexico and Mississippi river plume. ISME J. 2007;2:132–144. doi: 10.1038/ismej.2007.102. [DOI] [PubMed] [Google Scholar]
- 22.Bongiorni L, Magagnini M, Armeni M, Noble RT, Danovaro R. Viral production, decay rates, and life strategies along a trophic gradient in the north Adriatic Sea. Appl Environ Microbiol. 2005;71:6644–6650. doi: 10.1128/AEM.71.11.6644-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raven JA, Andrews M, Quigg A. The evolution of oligotrophy: Implications for the breeding of crop plants for low input agricultural systems. Ann Appl Biol. 2005;146:261–280. [Google Scholar]
- 24.Kirchman D. Limitation of bacterial growth by dissolved organic matter in the subarctic Pacific. Mar Ecol Prog Ser. 1990;62:47–54. [Google Scholar]
- 25.Morris RM, Longnecker K, Giovannoni SJ. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol. 2006;8:1361–1370. doi: 10.1111/j.1462-2920.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 26.Moran MA, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 27.DeLong EF. Microbial community genomics in the ocean. Nat Rev Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- 28.Ishoey T, Woyke T, Stepanauskas R, Novotny M, Lasken RS. Genomic sequencing of single microbial cells from environmental samples. Curr Opin Microbiol. 2008;11:198–204. doi: 10.1016/j.mib.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker A, Parkhill J. Single-cell genomics. Nat Rev Microbiol. 2008;6:176–177. doi: 10.1038/nrmicro1862. [DOI] [PubMed] [Google Scholar]
- 30.Karl D. Microbial oceanography: Paradigms, processes, and promise. Nat Rev Microbiol. 2007;5:759–769. doi: 10.1038/nrmicro1749. [DOI] [PubMed] [Google Scholar]
- 31.Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauro FM, Bartlett DH. Prokaryotic lifestyles in deep sea habitats. Extremophiles. 2008;12:15–25. doi: 10.1007/s00792-006-0059-5. [DOI] [PubMed] [Google Scholar]
- 34.Rusch DB, et al. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 36.Noguchi H, Park J, Takagi T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kjelleberg S, Hermansson M, Marden P, Jones GW. The transient phase between growth and nongrowth of heterotrophic bacteria, with emphasis on the marine environment. Annu Rev Microbiol. 1987;41:25–49. doi: 10.1146/annurev.mi.41.100187.000325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information