Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells (original) (raw)
Abstract
Yeast and human Eme1 protein, in complex with Mus81, constitute an endonuclease that cleaves branched DNA structures, especially those arising during stalled DNA replication. We identified mouse Eme1, and show that it interacts with Mus81 to form a complex that preferentially cleaves 3′-flap structures and replication forks rather than Holliday junctions in vitro. We demonstrate that _Eme1_–/– embryonic stem (ES) cells are hypersensitive to the DNA cross-linking agents mitomycin C and cisplatin, but only mildly sensitive to ionizing radiation, UV radiation and hydroxyurea treatment. Mammalian Eme1 is not required for the resolution of DNA intermediates that arise during homologous recombination processes such as gene targeting, gene conversion and sister chromatid exchange (SCE). Unlike Blm-deficient ES cells, increased SCE was seen only following induced DNA damage in Eme1-deficient cells. Most importantly, Eme1 deficiency led to spontaneous genomic instability. These results reveal that mammalian Eme1 plays a key role in DNA repair and the maintenance of genome integrity.
Keywords: DNA repair/genomic instability/homologous recombination/inter-strand DNA cross-links/Mus81–Eme1
Introduction
Eme1 (essential meiotic endonuclease 1) was discovered in Schizosaccharomyces pombe by virtue of its interaction with Mus81 (methyl methanesulfonate-sensitive UV- sensitive 81) (Boddy et al., 2001). The ‘fuss’ about these molecules stemmed from the ability of the Mus81–Eme1 complex to function as a heterodimeric endonuclease, cleaving branched DNA structures such as replication forks (RFs), 3′ DNA flaps and Holliday junctions (HJs) (Haber and Heyer, 2001). Yeast Eme1 by itself has no endonuclease activity; however, its interaction with Mus81 is essential for the endonucleolytic activity of Mus81 (Boddy et al., 2001). Mus81 is related to the XPF family of endonucleases. This family shares a similar active site, constituted by the amino acids VERKxxxD (Boddy et al., 2000; Enzlin and Scharer, 2002). The Mus81–Eme1 complex resembles the Rad1–Rad10 protein complex that is involved in nucleotide excision repair (NER), where only one partner (yeast Rad1, human Xpf) possesses endonucleolytic activity. The functional binding partner of Mus81 in Saccharomyces cerevisiae is Mms4 (methyl methanesulfonate-sensitive 4), which shares weak similarity with S.pombe Eme1 (Boddy et al., 2001; Kaliraman et al., 2001; Mullen et al., 2001). Human Eme1 was subsequently identified and, in complex with human Mus81, cleaves RFs, 3′ DNA flaps and HJs (Ciccia et al., 2003; Ogrunc and Sancar, 2003).
The physical interaction of yeast Mus81 with Cds1 (Rad53/Chk2) (Boddy et al., 2000) and Rad54 (Interthal and Heyer, 2000), and a functional link with RecQ helicase Sgs1 (Mullen et al., 2001), indicated potential roles for the Mus81–Eme1 complex in cell cycle checkpoints, as well as DNA repair, recombination and replication processes. mus81, eme1 or mms4 mutants in yeast showed sensitivity to UV radiation, MMS and camptothecin (CPT), but not to ionizing radiation (IR). Since UV, MMS and CPT interfere with yeast DNA replication, the sensitivity of the mutants was attributed to an inability to process DNA intermediates arising at sites of stalled DNA replication (Xiao et al., 1998; Boddy et al., 2000, 2001; Interthal and Heyer, 2000; de los Santos et al., 2001; Kaliraman et al., 2001; Mullen et al., 2001; Bastin-Shanower et al., 2003). Rqh1 and Sgs1 helicases, in their respective fission and budding yeast, are known to play a role in the restart of stalled DNA replication. The requirement for Mus81 for the viability of rqh1/_sgs1_-null yeast indicated that these molecules are mechanistically distinct, yet functionally related, providing further evidence for the involvement of Mus81–Eme1 in the restart of stalled DNA replication (Kaliraman et al., 2001; Mullen et al., 2001; Doe et al., 2002; Fabre et al., 2002). DNA replication can stall due to lack of nucleotides, lack of replication checkpoints, and obstructions caused by DNA lesions or protein complexes (McGlynn and Lloyd, 2002).
That Eme1 could play a role in homologous recombination (HR) was expected from yeast studies, where loss of Mus81, Eme1 or Mms4 led to meiotic failure, as seen by a severe reduction in sporulation and spore viability, though some yeast strains displayed a moderate reduction (Boddy et al., 2001; de los Santos et al., 2001; Kaliraman et al., 2001). This defect was traced to DNA processing after the meiosis-induced double-strand breaks (DSBs) were initiated, resulting in improper segregation of chromosomes (Boddy et al., 2001; de los Santos et al., 2001; Kaliraman et al., 2001). This trend was also observed in vegetative yeast cells, where eliminating HR could restore viability to mus81 sgs1 or mms4 sgs1 double mutants, proving that unprocessed HR-generated DNA intermediates led to toxicity in these mutants (Fabre et al., 2002; Bastin-Shanower et al., 2003).
Analysis of various branched substrates showed that the Mus81-associated endonuclease activity from human extracts, and prepared as recombinant proteins in bacteria, could cleave RFs and 3′ DNA flaps more efficiently than HJs (Kaliraman et al., 2001; Constantinou et al., 2002; Doe et al., 2002; Ciccia et al., 2003; Whitby et al., 2003). The replication fork and 3′-flap DNA structures that are cleaved well by Mus81–Eme1 are speculated to form when DNA replication stalls (Whitby et al., 2003), and it is postulated that the 3′-flap can occur during synthesis-dependent strand annealing, also referred to as strand displacement and annealing (de los Santos et al., 2001; Haber and Heyer, 2001).
Despite the many studies characterizing the function of Mus81 and Eme1/Mms4 in yeast, the biological function of these molecules in mammals is unknown. In this study, we describe the identification and characterization of mouse Eme1, giving evidence for its role in DNA repair and maintenance of genomic stability.
Results
Cloning of mouse Eme1
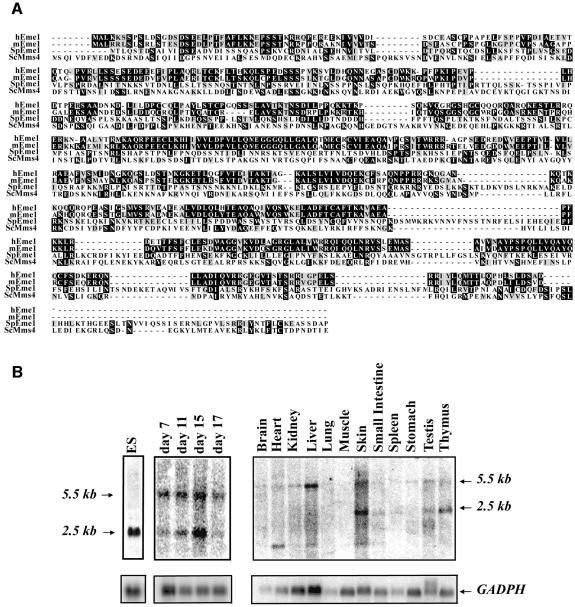
Sequence similarity searches of the DNA databases identified mouse open reading frames (ORFs) with similarity to fission yeast Eme1. The mouse cDNA was predicted to encode a 570 amino acid protein, which is the same size as the human counterpart, but smaller than the 738 amino acid yeast Eme1 (Boddy et al., 2001). The identity of the mouse protein to the S.pombe Eme1 molecule was 19%, and the similarity was 36% (Figure 1A). Comparison of mouse Eme1 with human Eme1 showed 66% identity and 76% similarity. Based on NCBI and Ensembl searches, the mouse Eme1 gene is localized to chromosome 11 band C and is encoded by nine exons spanning a genomic DNA region of ∼9 kb.
Fig. 1. Identification of mammalian Eme1. (A) Alignment of mouse, human and S.pombe Eme1 proteins with S.cerevisae Mms4. Amino acid identities and similarities are highlighted in black and gray, respectively. (B) Mouse Eme1 is mainly expressed in proliferative tissues. Radiolabeled Eme1 full-length cDNA was used to probe northern blots of mouse ES cells (left panel), mouse embryos at day 7, 11, 15 and 17 of gestation (middle panel) and mouse adult tissues (right panel). Northern blots were subsequently probed with a GADPH cDNA to assess loading.
In vivo expression of Eme1
Northern blot analysis of Eme1 expression in embryonic stem (ES) cells, mouse embryos and adult tissues was performed using the full-length mouse Eme1 cDNA as a probe (Figure 1B). A major Eme1 transcript of 2.5 kb was detected in ES cells (Figure 1B, left panel), while two major Eme1 transcripts of 2.5 and 5.5 kb were detected at days 7, 11, 15 and 17 of mouse embryonic development (Figure 1B, middle panel). In adult tissues (Figure 1B, right panel), weak expression of the 5.5 kb Eme1 transcript was detected in all tissues tested, but the 2.5 kb transcript was predominantly seen in skin, testis and thymus. Another smaller Eme1 transcript was also detected in some adult tissues such as heart, liver and skin. Together, these results demonstrate that Eme1 is expressed in many adult tissues and at various stages of embryonic development.
Mouse Eme1 and Mus81 interact to form a structure-specific endonuclease
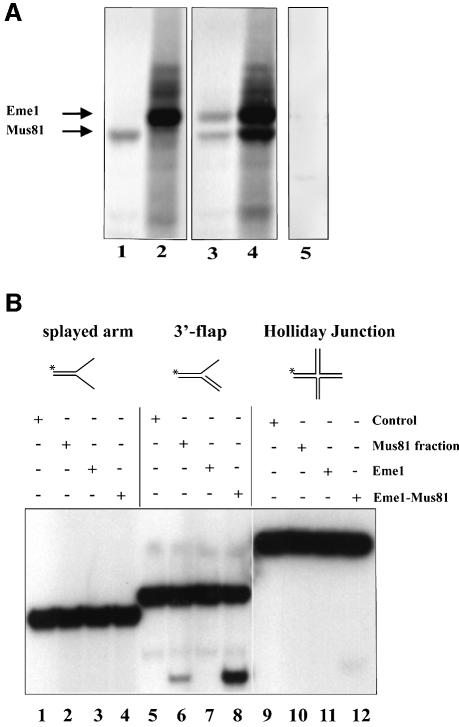
Using Flag-tagged mouse Mus81 and hemagglutinin (HA)-tagged mouse Eme1 proteins translated in vitro (Figure 2A), and transiently transfected into 293T human embryonic kidney (HEK) cells (data not shown), we demonstrated that these proteins interact with each other, as expected from their yeast and human counterparts (Figure 2A; lanes 3 and 4).
Fig. 2. Endonuclease activity of the mouse Mus81–Eme1 complex. (A) Interaction of mouse Eme1 with Mus81. In vitro translation reactions containing the templates Mus81-Flag (lane 1), Eme1-HA (lane 2), both Mus81-Flag and Eme1-HA (lanes 3 and 4) or no template (lane 5) were subject to immunoprecipitation (IP). The IP antibodies were anti-Flag (lanes 1, 3 and 5) and anti-HA (lanes 2 and 4). In co-translation reactions, both Mus81 and Eme1 were immunoprecipitated by anti-Flag and anti-HA antibodies (lanes 3 and 4), demonstrating the physical interaction of these proteins. (B) Mus81–Eme1 protein complex has DNA structure-specific endonuclease activity. Three branched DNA substrates, splayed-arm (lanes 1–4), 3′-flap (lanes 5–8) and HJ (lanes 9–12), were assessed for cleavage by immunocomplex negative control (lanes 1, 5 and 9), human Mus81 fraction (lanes 2, 6 and 10), mouse Eme1 immunocomplex (lanes 3, 7 and 11) and mouse Mus81–Eme1 immunocomplex (lanes 4, 8 and 12). Cleavage reactions were resolved by 10% neutral PAGE and visualized by phosphor imager analysis. Mouse Mus81–Eme1 cleaved the 3′-flap structure well (lane 8), but had only faint endonuclease activity against the HJ substrate tested (lane 12).
To determine whether mouse Mus81–Eme1 complex exhibited endonuclease activity, the ability of the Mus81–Eme1 pull-down complexes to cleave splayed-arm (Figure 2B, lanes 1–4), 3′-flap (lanes 5–8) and HJ (lanes 9–12) substrates was analyzed. We found that Mus81–Eme1 efficiently cleaved the 3′-flap structure (lane 8). Similar results were obtained with a structure that mimics an RF (data not shown). In contrast, little or no cleavage by Mus81–Eme1 was seen with the splayed-arm or HJ structure (lanes 4 and 12), as previously observed with the yeast and human complexes (Ciccia et al., 2003; Whitby et al., 2003). As a positive control in these reactions, we used a fraction containing Mus81 that had been prepared from fractionated HeLa cells (Constantinou et al., 2002) (Figure 2B, lane 6).
Generation of Eme1-deficient ES cells
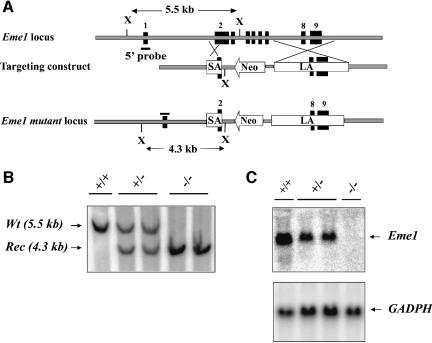
In order to investigate the cellular function of murine Eme1, we generated Eme1-deficient ES cells. A gene targeting construct (Figure 3A) was designed such that proper integration of the construct in the Eme1 genomic locus should result in the replacement of Eme1 exon 2 (partially) and exons 3–7 (completely) by the neomycin resistance (Neo) gene. Two Eme1+/– clones were reselected in high concentrations of G418 to obtain _Eme1_–/– clones (Figure 3B). Northern blot analysis confirmed the generation of _Eme1_-null mutation in these clones, as demonstrated by the complete loss of Eme1 transcript in the homozygous clones (Figure 3C). No difference in viability or growth rate was observed between _Eme1_–/– and wild-type ES cells (data not shown). Therefore, mammalian Eme1 is dispensable for the viability and growth of ES cells, as was observed for S.pombe and S.cerevisiae.
Fig. 3. Generation of Eme1+/– and _Eme1_–/– ES cells. (A) Schematic representations of the Eme1 locus, the gene-targeting construct and the targeted Eme1 allele. Exons are denoted by solid black boxes. SA, short arm; LA, long arm; Neo, neomycin resistance gene; X, _Xba_I site. (B) Southern blot analysis of wild-type, Eme1+/– and _Eme1_–/– ES cells. Probing Southern blots of _Xba_I-digested ES cell DNA with a 5′-flanking Eme1 probe differentiates the wild-type allele (5.5 kb) from the recombined allele (4.3 kb). (C) Northern blot analysis showing loss of Eme1 mRNA in _Eme1_–/– ES cells. A 20 µg aliquot of total RNA from the indicated Eme1 ES genotypes was northern blotted and probed with a 32P-radiolabeled full-length Eme1 cDNA. The northern blot was subsequently probed with a GAPDH cDNA to assess loading of mRNA.
Loss of Eme1 sensitizes ES cells to DNA damage
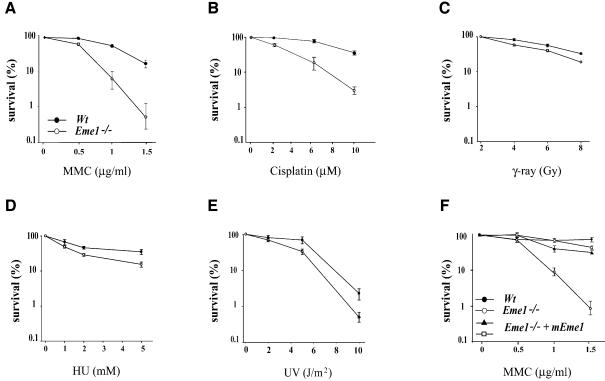
mus81, eme1 and mms4 mutation in yeast sensitized cells to UV radiation and hydroxyurea (HU), with little or no sensitivity to IR (Boddy et al., 2000; Interthal and Heyer, 2000; Mullen et al., 2001). In order to assess the role of the mammalian Eme1 in DNA repair, we performed colony survival assays on _Eme1_–/– and wild-type ES cells following exposure to DNA-damaging agents [IR, UV, HU, cisplatin and mitomycin C (MMC)]. As depicted in Figure 4, loss of Eme1 function led to a dramatic increase in the sensitivity of ES cells to increasing concentrations of MMC and cisplatin (Figure 4A and B), agents that cause interstrand cross-links (ICLs) (Dronkert and Kanaar, 2001). _Eme1_–/– ES cells were also found to be sensitive, though to a lesser extent, to IR, UV and HU treatments (Figure 4C–E), in comparison with the wild-type control. The sensitivity of Eme1+/– ES cells to these agents was comparable with that of wild-type cells (data not shown).
Fig. 4. Loss of Eme1 sensitizes ES cells to DNA damage. (A–E) Wild-type (solid circles) and _Eme1_–/– (open circles) ES colony survival following DNA damage by MMC, cisplatin, IR, HU and UV treatment. Doses of the individual treatments are plotted on the _x_-axis, while the _y_-axis denotes the corresponding fold sensitivity of colonies over untreated controls. _Eme1_–/– ES cells were extremely sensitive to MMC and cisplatin treatments, and only mildly sensitive to the other agents. (F) MMC sensitivity of _Eme1_–/– ES clones complemented with HA-tagged Eme1 cDNA (closed triangle and open square). Stable transfection of Eme1-HA cDNA in Eme1-deficient ES cells restores MMC sensitivity to near wild-type levels.
Reconstitution of Eme1 in the _Eme1_–/– ES cells was carried out to determine whether the remarkable sensitivity to MMC was specifically caused by the loss of Eme1. Following electroporation of a plasmid construct expressing HA-tagged Eme1 into _Eme1_–/– ES cell clones, colonies expressing HA-Eme1 were identified by immunoprecipitation and western blotting (data not shown). Two HA-Eme1 reconstituted _Eme1_–/– ES clones from two different _Eme1_–/– ES parental clones showed rescue to near wild-type levels when tested for MMC sensitivity (Figure 4F). This complementation demonstrates that the observed phenotype is caused by the specific loss of Eme1.
Thus, mammalian Eme1 is essential for the repair of MMC- and cisplatin-induced DNA damage in ES cells, and marginally involved in the repair of IR, UV and HU damage.
DNA damage-activated cell cycle checkpoints are intact in Eme1–/– ES cells
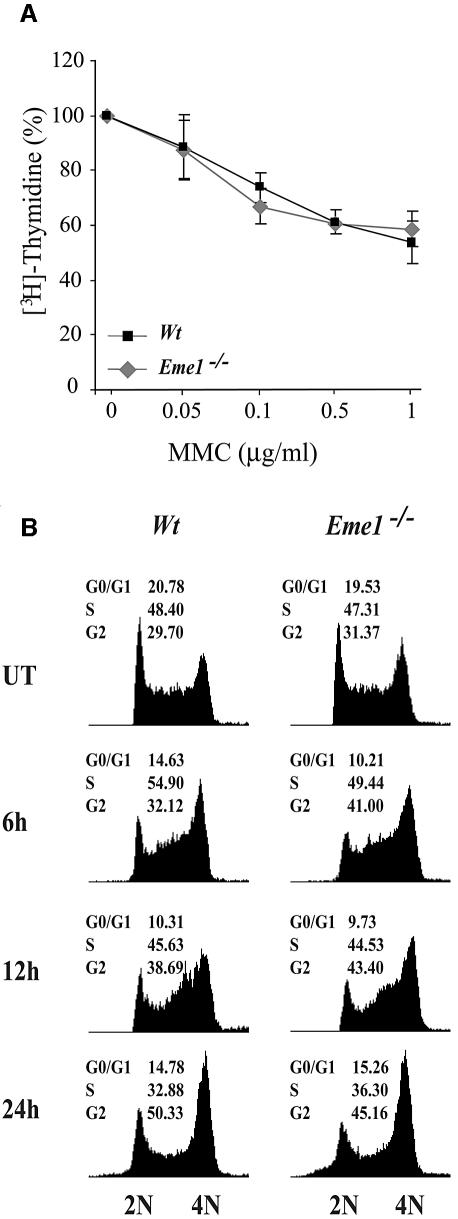
Cell cycle checkpoints ensure proper DNA repair, and hence cell survival, by preventing cells with damaged DNA from dividing until the damage has been repaired (Kaufmann, 1995; Zhou and Elledge, 2000). The interaction of yeast and human Mus81 with the cell cycle checkpoint protein Chk2, and the increased abundance of Mus81 in human cells exposed to agents that damage DNA or block replication, raised the possibility of a cell cycle checkpoint role for Mus81–Eme1 (Boddy et al., 2000; Chen et al., 2001). Intra-S-phase checkpoints were analyzed by [3H]thymidine incorporation into the DNA of wild-type and _Eme1_–/– ES cells following MMC treatment, and compared with untreated controls (Figure 5A). Increasing doses of MMC resulted in decreased DNA synthesis that was equivalent in both wild-type and _Eme1_–/– ES cells, showing no contribution of Eme1 to the cell cycle replication checkpoint. Moreover, fluorescence-activated cell sorting (FACS) analysis of propidium iodide-stained DNA content for cell cycle distribution of untreated and MMC-treated (0.1, 0.5 and 1 µg/ml) wild-type and _Eme1_–/– ES cells over a time course (6, 12 and 24 h) was comparable in both genotypes (Figure 5B; data not shown). Also, there was no difference in the cell cycle profile of these cells following 24 h of IR, UV and HU treatments (data not shown). These results, again, showed no role for Eme1 in regulating the cell cycle.
Fig. 5. Intact intra-S-phase checkpoint and G2–M cell cycle arrest in the absence of Eme1. (A) The MMC-induced intra S-phase checkpoint is not affected in _Eme1_–/– ES cells. The proportion of [3H]thymidine incorporation in untreated wild-type (black square) and _Eme1_–/– (gray diamond) cells was set as 100%. The decreased proportion of [3H]thymidine incorporation following increasing doses of MMC treatments was similar for both wild-type and _Eme1_–/– ES cells. (B) G2–M checkpoint activation of wild-type and _Eme1_–/– ES cells following MMC treatment. Wild-type (left panel) and _Eme1_–/– (right panel) ES cells were subjected to 1 µg/ml of MMC for 1 h and returned to regular medium for 6, 12 and 24 h, following which time they were stained with propidium iodide and analyzed by FACS. Both genotypes displayed similar cell cycle profiles, with cells progressively accumulating in the G2/M phase.
Eme1 is not essential for gene targeting
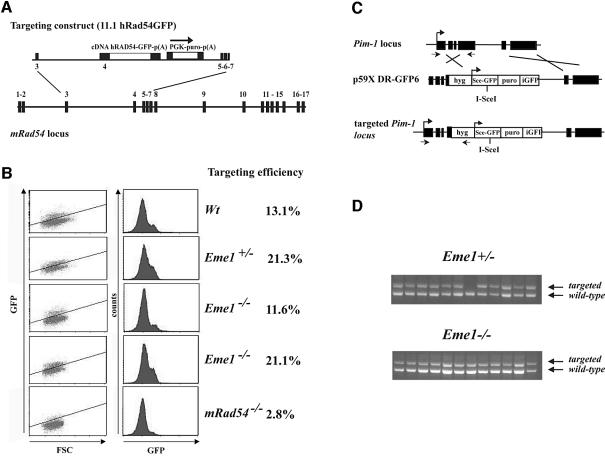
We sought to determine if mammalian Eme1 would have a role in HR, as was demonstrated for its yeast counterpart, by analyzing HR-mediated gene targeting. Although the precise mechanism for gene targeting has not been established, HR between the exogenous DNA sequence and the chromosome is expected in the regions of flanking homology followed by resolution of DNA intermediates (Niedernhofer et al., 2001). The efficiency of gene targeting between wild-type/Eme1+/– and _Eme1_–/– ES cells was assessed at two independent genomic loci, Rad54 (J.Essers and R.Kanaar, in preparation) and Pim1 (te Riele et al., 1990; Moynahan et al., 2001). The Rad54 targeting vector was designed with a promoterless human Rad54 cDNA fused to green fluorescent protein (GFP), such that expression could occur only following integration at its specific locus (Figure 6A). Upon puromycin selection, while _Rad54_–/– ES cells showed reduced GFP+ cells (2.8%), comparable numbers of wild-type (13.1%), Eme1+/– (21.3%) and _Eme1_–/– (11.6 and 21.1%) ES clones were GFP+ (Figure 6B), showing no loss of HR-mediated gene targeting potential in the absence of Eme1. Southern analysis confirmed that GFP+ cells had proper integration of the targeting construct (data not shown). The use of a high efficiency Pim1 targeting construct (Figure 6C) and the expression of its selection marker hygromycin (_hyg_R), being dependent on specific integration at the Pim1 locus, led to similar numbers (>90% of the _hyg_R) of Eme1+/– and _Eme1_–/– clones being correctly gene targeted (Figure 6D). These findings indicate that Eme1 is not required for the resolution of DNA intermediates that arise during HR-mediated gene targeting.
Fig. 6. Eme1 does not impair homologous recombination processes such as gene targeting. (A) Schematic of the gene targeting strategy at the Rad54 locus. The Rad54 targeting construct (11.1hRAD54-GFP) and gene locus are depicted. Expression of Rad54–GFP is dependent on proper gene targeting at the Rad54 locus. (B) FACS analysis of the percentage of GFP+ cells after puromycin selection. Whereas gene targeting is clearly impaired in _Rad54_–/– ES cells, levels of GFP expression in Eme1+/– and _Eme1_–/– were similar to the wild-type control. (C) Gene targeting strategy at the Pim1 locus. (D) PCR analysis of gene-targeted clones. Eme1+/– and _Eme1_–/– ES cells were electroporated with the linear p59XDR-GFP6 targeting construct that favors gene targeting by selecting predominantly for survival of colonies that had integration at the Pim-1 locus. Homologous recombination-mediated gene targeting was tested by PCR, and showed no difference between the two genotypes.
Eme1 is not essential for gene conversion repair of DSBs
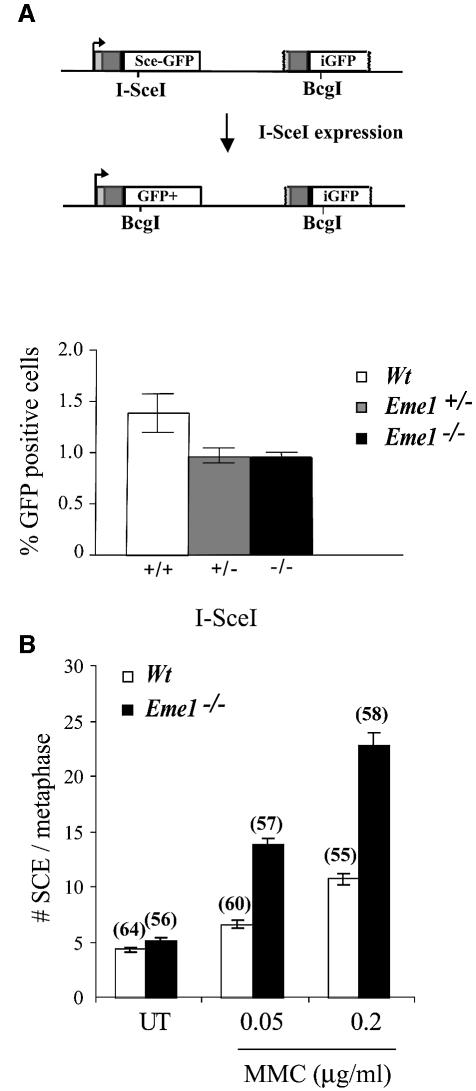
Continuing our investigation into a role for Eme1 in HR, we assessed HR-mediated gene conversion by analyzing the ability of wild-type, Eme1+/– and _Eme1_–/– ES cells to repair a DSB by this process. Gene conversion is a Rad51-mediated invasion of the 3′-single-stranded tail of the broken DNA into homologous regions present on either the same or different chromosomes (Johnson and Jasin, 2001). A single DSB was induced by transient expression of _Sce_I at the Pim1 locus, targeted with a construct bearing a _Sce_I site and two non-functional GFP ORFs (Figure 7A, upper panel) (Moynahan et al., 2001). Repair of the break using the homologous downstream iGFP gene can occur by gene conversion without crossing-over. A gene conversion event results in the conversion of the I-_Sce_I restriction site to the native _Bcg_I GFP sequence, and restoration of GFP expression. Repair by gene conversion was scored by the number of GFP+ cells using flow cytometry, and showed no significant difference between wild-type, Eme1+/– or _Eme1_–/– cells (Figure 7A, lower panel). The minor difference between wild-type and Eme1+/– cells was not statistically significant (p = 0.12). This finding indicates that Eme1 does not play an essential role in the resolution of DNA intermediates formed during HR-mediated gene conversion.
Fig. 7. Induced DSB repair and sister chromatid exchange in the absence of Eme1. (A) Upper panel: schematic representation of the generation of a functional GFP gene by HR-mediated gene conversion. Transient I-_Sce_I expression induces a DSB in a stably integrated construct bearing two non-functional GFP genes in tandem. HR- mediated gene conversion without crossing-over replaces the I-_Sce_I with the Bcg_I site, and also restores GFP expression. Lower panel: quantification of GFP+ cells following I-Sce_I-induced DSB repair. Wild-type, Eme1+/– and _Eme1_–/– ES clones were analyzed by FACS to detect the proportion of GFP+ cells, indicative of successful gene conversion at the non-functional GFP locus. Wild-type, Eme1+/– and _Eme1_–/– ES clones showed no difference in the number of GFP+ cells, indicating equivalent HR-mediated gene conversion following an induced DSB. (B) Increased MMC-induced SCE in the absence of Eme1. The frequency of spontaneous SCE was similar between wild-type and _Eme1_–/– ES cells. However, a 2-fold increase of MMC- induced SCE was observed in Eme1-deficient ES cells by comparison with wild-type cells. The number of metaphases analyzed is given in parentheses above the corresponding histogram.
Loss of Eme1 leads to increased sister chromatid exchange (SCE) upon DNA damage
Increased spontaneous SCE is a hallmark of defects in BLM helicase, a member of the RecQ family of helicases that are involved in the restart of stalled DNA replication, a function also attributed to Mus81–Eme1 (Chakraverty and Hickson, 1999; Boddy et al., 2001; Kaliraman et al., 2001). In mitotic cells, SCE is largely a repair response to DNA damage, primarily DNA breaks that occur during DNA replication (Chakraverty and Hickson, 1999; Sonoda et al., 1999; Wang et al., 2000). We assessed SCE in wild-type and _Eme1_–/– ES cells as a measure of spontaneous and MMC-induced DNA breaks. Both wild-type and _Eme1_–/– ES cell clones had similar levels of spontaneous SCE per metaphase (4.4 ± 0.23 and 5.2 ± 0.23, respectively) (Figure 7B). Following MMC treatment, however, the frequency of SCE in _Eme1_–/– ES cells was twice that of wild-type cells at both concentrations tested (Figure 7B). For wild-type versus _Eme1_–/– ES cell clones, at 0.05 µg/ml MMC, the number of SCE per metaphase was 6.7 ± 0.37 versus 13.98 ± 0.48, and at 0.2 µg/ml MMC it was 10.8 ± 0.51 versus 22.96 ± 1.12 (Figure 7B). This finding suggests that there is a greater number of DNA strand breaks occurring in the absence of Eme1, which initiate SCE to repair the damage.
SCE is dependent on HR (Sonoda et al., 1999; Wang et al., 2000). Comparable rates of spontaneous SCE in wild-type and _Eme1_–/– ES cells, and a 2-fold increase in MMC-treated _Eme1_–/– ES cells, rule out an essential role for Eme1 in the HR process that mediates SCE.
Eme1 deficiency results in chromosomal instability
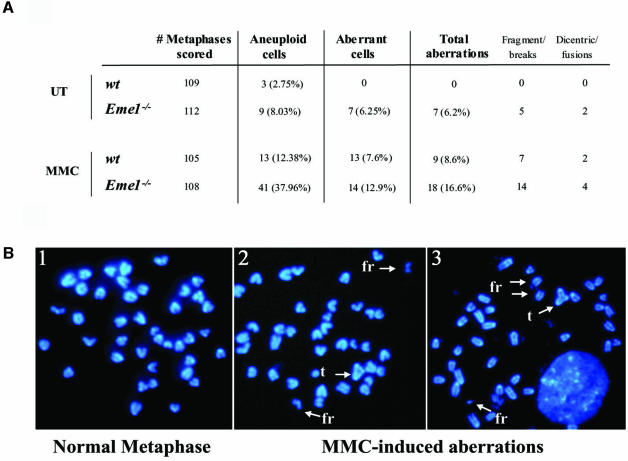
The hypersensitivity of _Eme1_–/– ES cells to DNA damage indicates defective DNA repair, a process that could lead to accumulation of chromosomal abnormalities and genomic instability (Rouse and Jackson, 2002). We therefore assessed the level of spontaneous genomic instability in three _Eme1_–/– ES clones, relative to several wild-type ES clones. Examination of 4′,6-diamidino-2-phenylindole (DAPI)-stained metaphase spreads revealed increased levels of chromosomal abnormalities including breaks, fragments, chromosomal fusions and dicentric chromosomes in the Eme1-deficient ES cells (Figure 8A). Overall, 6% of metaphase spreads from _Eme1_–/– ES cells showed aberrations, compared with none in wild-type metaphase spreads (p < 0.01). Moreover, increased aneuploidy was also seen in _Eme1_–/– ES cells compared with wild-type controls (Figure 8A).
Fig. 8. Increased genomic instability in Eme1-deficient ES cells. (A) Spontaneous and MMC-induced chromosomal instability in Eme1-deficient ES cells. Metaphase spreads of untreated and MMC-treated wild-type and _Eme1_–/– ES cells were analyzed for chromosomal instability. Increased spontaneous aneuploidy is observed in _Eme1_–/– ES cells, with other aberrations represented by breaks, triradials and chromosomal fusions. (B) MMC treatment aggravates chromosomal aberrations in _Eme1_–/– ES cells. A normal metaphase spread from untreated _Eme1_–/– ES cells (1) and two metaphase spreads from MMC-treated _Eme1_–/– ES cells (2 and 3) showing MMC-induced accumulation of fragments (fr) and triradials (t).
As _Eme1_–/– ES cells exhibited a profound sensitivity to MMC, we next analyzed the effect of Eme1 deficiency on the accumulation of chromosomal aberrations induced following MMC treatment. Wild-type and _Eme1_–/– ES cells were subjected to 40 ng/ml of MMC for 1 h, and cultured 24 h before preparation of metaphase spreads. There were 2-fold more chromosomal aberrations in the Eme1-deficient ES cells compared with wild-type ES cells (16.6% versus 8.6%; p < 0.05) following MMC treatment (Figure 8A). Some of the chromosomal abnormalities (fragments and triradial structures) observed in _Eme1_–/– cells following MMC-induced damage are shown in Figure 8B. Interestingly, MMC treatment greatly increased the proportion of aneuploid cells in the absence of Eme1 (38% for _Eme1_–/– versus 12.4% for wild-type; Figure 8A). These results highlight an important role for Eme1 in maintaining genomic stability especially following DNA damage caused by MMC.
Discussion
The inability of cells to induce cell cycle arrest, apoptosis or DNA repair efficiently often leads to various disorders in humans, from immunodeficiencies to cancer (Dasika et al., 1999; Bassing et al., 2002). Previous studies established the importance of the yeast Mus81–Eme1 complex in DNA repair (Boddy et al., 2000; Interthal and Heyer, 2000; Mullen et al., 2001). In this study, we report the biological functions of mammalian Eme1, by characterizing the phenotype of ES cells lacking this gene.
Role of Eme1 in DNA repair
The hypersensitivity of _Eme1_–/– ES cells to DNA damage caused by MMC and cisplatin, as opposed to the milder sensitivity to IR, UV and HU, suggests a role for Eme1 in ICL DNA repair. ICLs can block DNA replication and transcription, thereby leading to accumulation of DNA intermediate structures. It is likely that lack of cleavage, or processing of these intermediates in _Eme1_–/– ES cells, results in increased cell death caused by collapse of replication forks, or debilitating genomic instability. It is possible that damage by IR, UV and HU is quickly repaired before it hinders DNA replication, thereby not requiring Mus81–Eme1 function in DNA repair, accounting for the corresponding reduced sensitivity.
Eme1 is not required for homologous recombination-mediated gene targeting and gene conversion repair of DSBs
Yeast Mus81, Eme1 and Mms4 have been implicated in HR in both vegetative and meiotic cells (Boddy et al., 2001; de los Santos et al., 2001; Kaliraman et al., 2001; Bastin-Shanower et al., 2003). Also, yeast Mus81 was reported to interact with Rad54, a molecule important in HR (Interthal and Heyer, 2000). These observations indicated a possible role for mammalian Eme1–Mus81 in HR, a process that generates several DNA intermediate structures. Moreover, Mus81 is related to Xpf, a component of the Ercc1–Xpf endonuclease, and both proteins have the same active site (Boddy et al., 2000; Interthal and Heyer, 2000). _Ercc1_–/– mouse ES cells displayed a complete inability to integrate a gene-targeting construct in three different genomic loci (Niedernhofer et al., 2001), raising the possibility that the related Mus81–Eme1 endonuclease may also have a role in the HR process of gene targeting. The lack of a defect in HR-mediated gene targeting and DSB repair by gene conversion in _Eme1_–/– ES cells demonstrates no essential role for Eme1 in the processing of DNA intermediates that could be generated during these specific HR processes.
Increased SCE in the absence of Eme1 is dependent on induced DNA damage
Increased SCE and hyper-recombination are hallmarks of defects in the RecQ family of helicases, as seen with mutated BLM and sgs1/rqh1 (Chakraverty and Hickson, 1999). Mutation of BLM in mouse ES cells led to a 10-fold increase in SCE (Luo et al., 2000). Wang et al. (2000) proposed that BLM decreases DNA strand breaks that occur during DNA replication. Since these helicases are also involved in the restart of stalled DNA replication, and hence have a functional overlap with Mus81–Eme1 (Kaliraman et al., 2001; Mullen et al., 2001; Doe et al., 2002; Fabre et al., 2002), we investigated whether loss of Eme1 would affect SCE frequency. Unlike loss of RecQ helicases, deletion of Eme1 had no effect on spontaneous rates of SCE, underscoring mechanistic differences between these two groups of molecules. It can be speculated that the RecQ helicases have a greater role in restart of stalled DNA replication, or that their substrates far outnumber those that are responsive to Mus81–Eme1. Also, RecQ helicases may have other roles in addition to restart of stalled DNA replication that could account for the increased SCE seen in their absence. Upon MMC treatment, however, the 2-fold increase in SCE frequency of _Eme1_–/– ES cells over wild-type, could be indicative of DNA breaks occurring presumably at unprocessed branched DNA intermediates, or of failed replication restart.
Kaliraman et al. (2001) speculated that the Mus81-associated endonuclease might be responsible for the increased SCE frequency in cells deficient for RecQ helicases, and the increased replication-dependent SCE seen following UV damage. If Mus81-associated endonuclease activity is dependent exclusively on Eme1, and was directly responsible for either spontaneous or DNA damage-induced SCE, then inactivation of Eme1 should prevent the well-documented increase in MMC-induced SCE frequency (Sonoda et al., 1999; Wang et al., 2003). Since this is not the case in _Eme1_–/– ES cells, where MMC treatment leads to a 2-fold increase in SCE over wild-type, other molecules must be responsible in the absence of BLM or Eme1 for inducing SCE. However, inactivation of both BLM and Eme1/Mus81 would be required to prove definitively that Mus81–Eme1 does not contribute to the enhanced SCE in BLM-defective cells.
Role of Eme1 in maintaining genomic stability
Genetic instabilities, including changes to chromosome numbers and chromosomal aberrations, are frequent in human cancers and are thought to be required for the generation of the multiple genetic hits necessary for malignant transformation (Rouse and Jackson, 2002). Genomic instability is also a hallmark of impaired DNA damage repair and has been associated with mutations that impair DNA repair, including mutations in BRCA1, BRCA2, Ku80, XRCC4, NBS1 and genes involved in Fanconi anemia (Sekiguchi et al., 2001; Roth, 2002; D’Andrea and Grompe, 2003; Rooney et al., 2003). The occurrence of spontaneous genomic instability in _Eme1_–/– ES cells signifies a requirement for Eme1 in maintaining genomic integrity. This Eme1 function is probably due to its role in constituting a functional heterodimeric endonuclease with Mus81. Based on the substrates that the Mus81–Eme1 complex can cleave in vitro, we speculate that spontaneous genomic instability in _Eme1_–/– ES cells is due to a lack of processing of DNA intermediates that arise during stalled DNA replication, and in DNA repair events that generate 3′-flap structures. The lack of processing of these DNA intermediates in _Eme1_–/– ES cells could result in chromosomal breaks and fusions, chromosome mis-segregation and aneuploidy. The increased genomic instability in MMC-treated _Eme1_–/– ES cells is likely to result from the increased number of unresolved Mus81–Eme1 substrates in these cells. The increased genomic instability in ES cells deficient for Eme1 demonstrates the fundamental role that this mammalian protein plays in maintaining genomic integrity.
Conclusions
We demonstrate that the endonucleolytic activity of the mouse Mus81–Eme1 complex cleaves 3′-flap and RFs prominently, and HJs weakly. The mild sensitivity of _Eme1_–/– ES cells to IR, UV and HU, and the overwhelming sensitivity to MMC and cisplatin, unequivocally implicate mammalian Eme1 as a DNA repair protein. The occurrence of chromosomal aberrations in the absence of Eme1 highlights the importance of the Mus81–Eme1 complex in processing spontaneous intermediate DNA structures, and thereby suppressing genomic instability. The human Eme1 gene localizes to a region (17q21.33) that is associated with leukemia, and other forms of cancer. Whether Eme1 is a tumor suppressor is a question that must be addressed, especially given our finding that loss of Eme1 leads to genomic instability.
Materials and methods
Cloning of mouse Eme1
Mouse Eme1 (NCBI accession No. AK078516) was identified by performing protein BLAST searches using the S.pombe Eme1 protein sequence. Eme1 was amplified by RT–PCR from a murine brain cDNA library (Clontech) with the following primers: 5′-GTCATATGG CTCTAAGAAGGTTATCCCT-3′ and 5′-AAGTCGACTCAGTCA ACACTGTCTAAGATGAG-3′, TA-cloned into a pCR2.1-TOPO (Invitrogen Life Technologies), and sequenced.
Northern analysis
A 20 µg aliquot of total RNA was separated on 1% agarose, 2.2 M formaldehyde gels in 1× MOPS buffer and transferred to Nylon membranes (Amersham Pharmacia). Multiple Choice™ northern blots (Origene) and mouse embryo MTN blot (Clontech) were also used to analyze Eme1 mRNA expression. The blots were probed with 32P-labeled full-length Eme1 and GADPH cDNA, and exposed on a phosphor imager (Molecular Dynamics).
In vitro translation and co-immunoprecipitation
N-terminal tags were added to coding sequences as follows: Flag tag to Mus81, and HA tag to Eme1 to generate pcDNAMus81-Flag and pcDNAEme1-HA, respectively. Mus81-Flag and Eme1-HA were expressed individually, or co-expressed, in the presence of [35S]methionine using the TNT coupled reticulocyte lysate systems (Promega). Each 50 µl in vitro translation reaction was suspended in 1 ml of co-immunoprecipitation buffer [50 mM Tris pH 8, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.2% NP-40, 50 µg/ml Pefabloc and 1 protease inhibitor tablet (Roche)/10 ml]. Immunoprecipitations were carried out with 1 µg of anti-Flag (Sigma) or anti-HA antibody for 1 h at 4°C. Immunoprecipitates were collected on protein G–Sepharose beads, washed and boiled in Laemmli buffer for 3 min. Samples were run on 9% SDS–polyacrylamide gels, and the gels were dried and exposed on a phosphor imager.
In vitro endonuclease cleavage assays
DNA substrates were prepared as described previously (Constantinou et al., 2002). The HJ was the immobile junction X0 (Benson and West, 1994). Reactions were performed in phosphate buffer [60 mM Na2HPO4/NaH2PO4 pH 7.4, 5 mM MgCl2, 1 mM dithiothreitol (DTT) and 100 µg/ml bovine serum albumin (BSA)], with immunoprecipitations (3 µl) prepared from in vitro translation lysates containing no template, Eme1 or Mus81–Eme1. The Mus81 HeLa cell fraction previously described was used as a positive control (Constantinou et al., 2002). 32P-labeled DNA products were visualized by autoradiography after electrophoresis through 10% polyacrylamide.
Generation of Eme1+/– and Eme1–/– ES cells
A targeting vector was designed such that homologous recombination would replace a 3.6 kb genomic fragment containing Eme1 exons 2–7 with the PGKneo resistance expression cassette. The targeting vector was linearized with _Kpn_I and electroporated into E14K ES cells as previously described (Hakem et al., 1998). Cells were subsequently cultured in the presence of 300 µg/ml G418 (Sigma) for 10 days. Gene targeting at the Eme1 locus was identified by probing Southern blots of _Xba_I-digested DNA with a Eme1 5′-flanking cDNA. Proper integration was confirmed by Southern blots probed with a 3′-flanking cDNA and neomycin cDNA. Four correctly recombined Eme1+/– ES clones were identified. Two of these were cultured in increasing concentrations of G418. Many resistant colonies growing at a G418 concentration of 8.4 mg/ml had successfully replaced their remaining wild-type allele with the gene-targeted allele, generating _Eme1_–/– ES clones.
Clonogenic assays
ES cells were seeded onto gelatinized 6-well plates (2 × 103 cells/well) and subjected to DNA-damaging treatments 18 h later. Cells were treated with γ-rays (1, 2 and 4 Gy), UV (2, 3 and 10 J/m2), MMC (0.1, 0.5 and 1.5 µg/ml for 1 h), cisplatin (2, 6 and 10 mM for 2 h) and HU (1, 2.5 and 5 mM for 4 h). After 7–10 days in culture, colonies were stained with crystal violet and counted.
Cell cycle analysis
Cells were subjected to MMC treatment for 1 h (0.1, 0.5 and 1 µg/ml), washed and transferred to fresh media. Cells were then trypsinized after 6, 12 and 24 h of culture, and prepared as previously described (Hirao et al., 2000). Samples were analyzed using the Becton Dickinson FACScalibur instrument.
For analysis of intra S-phase checkpoints, cells plated in 96-well plates were treated with 0.05, 0.1, 0.5 and 1 µg/ml of MMC for 2 h, washed twice with phosphate-buffered saline (PBS), then pulsed with 5 µCi of [3H]thymidine (Amersham) per well for 2 h and harvested.
HR-mediated gene targeting
Gene targeting at the Rad54 locus was performed essentially as described (Essers et al., 1997, 2002). Wild-type, Eme1+/–_, Eme1_–/– and _Rad54_–/– ES cells were transfected with a RAD54-GFP knock-in construct. One week after selection in puromycin, single-cell suspensions of surviving colonies were made by trypsinization and analyzed by FACS on a green fluorescence (FL1-H) versus forward scatter (FSC-H) plot. GFP+ and GFP– cells appeared in separate populations, above and below the diagonal line, respectively (J.Essers and R.Kanaar, unpublished results). Results were also plotted in a fluorescence (FL1-H) histogram.
Gene targeting efficiency of Eme1+/– and _Eme1_–/– ES cells was also assessed on the Pim-1 locus after electroporation of p59xDR-GFP6 as previously described (Moynahan et al., 2001). Gene targeting efficiency was evaluated by PCR using the following primers: Pim1Ex1F, sense 5′-AAGATCAACTCCCTGGCCCACCTGCG-3′ and antisense Pim1Ex4R 5′-TGTTCTCGTCCTTGATGTCG-3′; and Hyg3A 5′-CCGCTCGTC TGGCTAAGAT-3′. PCR amplified a 786 bp fragment at the endogenous Pim1 locus and a 958 bp fragment at the p59xDR-GFP targeted allele.
I-SceI-induced DNA DSB repair
Pim1_-targeted wild-type, Eme1_+/– and _Eme1_–/– cells were electroporated with the I-_Sce_I expression vector, mock DNA or the positive control plasmid pNZE-CAG (Pierce et al., 1999) as previously described (Richardson et al., 1998). Cells were analyzed by FACS after 48 h. Three independent wild-type, Eme1+/– and _Eme1_–/– clones were analyzed. The proportion of GFP+ cells between wild-type, Eme1+/– and _Eme1_–/– clones was assessed using the unpaired _t_-test. The tests were two-sided and the cut-off for significance was 0.05.
Sister chromatid exchange
Wild-type and _Eme1_–/– ES cells were treated with MMC (0.05 and 0.2 µg/ml) for 1 h, and grown in fresh medium containing 10 µM of bromodeoxyuridine (BrdU) for 36 h. Untreated and treated cells were then incubated with colcemid (0.1 µg/ml) for 4 h, harvested and treated in hypotonic buffer (0.075 M KCl) at 37°C for 15 min. After fixation in ice-cold methanol:acetic acid (3:1) buffer, cells were dropped onto glass slides. After denaturation, slides were incubated with anti-BrdU antibody (BD Biosciences), and subsequently with anti-mouse IgG–fluorescein isothiocyanate (FITC) antibody (eBioscience). Slides were dehydrated in ethanol and embedded with mounting medium (Vector Labs) containing 0.3 µg/ml propidium iodide. Analysis was performed under a Zeiss Axioplan 2 Imaging microscope (Carl Zeiss, Germany) equipped with a CCD camera. The number of metaphases analyzed is as follows for wild-type and _Eme1_–/– cells, respectively: untreated, 64 and 56; 0.05 µg/ml MMC, 60 and 57; 0.2 µg/ml MMC, 55 and 58.
Genomic stability assays
Wild-type and _Eme1_–/– ES cells were treated with MMC (40 ng/ml) for 1 h, and cultured for an additional 24 h. Cells were processed as described above. Slides were stained with DAPI (Sigma), and chromosome number and gross chromosomal rearrangements were determined in 100 metaphase cells per sample from each cell type. The slides were observed under a Leica DMIRB fluorescence microscope (Germany) equipped with digital camera (Leica DC 300RF). Images were acquired using Leica Image Manager software.
Acknowledgments
Acknowledgements
We thank Anne Hakem, Leonardo Salmena and Ashwin Pamidi for critically reviewing the manuscript, Paul Sadowski and Linda Lee for helpful discussions, and Eva Migon, Elzbieta Matysiak-Zablocki, Ellen van Drunen, Denis Bouchard and Shili Duan for technical support. M.P.H. acknowledges the support in the form of start-up grants by the National University of Singapore and Oncology Research Institute, NUMI. This research was supported by an AMDI transition fund awarded to R.H. by the University Health Network, Toronto.
References
- Bassing C.H., Swat,W. and Alt,F.W. (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109, S45–S55. [DOI] [PubMed] [Google Scholar]
- Bastin-Shanower S.A., Fricke,W.M., Mullen,J.R. and Brill,S.J. (2003) The mechanism of Mus81–Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1–Rad10. Mol. Cell. Biol., 23, 3487–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson F.E. and West,S.C. (1994) Substrate specificity of the Escherichia coli RuvC protein. Resolution of three- and four-stranded recombination intermediates. J. Biol. Chem., 269, 5195–5201. [PubMed] [Google Scholar]
- Boddy M.N., Lopez-Girona,A., Shanahan,P., Interthal,H., Heyer,W.D. and Russell,P. (2000) Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol., 20, 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard,P.H., McDonald,W.H., Shanahan,P., Yates,J.R.,3rd and Russell,P. (2001) Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell, 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication. BioEssays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- Chen X.B. et al. (2001) Human Mus81-associated endonuclease cleaver Holliday junctions in vitro. Mol. Cell, 8, 1117–1127. [DOI] [PubMed] [Google Scholar]
- Ciccia A., Constantinou,A. and West,S.C. (2003) Identification and characterization of the human Mus81–Eme1 endonuclease. J. Biol. Chem., 278, 25172–25178. [DOI] [PubMed] [Google Scholar]
- Constantinou A., Chen,X.B., McGowan,C.H. and West,S.C. (2002) Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J., 21, 5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea A.D. and Grompe,M. (2003) The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer, 3, 23–34. [DOI] [PubMed] [Google Scholar]
- Dasika G.K., Lin,S.C., Zhao,S., Sung,P., Tomkinson,A. and Lee,E.Y. (1999) DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene, 18, 7883–7899. [DOI] [PubMed] [Google Scholar]
- de los Santos T., Loidl,J., Larkin,B. and Hollingsworth,N.M. (2001) A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics, 159, 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Ahn,J.S., Dixon,J. and Whitby,M.C. (2002) Mus81–Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem., 277, 32753–32759. [DOI] [PubMed] [Google Scholar]
- Dronkert M.L. and Kanaar,R. (2001) Repair of DNA interstrand cross-links. Mutat. Res., 486, 217–247. [DOI] [PubMed] [Google Scholar]
- Enzlin J.H. and Scharer,O.D. (2002) The active site of the DNA repair endonuclease XPF–ERCC1 forms a highly conserved nuclease motif. EMBO J., 21, 2045–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- Essers J., Hendriks,R.W., Wesoly,J., Beerens,C.E., Smit,B., Hoeijmakers,J.H., Wyman,C., Dronkert,M.L. and Kanaar,R. (2002) Analysis of mouse Rad54 expression and its implications for homologous recombination. DNA Repair, 1, 779–793. [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan,A., Heyer,W.D. and Gangloff,S. (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4 and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA, 99, 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. and Heyer,W.D. (2001) The fuss about Mus81. Cell, 107, 551–554. [DOI] [PubMed] [Google Scholar]
- Hakem R. et al. (1998) Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell, 94, 339–352. [DOI] [PubMed] [Google Scholar]
- Hirao A., Kong,Y.Y., Matsuoka,S., Wakeham,A., Ruland,J., Yoshida,H., Liu,D., Elledge,S.J. and Mak,T.W. (2000) DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science, 287, 1824–1827. [DOI] [PubMed] [Google Scholar]
- Interthal H. and Heyer,W.D. (2000) MUS81 encodes a novel helix–hairpin–helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet., 263, 812–827. [DOI] [PubMed] [Google Scholar]
- Johnson R.D. and Jasin,M. (2001) Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans., 29, 196–201. [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Mullen,J.R., Fricke,W.M., Bastin-Shanower,S.A. and Brill,S.J. (2001) Functional overlap between Sgs1–Top3 and the Mms4–Mus81 endonuclease. Genes Dev., 15, 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann W.K. (1995) Cell cycle checkpoints and DNA repair preserve the stability of the human genome. Cancer Metastasis Rev., 14, 31–41. [DOI] [PubMed] [Google Scholar]
- Luo G., Santoro,I.M., McDaniel,L.D., Nishijima,I., Mills,M., Youssoufian,H., Vogel,H., Schultz,R.A. and Bradley,A. (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet., 26, 424–429. [DOI] [PubMed] [Google Scholar]
- McGlynn P. and Lloyd,R.G. (2002) Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol., 3, 859–870. [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Pierce,A.J. and Jasin,M. (2001) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell, 7, 263–272. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman,V., Ibrahim,S.S. and Brill,S.J. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics, 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L.J., Essers,J., Weeda,G., Beverloo,B., de Wit,J., Muijtjens,M., Odijk,H., Hoeijmakers,J.H. and Kanaar,R. (2001) The structure-specific endonuclease Ercc1–Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J., 20, 6540–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrunc M. and Sancar,A. (2003) Identification and characterization of human MUS81–MMS4 structure-specific endonuclease. J. Biol. Chem., 278, 21715–21720. [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Johnson,R.D., Thompson,L.H. and Jasin,M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev., 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C., Moynahan,M.E. and Jasin,M. (1998) Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev., 12, 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S. et al. (2003) Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med., 197, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.B. (2002) Amplifying mechanisms of lymphomagenesis. Mol. Cell, 10, 1–2. [DOI] [PubMed] [Google Scholar]
- Rouse J. and Jackson,S.P. (2002) Interfaces between the detection, signaling and repair of DNA damage. Science, 297, 547–551. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J. et al. (2001) Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl Acad. Sci. USA, 98, 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Maandag,E.R., Clarke,A., Hooper,M. and Berns,A. (1990) Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature, 348, 649–651. [DOI] [PubMed] [Google Scholar]
- Wang W., Seki,M., Narita,Y., Sonoda,E., Takeda,S., Yamada,K., Masuko,T., Katada,T. and Enomoto,T. (2000) Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J., 19, 3428–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. (2003) Functional relation among RecQ family helicases RecQL1, RecQL5 and BLM in cell growth and sister chromatid exchange formation. Mol. Cell. Biol., 23, 3527–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M.C., Osman,F. and Dixon,J. (2003) Cleavage of model replication forks by fission yeast mus81–eme1 and budding yeast mus81–mms4. J. Biol. Chem., 278, 6928–6935. [DOI] [PubMed] [Google Scholar]
- Xiao W., Chow,B.L. and Milo,C.N. (1998) Mms4, a putative transcriptional (co)activator, protects Saccharomyces cerevisiae cells from endogenous and environmental DNA damage. Mol. Gen. Genet., 257, 614–623. [DOI] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge,S.J. (2000) The DNA damage response: putting checkpoints in perspective. Nature, 408, 433–439. [DOI] [PubMed] [Google Scholar]