MyosinV controls PTEN function and neuronal cell size (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 1.
Published in final edited form as: Nat Cell Biol. 2009 Sep 20;11(10):1191–1196. doi: 10.1038/ncb1961
Abstract
The tumour suppressor PTEN can inhibit proliferation and migration as well as control cell growth in different cell types1. PTEN functions predominately as a lipid phsophatase, converting PI(3,4,5)P3 to PI(4,5)P2, thereby antagonizing PI3K (Phosphoinositide 3-kinase) and its established downstream effector pathways2. However, much is unclear concerning the mechanisms that regulate PTEN movement to the cell membrane necessary for PTEN’s activity towards PI(3,4,5)P33-5.
Here we show a requirement for functional motor proteins in the control of PI3K signalling, involving a previously unknown association between PTEN and MyosinV. FRET measurements revealed that PTEN interacts directly with MyosinV, dependent on PTEN phosphorylation mediated by CK2 and/or GSK3. Inactivation of MyosinV-transport function in neurons increased cell size, which – in line with known attributes of PTEN-loss6, 7 - required PI3K and mTor. Our data demonstrate a myosin-based transport mechanism regulating PTEN function, providing new insights into the signalling networks regulating cell growth.
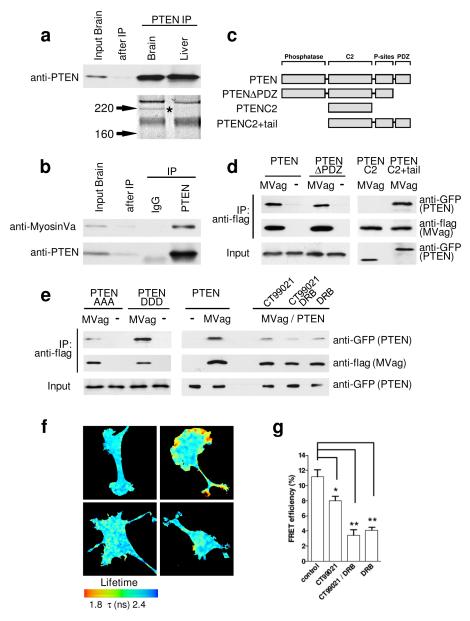
Although sub-cellular distribution of PTEN has been reported to regulate important aspects of PI3K signalling in Dictyostelium, epithelial cells, neutrophils and neurons8-11, the determining cellular machinery remains to be established. We set out to identify proteins involved in regulating the sub-cellular localisation and/or function of PTEN in neurons. Following PTEN immunoprecipitation from brain and liver, PAGE revealed a prominent brain-specific band of approximately 205 kD that was identified as MyosinVa by mass spectrometry (Fig. 1a). The PTEN:MyosinVa association was confirmed using co-immunoprecipitation from brain lysates (Figure 1b). MyosinVa is a plus-end directed, processive motor, which transports mRNA, proteins and multiple organelles along actin filaments into the cell periphery, predominantly through interactions with its globular, cargo-binding domain12. We co-expressed flag-tagged MyosinVa globular domain (MVag) with GFP-PTEN in HEK293 cells and co-immunoprecipitated PTEN with MyosinVa (Fig. 1d). To identify the region within PTEN that mediates the association, flag-MVag was co-expressed with different GFP-tagged PTEN deletion mutants (Fig. 1c, d). The results indicated that the interaction required PTEN aa354-399, which contains a cluster of phosphorylation sites regulated by CK2 and GSK3 (Fig. 1c, d)13, 14. To test if the interaction depends on C-terminal PTEN phosphorylation we used mutant PTEN constructs in which Ser380, Thr382, and Thr383 are mutated to alanines (PTEN AAA) or aspartic acid (PTEN DDD) and found that PTEN:MyosinVa appears to be greatly reduced in PTEN AAA, whilst PTEN DDD was as efficient in associating with MyosinVa as wild type PTEN (Fig, 1e). We then applied inhibitors specific for CK2 (DRB) and/or GSK3 (CT99021) to GFP-PTEN/flag-MVag expressing cells and performed immunoprecipitations as before. The PTEN:MyosinVa association was sensitive to individual inhibition of CK2 or GSK3, and reduced further when both kinase inhibitors were present (Fig. 1e). Thus, the PTEN:MyosinVa interaction depends on the phosphorylation of residues at the PTEN C-terminus, a mechanism previously demonstrated to regulate binding of PTEN with neutral endopeptidase15.
Figure 1. Identification and characterisation of the PTEN:MyosinVa interaction.
a, PTEN was immunoprecipitated (IP) from E18 rat brain and liver homogenate. Coomassie-stained gel of PTEN IP shows a brain-specific band of approximately 205 kD (star), which was identified as MyosinVa by tandem mass spectrometry. b, E18 rat brain extract was incubated with anti-PTEN antibody or control IgG; IPs were analysed with indicated antibodies. c, Domain structure of PTEN and PTEN-deletion constructs. d, HEK293 cells were co-transfected with GFP-PTEN (or indicated GFP-PTEN deletion constructs) and flag-MyosinVa globular domain (MVag). Following anti-flag immunoprecipitation, co-precipitates were analysed with anti-GFP (for PTEN) and anti-flag antibodies (for MVag). The empty flag vector (−) was used in control experiments. The PTEN:MyosinVa interaction is independent of the PTEN PDZ binding site (aa 400-403). The PTEN C2 domain alone does not support interaction (aa 182-353), whilst the PTEN C2 domain plus the C-terminal tail (aa 182-403) shows interaction with MVag. e, The PTEN:MyosinV interaction is regulated by phosphorylation. Left; alanine substitutions of S380, T382, T383 (PTEN AAA) reduces the PTEN:MyosinVa interaction in comparison to substitution of the amino acids to aspartic acids (PTEN DDD). Right; HEK293 cells co-expressing GFP-PTEN and MVag were treated with the CK2 inhibitor 5,6-Dichlorobenzimidazole Riboside (DRB; 10 μM) or the GSK3 inhibitor CT99021 (2 μM), before co-immunoprecipitation as described in d. f, Analysis of the PTEN:MyosinVa interaction by FRET in PC12 cells. Images show the lifetime maps of spatial FRET across cells using a pseudocolour scale (blue, normal GFP lifetime; red, FRET). GFP-PTEN alone (top left) demonstrates normal GFP lifetime in the absence of acceptor, all other images show co-expression of GFP-PTEN and mCherry-MVag. In the lower panels, cells were treated with DRB (left), or DRB and CT99021 (right). g, Bar graph representing the average FRET efficiency of 12 cells (4 cells in 3 independent experiments) ± sem. *p<0.02; **p<0.002.
To analyse localisation and regulation of the protein:protein interaction in intact cells we measured fluorescence resonance energy transfer (FRET) between PTEN and MyosinVa. We expressed GFP-PTEN and mCherry-MVag in PC12 cells and analysed FRET by fluorescence lifetime imaging microscopy (FLIM)16. Data demonstrated reproducible spots in GFP lifetime reduction, indicative of direct PTEN:MyosinVa interaction that were confined to small patches in the cell periphery (Fig. 1f). In agreement with our co-immunoprecipitation experiments we found that treatment with DRB and/or CT99021 reduced PTEN:MyosinVa FRET efficiencies (Fig. 1f, g). Thus, PTEN interacts directly with MyosinVa in a spatially regulated manner, dependent on the activity of CK2 and GSK3.
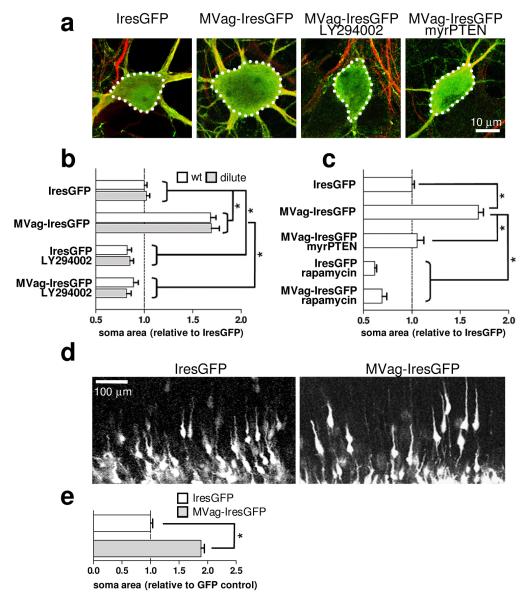
MyosinVa is highly enriched in the nervous system (Supplementary Fig. 1), where it localizes within neuronal growth cones and dendritic spines17-19. In humans, mutations in MyosinVa cause Griscelli syndrome type 1 and Elejalde syndrome, which are both characterized by hypopigmentation of the skin and hair, as well as prepubescent onset of severe neurological abnormalities, including seizures and mental retardation20. Similarly, mice lacking functional MyosinVa due to homozygous deletion of dilute lethal Myo5ad-1 - hereafter referred to as dilute lethal mice - show neurological abnormalities17. We utilized dilute lethal mice to test the possibility that MyosinVa mediates PTEN movement toward the membrane. In this case MyosinVa deficiencies would phenocopy loss of PTEN, potentially generating neurons in the cerebral cortex and hippocampus with enlarged soma, ectopic processes and increased numbers of synapses6, 7, 21, 22. However, gross morphology of neurons in the cortex and the hippocampus at postnatal day (P) 16 was normal in dilute lethal mice (Supplementary Fig. 2). In addition to MyosinVa, two other class members, MyosinVb and MyosinVc, have been identified23, 24 and co-immunoprecipitation experiments reveals interactions with PTEN (Supplementary Fig. 3). MyosinVb, but not MyosinVc, is expressed in the developing brain (Supplementary Fig. 4), raising the possibility for functional compensation following loss of MyosinVa. Therefore, we exploited the MyosinVa globular domain, which is known to block MyosinVa-mediated transport25, and, when present in excess, should compete for PTEN interactions to other MyosinVs. IresGFP or MVag-IresGFP were expressed in hippocampal neurons cultured from wt mice and dilute lethal littermates at 7 days in vitro (DIV) and neuronal morphology was assessed at 14 DIV. MVag-IresGFP induced a significant increase in neuronal soma size (Fig. 2a, b), which was antagonised by the PI3K inhibitor LY294002 (Fig. 2a, b). This result provides functional evidence that MyosinV regulates PI3K signalling, possibly by coordinating membrane transport of PTEN. To test this idea, we forced PTEN to the membrane by myristoylation (myrPTEN), which uncouples MyosinV-mediated transport from PTEN function. Expression of myrPTEN inhibited MVag’s effect on soma size (Fig. 2c). We then tested the effect of the mTor inhibitor rapamycin, an established downstream signalling component of neuronal hypertrophy in PTEN deficient brains6. Rapamycin treatment reduced soma size in IresGFP and also in MVag expressing neurons (Fig. 2c). We also complemented the dominant negative MVag approach by silencing MyosinVb in neurons. We co-transfected hippocampal neurons cultured from wt mice and dilute lethal littermates with IresGFP in the presence of unspecific control siRNA or MyosinVb specific siRNA, which resulted in a small but significant increase in soma sizes in neurons obtained from dilute lethal mice, only (Supplementary Fig. 5). These data demonstrate that loss of MyosinV transport function through overexpression of MVag – as well as through functional inactivation of the two main MyosinVs in hippocampal neurons - mimic PTEN deficiencies with respect to soma size. Collectively, these results are consistent with a model in which MyosinV-transport regulates PI3K signalling through interaction with PTEN.
Figure 2. MyosinV regulates neuronal soma size through PI3K signalling.
Hippocampal neurons from E15.5 wild type (wt) or dilute lethal (dilute) mice were cultured for 7 days in vitro (DIV) and transfected with different constructs before fixation at 14 DIV. Where indicated, LY294002 (10 μM) or Rapamycin (200 nM) was applied 2 hours after transfection. a, Confocal images of neurons labelled with anti-Map-2 antibody (red); GFP (green) visualizes Ires-GFP or MVag-IresGFP expression. b, c, Quantification of neuronal soma size. For measurements, images were taken on an epifluorescence microscope, and examined using SimplePCI software after manually delineating the cell margins. Each data point represents the relative neuronal soma area compared to wt IresGFP expressing neurons ± sem, which was set to 1. n=4 independent experiments (n=6 for wt and dilute neurons, Ires-GFP or MVag-IresGFP), in each experiment at least 30 neurons were assessed. *p<0.001 (Student’s T-test). d, Expression of MVag in cortical neurons in vivo increases neuronal soma size. Coronal sections of E15.5 mouse cortices electroporated with control GFP (left) or GFP/MVag (right) at E13.5. e, Quantification of neuronal soma size in vivo. Only neurons undergoing radial migration were evaluated. n=3 independent control experiments and 4 MVag experiments, in each experiment at least 70 neurons were assessed. *p<0.001
To study if MyosinV controls neuronal cell size in vivo, we injected the constructs expressing MVag and GFP, or GFP alone, into the lateral ventricle of E13.5 mouse embryos and performed in utero electroporation26. After normal embryonic development in vivo for two days, many GFP expressing neurons adopted morphologies typical for their movement along the radial fibre scaffold, with a leading process extending toward the cortical plate and an axon trailing toward the ventricular zone (Fig. 2d). MVag expression permitted radial orientation in the majority of neurons, although the calibre of the leading process appeared to be enlarged (Fig. 2d). Most notably, MVag expressing neurons showed a significant increase in the size of their somas (Fig. 2d, e). Thus, functional MyosinVs are also essential for proper control of neuronal cell size in vivo.
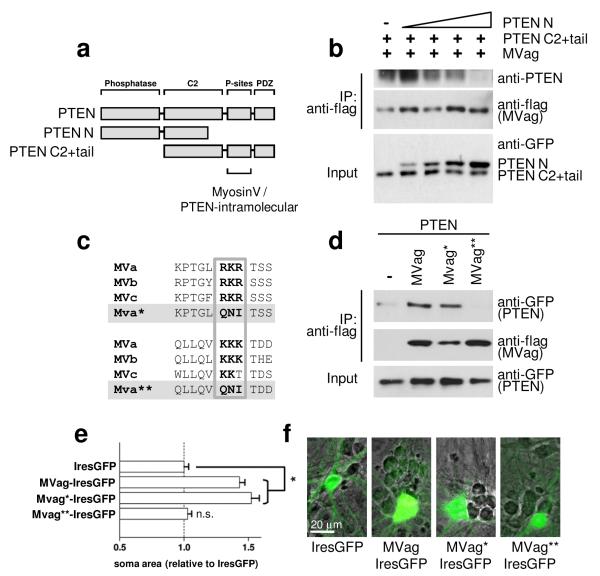
C-terminal PTEN phosphorylation has previously been shown to induce a conformational switch of the protein, keeping the enzyme in a ‘closed’ formation with decreased membrane localisation and biological activity27, 28. Our results demonstrating that PTEN binds preferentially with MyosinVa in its phosphorylated state may indicate competitions between this intramolecular PTEN interaction and an interaction with MyosinV. In order to test this idea we examined if the association of the PTEN C2+tail mutant with flag-MVag (see Fig.1d) can be blocked by an N-terminal fragment of PTEN (PTEN N, aa 1-317) (Fig. 3a). We co-expressed MVag, the GFP-PTEN C2+tail mutant and increasing levels of PTEN N in HEK293 cells, and co-immunoprecipitated the PTEN tail mutant with MyosinVa. While PTEN N failed to interact with MVag (not shown here), increasing levels of this mutant competed for the PTEN tail association with the Myosin globular domain (Fig. 3b). Phosphorylation-dependent interactions of the PTEN C-terminal tail have been reported to require a cluster of three positively charged amino acids15, 27. We identified two similar clusters with high degree in conservation amongst all MyosinV class members (Fig. 3c), and mutated them individually to uncharged aa glutamine, asparagine, and isoleucine (MVa* RKR↦QNI; MVa** KKK↦QNI), as previously15. Only the second cluster (KKK) was essential for MVag’s association to PTEN in immunoprecipitation experiments, and – significantly – also for increases in neuronal soma size of native MVag-IresGFP (Fig. 3d-f). These experiments indicate that PTEN:MyosinV is based on electrostatic interactions between PTEN and a positively charged region located in the globular domain of MyosinV, which is in competition with PTEN’s closed conformation.
Figure 3.
The PTEN:MyosinV interaction competes with PTEN’s closed conformation, and is mediated by a positively charged region located in the globular domain of MyosinV. a, Domain structure of PTEN and PTEN-deletion constructs used in co-immunoprecipitation experiments. C-terminal PTEN phosphorylation sites (P-sites) involved in mediating intramolecular PTEN and MyosinV binding are indicated. b, the PTEN:MyosinVa interaction can be competed for by the PTEN N-terminal region. HEK293 cells were co-transfected with flag-MyosinVa globular domain (MVag) and the GFP-PTEN C2+tail mutant, in the presence of increasing amounts of GFP-PTEN N. Following anti-flag immunoprecipitation, precipitates were analysed with anti-flag antibodies (for MVag) and with an anti-PTEN antibody recognising the PTEN C-terminus, only. c, Alignment of positively charged amino acid clusters present in the globular domains of MyosinVa, MyosinVb and MyosinVc. MVa* and MVa** denotes MVa RKR↦QNI and MVa KKK↦QNI mutants, respectively. d, Essential requirement of the KKK cluster for the PTEN:MyosinV interaction. Flag-MVag, flag-MVag* or flag-MVag** were co-expressed with GFP-PTEN as before and following anti-flag immunoprecipitation, precipitates were analysed with anti-flag antibodies (for MVags). The empty flag vector (−) was used in control experiments. e, Requirement of the KKK cluster for MVag induced increases in cell size (Fig. 2). Hippocampal neurons were cultured and transfected with control IresGFP, MVag, MVag*, or MVag** as before. Each data point represents the relative neuronal soma area compared to control (IresGFP) expressing neurons ± sem, n=3 independent experiments, in each experiment at least 40 neurons were assessed. *p<0.0001. f, Hippocampal neurons from E15.5 mice were cultured for 7 days in vitro (DIV) and transfected with MVag-IresGFP before fixation at 14 DIV. Phase contrast image was overlaid with GFP to visualise neuronal soma increase in MVag expressing, in comparison to non-expressing, neurons in the same culture.
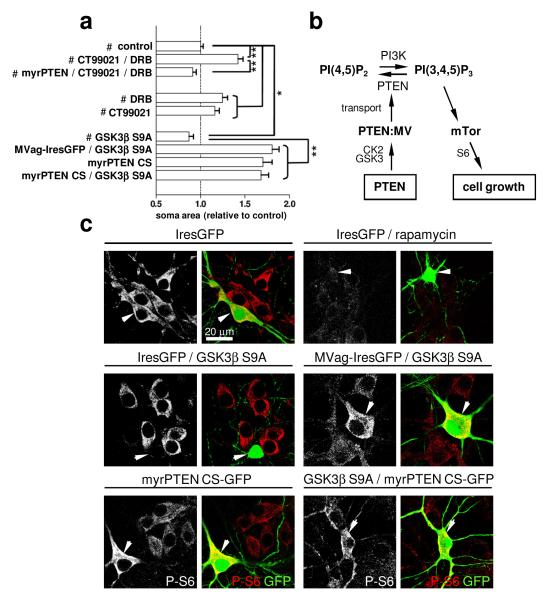
We next investigated potential upstream regulators of the PTEN:MyosinV interaction in the control of neuronal cell size. We showed that the PTEN:MyosinVa interaction is antagonized following inhibition of GSK3 and CK2 (Fig. 1), which indicates that these kinases may negatively regulate neuronal soma size upstream of PTEN transport. To test this hypothesis, we applied DRB and CT99021 to hippocampal neurons for 7 days, before analysis of soma size. Inhibition of CK2 and GSK3 led to soma size increases in IresGFP expressing neurons, but not in neurons expressing myrPTEN (Fig. 4a). In mice, neuron-specific overexpression of constitutive-active GSK3 decreases the brain volume, resulting in microcephaly due to reduced sizes of neurons29. Correspondingly, we find that somas of hippocampal neurons were significantly smaller following overexpression of the constitutive-active GSK3βS9A mutant (Fig. 4a). In contrast, perturbation of MyosinV-transport by co-expressing MVag with GSK3β S9A maintained MVag-enlarged neuronal soma size, which places GSK3 activity upstream of MyosinV transport and cell size regulation. To further substantiate this signalling relationship, we co-expressed a myristoylated phosphatase-dead PTEN mutant (myrPTEN C124S) with GSK3β S9A. Expression of myrPTEN C124S alone increased neuronal soma size (Fig. 4a), and we reasoned that in combination with GSK3β S9A, myrPTEN C124S signifies unequivocally whether GSK3 operates upstream or downstream of PTEN:MyosinV in controlling soma size. The myrPTEN C124S phenotype was dominant (Fig. 4a), which highlights a requirement for functional PTEN to decrease growth resulting from overactivation of GSK3.
Figure 4. Neuronal soma size is controlled by GSK3 and CK2 upstream of PTEN and MyosinV.
Hippocampal neurons were cultured and transfected as described in Figure 2. Control IresGFP (#) was expressed in all indicated treatments. CT99021 (2 μM) or DRB (10 μM) was applied 2 hours after transfection. a, Quantification of neuronal soma size at 14 DIV. Each data point represents the relative neuronal soma area compared to control (IresGFP) expressing neurons ± sem, n=3 independent experiments, in each experiment ≥ 40 neurons were assessed. *p<0.03, **p<0.0001. b, Schematic illustrating the signalling relationship. c, Neuronal cultures were labelled with anti-PS6 antibody (left; shown in red in the merge); green neurons in the merged image (arrowhead) visualize neurons expressing indicated constructs.
We next assessed whether the presented evidence for GSK3 in controlling neuronal soma size by regulating the PTEN:MyosinV interaction can be correlated with the activity of the growth-control signalling machinery. Cell size enlargements caused by PTEN-deficiencies require mTor6, and correlate with elevated phosphorylation of its effector ribosomal protein S67. Hippocampal neurons maintain robust pS6 signals, which remain unaffected by expression of IresGFP (Fig. 4c). As expected, rapamycin treatment resulted in decreased pS6-labelling, confirming the specificity of this approach. Similarly, GSK3β S9A expression was accompanied by decreased pS6-labelling, whilst co-expression of GSK3β S9A with MVag antagonised GSK3-induced reduction in pS6 (Fig. 4c). Neurons expressing myrPTEN C124S, or myrPTEN C124S in combination with GSK3β S9A both exhibited marked amplification of pS6 in comparison to non-transfected neurons (Fig. 4c). Collectively, these results establish a causal link between GSK3 as upstream regulators of the PTEN:MyosinV interaction in the control of neuronal soma size and mTor signalling.
We have demonstrated a transport mechanism that controls PI3K signalling and neuronal soma size, which involves direct interaction of MyosinV with the tumour suppressor PTEN. This interaction is likely to permit a degree of cellular signalling integration, since PTEN associates with MyosinV – as well as antagonises PI3K signalling and functions – when GSK3 is active. Consistent with an established role for PI3K in negatively regulating GSK3 itself, our identification of a mechanism by which GSK3 phosphorylation directs PTEN function provides a possible novel positive feedback system that operates in concert with CK2 (Fig. 4b). Such organization offers a powerful way to amplify or antagonise PI3K signalling thresholds required for neurons to respond to extracellular signals involved in determining and maintaining proper neuronal soma size in the brain.
Methods
In utero electroporation
For in utero electroporation, timed pregnant E13.5 CD1 mice were used (the morning of the vaginal plug was E0.5). Experiments were performed following institutional guidelines and approved protocols. pCAβ-IRES-eGFPm5-grip was used as a control for pCAβ-MVag-IRES-eGFPm5-grip. One μl of purified DNA (1-2μg/μl) mixed with 0.005% Fast Green was injected in utero into the lateral ventricle and electroporated into the neocortical ventricular zone according to published protocols26. Brains were PFA-fixed and vibratome-sectioned for analyses by confocal microscopy.
FRET measurements by Fluorescence Lifetime Imaging Microscopy (FLIM)
In order to determine the spatial interaction between PTEN and MyosinVA, PC12 cells were transfected with p-mCherry-MVag and/or PTEN-EGFP-c2, differentiated for 2 days with NGF (50 ng/ml) and fixed for FLIM experiments to measure Förster resonance energy transfer FRET. Time-domain FLIM experiments and data analysis were performed as described previously16.
Cloning
The globular domain from chick MyosinVa (MVag) was amplified by PCR (Proofstart;QIAGEN) (primers: 5′-GCCACCATGAGTCTGCAGCATGAGATCACC-3′ and 5′-GACACGTGATATGAAACCCAG-3′) and cloned into pCAβ-IRES-eGFPm5-grip, p-mCherry n1 (Clontech) or p3XFLAG-CMV™-7.1 (Sigma). Similarly, Mouse cDNA was used to clone the globular domain of MyosinVb (primers: 5′- ATGTCCACCATCAACGGCATTAAG-3′ and TTGACTTCATTGAGAAACTCCAG-3′) and MyosinVc (primers: 5′-ATGAGTGCTATTAACGGCATCAAGC-3′ and 5′- TTCAGTCTCCGGAGAAAGCCG-3′) into p3XFLAG-CMV™-7.1. Site directed mutagenesis of parental flag-MVag or MVag-IresGFP vectors were performed using the QuikChange II XL mutagenesis kit (Stratagene) according to the manufacturer’s protocols. The primers used were 5′-GGGTGAAACCAACAGGGCTGCAAAACATAACATCCAGCATTGCTGATGA-3′ and 5′-TCATCAGCAATGCTGGATGTTATGTTTTGCAGCCCTGTTGGTTTCACCC-3′ for the MVa RKR↦QNI mutation, and 5′-GCTGCACAGTTGTTGCAAGTGCAAAACATAACAGATGAAGATGCA-3′ and 5′-GCTTCTGCATCTTCATCTGTTATGTTTTGCACTTGCAACAACTGT-3′ for the MVa KKK↦QNI mutation. All mutations were validated by sequencing.
Immunoprecipitations and tandem mass spectrometry
Cortices or livers from E18 rat embryos were homogenized in lysis buffer (50 mM Tris pH 7.5; 150 mM NaCl; 1% Triton X-100; 2 μM NaOV and 500 μM NaF) containing protease inhibitors (Roche). Immunoprecipitations were performed overnight (4°C) using 5μg of anti-PTEN antibody (A2B1, Santa Cruz) or mouse control antibody and protein-G beads (PIERCE). After washing, beads were boiled in loading buffer and separated on 10% SDS-polyacrylamide gels, which were fixed and stained using Brilliant Blue G - Colloidal Concentrate (Sigma) according to manufacturer’s instructions. Selected protein bands were isolated, processed and sequenced by Proteome Sciences plc. In short, after in gel trypsin digestion, samples were analysed by LC/MS/MS, the mass spectral data was processed into peak lists and searched against the Swiss Prot and NCBI non-redundant databases using Mascot software (Matrix Science, UK). For co-immunoprecipitation experiments, HEK293 cells were transfected with different constructs using Lipofectamine 2000 and anti-FLAG® M2 Affinity Gel (Sigma) was used for isolating the 3xFlag-MVag complex. Western blot analysis of immunoprecipitates was performed using anti-flag (Sigma), anti-GFP (Abcam) or anti-PTEN antibodies (Santa Cruz or Cell Signaling).
Real time quantitative PCR (qPCR)
Cortices from E15.5 dilute lethal and wildtype littermates were used to isolate total RNA (Qiagen RNeasy mini kit), which was reversed transcribed into random primed cDNA (Reverse Transcription Kit, Promega) and served as template for all qPCR experiments. MyosinVa, MyosinVb, MyosinVc and PTEN mRNA levels were measured relative to Gapdh, Pgk1 and Hprt. Primers for qPCR analysis of the different mRNAs were ordered from QIAGEN (QuantiTect Primer Assays), Quantifast SYBR Green (QIAGEN) was used as a reporter for detection with the Light Cycler1.0 (Roche). All reactions were performed according to the manufacturer’s instructions.
Supplementary Material
Supplementary Material
Acknowledgements
This work was funded by the Biotechnology and Biological Science Research Council to BJE (BB/C514307/1), an MRC program grant to CPD (G9403619). MP is a Royal Society University Research Fellow. We thank John Hammer 3rd for providing the MyosinVa cDNA, Calum Sutherland for the GSK3 inhibitor CT99021, Eric Blanc for help with statistical analyses and Frank Gertler for comments on the manuscripts.
Footnotes
Author information The authors declare no competing financial interest.
References
- 1.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13:478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 3.Pinal N, et al. Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr Biol. 2006;16:140–149. doi: 10.1016/j.cub.2005.11.068. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi Y, Morales FC, Kreimann EL, Georgescu MM. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. Embo J. 2006;25:910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vazquez F, et al. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc Natl Acad Sci U S A. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon CH, Zhu X, Zhang J, Baker SJ. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci U S A. 2003;100:12923–12928. doi: 10.1073/pnas.2132711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadborn NH, et al. PTEN couples Sema3A signalling to growth cone collapse. J Cell Sci. 2006;119:951–957. doi: 10.1242/jcs.02801. [DOI] [PubMed] [Google Scholar]
- 9.Heit B, et al. PTEN functions to ‘prioritize’ chemotactic cues and prevent ‘distraction’ in migrating neutrophils. Nat Immunol. 2008;9:743–752. doi: 10.1038/ni.1623. [DOI] [PubMed] [Google Scholar]
- 10.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Belmonte F, et al. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 2007;128:383–397. doi: 10.1016/j.cell.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology (Bethesda) 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 13.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 14.Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J Biol Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 15.Sumitomo M, et al. Synergy in tumor suppression by direct interaction of neutral endopeptidase with PTEN. Cancer Cell. 2004;5:67–78. doi: 10.1016/s1535-6108(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 16.Parsons M, Messent AJ, Humphries JD, Deakin NO, Humphries MJ. Quantification of integrin receptor agonism by fluorescence lifetime imaging. J Cell Sci. 2008;121:265–271. doi: 10.1242/jcs.018440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 18.Wang FS, Wolenski JS, Cheney RE, Mooseker MS, Jay DG. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 19.Espreafico EM, et al. Primary structure and cellular localization of chicken brain myosin-V (p190), an unconventional myosin with calmodulin light chains. J Cell Biol. 1992;119:1541–1557. doi: 10.1083/jcb.119.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanal O, et al. An allelic variant of Griscelli disease: presentation with severe hypotonia, mental-motor retardation, and hypopigmentation consistent with Elejalde syndrome (neuroectodermal melanolysosomal disorder) J Neurol. 2000;247:570–572. doi: 10.1007/s004150070162. [DOI] [PubMed] [Google Scholar]
- 21.Kwon CH, et al. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 22.van Diepen MT, Eickholt BJ. Function of PTEN during the formation and maintenance of neuronal circuits in the brain. Dev Neurosci. 2008;30:59–64. doi: 10.1159/000109852. [DOI] [PubMed] [Google Scholar]
- 23.Zhao LP, et al. Cloning and characterization of myr 6, an unconventional myosin of the dilute/myosin-V family. Proc Natl Acad Sci U S A. 1996;93:10826–10831. doi: 10.1073/pnas.93.20.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez OC, Cheney RE. Human myosin-Vc is a novel class V myosin expressed in epithelial cells. J Cell Sci. 2002;115:991–1004. doi: 10.1242/jcs.115.5.991. [DOI] [PubMed] [Google Scholar]
- 25.Brown JR, Peacock-Villada EM, Langford GM. Globular tail fragment of myosin-V displaces vesicle-associated motor and blocks vesicle transport in squid nerve cell extracts. Biol Bull. 2002;203:210–211. doi: 10.2307/1543401. [DOI] [PubMed] [Google Scholar]
- 26.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 27.Rahdar M, et al. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci U S A. 2009;106:480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spittaels K, et al. Neonatal neuronal overexpression of glycogen synthase kinase-3 beta reduces brain size in transgenic mice. Neuroscience. 2002;113:797–808. doi: 10.1016/s0306-4522(02)00236-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material