Integrins (original) (raw)
Abstract
Integrins are cell adhesion receptors that are evolutionary old and that play important roles during developmental and pathological processes. The integrin family is composed of 24 αβ heterodimeric members that mediate the attachment of cells to the extracellular matrix (ECM) but that also take part in specialized cell-cell interactions. Only a subset of integrins (8 out of 24) recognizes the RGD sequence in the native ligands. In some ECM molecules, such as collagen and certain laminin isoforms, the RGD sequences are exposed upon denaturation or proteolytic cleavage, allowing cells to bind these ligands by using RGD-binding receptors. Proteolytic cleavage of ECM proteins might also generate fragments with novel biological activity such as endostatin, tumstatin, and endorepellin. Nine integrin chains contain an αI domain, including the collagen-binding integrins α1β1, α2β1, α10β1, and α11β1. The collagen-binding integrins recognize the triple-helical GFOGER sequence in the major collagens, but their ability to recognize these sequences in vivo is dependent on the fibrillar status and accessibility of the interactive domains in the fibrillar collagens. The current review summarizes some basic facts about the integrin family including a historical perspective, their structure, and their ligand-binding properties.
Electronic supplementary material
The online version of this article (doi:10.1007/s00441-009-0834-6) contains supplementary material, which is available to authorized users.
Keywords: Integrins, History, Ligands, α and β subunits, RGD, GFOGER, Collagen
History
Integrins are cell adhesion receptors that are evolutionary old (Johnson et al. 2009). Despite their long history, they have only been characterized at the molecular level for approximately 25 years. During this period, a large number of articles on the ever-increasing intricacies of integrin action has been published. To summarize this large amount of data on integrins “at a glance” is thus almost an impossible task. In the current review, we have however attempted to provide some of the basic facts about integrins. Recent excellent reviews on various aspects of integrins structure and function will be referred to in the text.
One reason for the difficulties encountered when trying to characterize the integrin family is that many of their ligands are large multi-adhesive extracellular matrix (ECM) molecules that, in addition to binding integrins, bind other proteins including ECM molecules, growth factors, cytokines, and matrix-degrading proteases. One successful approach that was instrumental in the identification of integrins took advantage of antibodies that blocked cell adhesion (Horwitz et al. 1985; Knudsen et al. 1985). In another approach, the mapping of the minimal cell adhesion site in fibronectin to the RGDS sequence gave rise to affinity chromatography protocols with increased specificity (Pierschbacher et al. 1983; Ruoslahti and Pierschbacher 1986). In these affinity protocols, the optimization of ion composition in the purification buffers was essential and resulted in the empirical finding that manganese ions (Mn2+) increased integrin affinity (Gailit and Ruoslahti 1988). In 1986, the antibody approach led to the expression cloning of cDNA encoding the chick integrin β1 subunit (Tamkun et al. 1986). The name “integrin” was given to denote the importance of these receptors for maintaining the integrity of the cytoskeletal-ECM linkage (Hynes 2004, Tamkun et al. 1986). In the 1980s, several groups were working on seemingly disparate cell surface proteins, which at the time were named position specific (PS) antigens in Drosophila (Leptin et al. 1987; Wilcox et al. 1984), very late antigens of activation (VLA) on immune cells (Hemler et al. 1985), cell surface receptors on lymphoid and myeloid cells (Springer et al. 1986), and platelet glycoproteins (Parise and Phillips 1985, 1986). With the cloning of the cDNAs encoding these proteins, it became clear that they were related to the fibronectin receptors isolated by using RGD peptides or cell adhesion blocking antibodies, and that they all belonged to what was to be called the integrin family of cell adhesion receptors (Hynes 2004; Fig. 1, see also Electronic Supplementary Material).
Fig. 1.

Integrin founding fathers. Erkki Ruoslahti (left) and Richard O. Hynes (right) contributed seminal data in the early days of cell adhesion research leading to the characterization of the integrin family
Structure
When integrins were being identified with antibodies to integrin β subunits, several proteins were co-immunoprecipitated, and the number of subunits that composed the functional receptors was by no means obvious. However, with antibodies to integrin α subunits, and with protocols using RGDS peptides enabling the affinity purification of pure receptors, it became clear that the functional receptors were heterodimers. Integrin heterodimers are composed of non-covalently associated α and β subunits (Hynes 2002). In vertebrates, the family is composed of 18 α subunits and 8 β subunits that can assemble into 24 different heterodimers (Takada et al. 2007). The integrins can be grouped into subgroups based on ligand-binding properties or based on their subunit composition (see Table 1, 2).
Table 1.
Characteristics of human integrin α subunits.Data are presented for the human integrin α chains and have been retrieved from original data submitted to the NCBI database (http://www.ncbi.nlm.nih.gov/sites/entrez) and original publications. For ligand specificity, see references in text (ICAM intercellular adhesion molecule, VCAM vascular cell adhesion molecule, VEGF vascular endothelial growth factor)
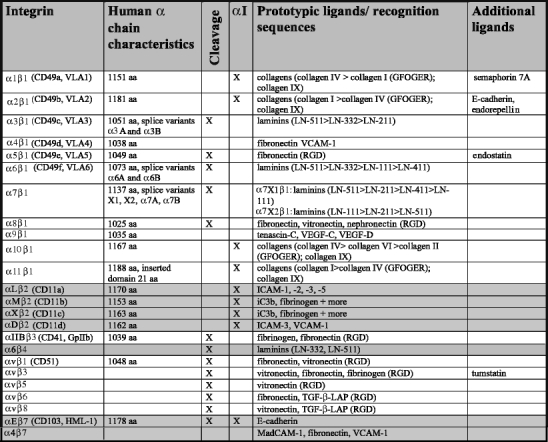
Table 2.
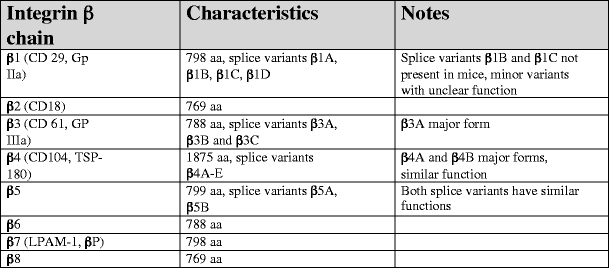
Characteristic of human integrin β subunits. Data are presented for the human integrin β chains and have been retrieved from original data submitted to NCBI database (http://www.ncbi.nlm.nih.gov/sites/entrez) and original publications (see text)
The β1 integrins, β2 integrins, and αv-containing integrins are the three largest groups in this kind of classification (Fig. 2, see also Electronic Supplementary Material). The α and β subunits show no homology to each other, but different α subunits have similarities among themselves, just as there are conserved regions in the different integrin β subunits.
Fig. 2.
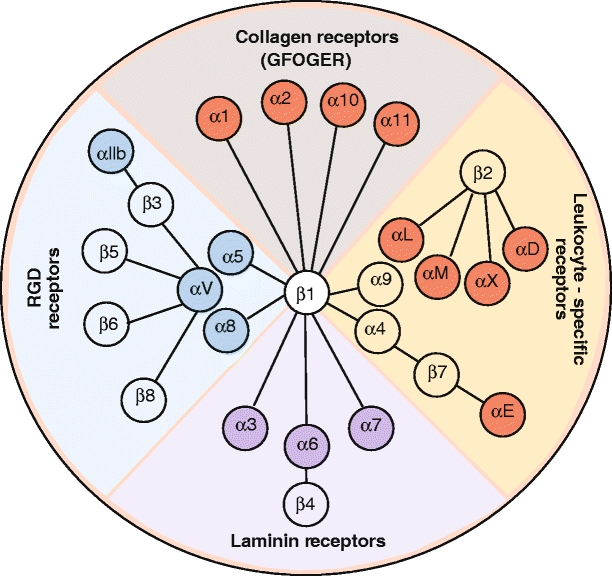
Representation of the integrin family. In vertebrates, the integrin family contains 24 heterodimers. Isolated species that have undergone genome duplication (e.g., Danio rerio) have more integrin family members. In higher vertebrates, the integrin family has 24 prototypical members
Integrin α subunits
The α subunit is composed of a seven-bladed β-propeller, which is connected to a thigh, a calf-1, and a calf-2 domain, together forming the leg structure that supports the integrin head (Fig. 3, see also Electronic Supplementary Material). The last three or four blades of the β-propeller contain EF-hand domains that bind Ca2+ on the lower side of the blades facing away from the ligand-binding surface. Ca2+ binding to these sites allosterically affects ligand binding (Humphries et al. 2003; Oxvig and Springer 1998).
Fig. 3.
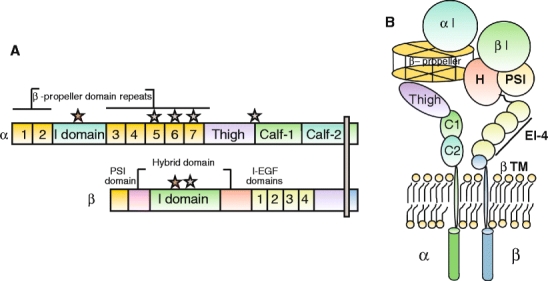
Representation of a prototypical αI-domain-containing integrin heterodimer. Nine out of the 18 integrin α chains contains an αI domain, as shown, but all integrins contain a βI domain in the β subunit. a Representation of the domains in αI domain-containing integrin (stars divalent cation-binding sites. b Representation of arrangement of domains in αI-domain-containing integrin kying in a membrane
Nine of the integrin α chains contain an I domain, also called the A domain, which is a domain of approximately 200 amino acids, inserted between blades 2 and 3 in the β-propeller (Larson et al. 1989). The αI first appeared in chordate integrins, and is thus absent in invertebrates but is present in vertebrates (Johnson et al. 2009). The αI domain is present in the β2 integrin subgroup of integrins, in the collagen-binding integrins belonging to the β1 subfamily (α1, α2, α10, and α11), and the αE integrin chain forming the αEβ7 heterodimer. The I domain assumes a Rossman fold with five β-sheets surrounded by seven α helices. Ligand binding occurs via a coordinating Mg2+ ion in the so-called metal-ion-dependent adhesion site (MIDAS) motif (Lee et al. 1995). The αI domains with the capacity to interact with collagens, in addition, contains a so-called αC helix (Emsley et al. 1997), which has been suggested to play a role in collagen binding.
The αI-domain-containing integrins show fairly high homology in their αI domains, but the α chain cytoplasmic domains are highly divergent, only sharing the GFFKR sequence or even the core GFFXR sequence in the membrane proximal region.
Relatively little is known about proteins interacting with the α-chain cytoplasmic tails.
An area between the hybrid domain in the β subunit and a surface in the β-propeller of the α subunit seems to be crucial for the integrin heterodimerization that occurs intracellularly prior to transport to the cell surface (Humphries 2000). Presumably, the specificity of chain selection lies in those sequences adjacent to these interacting surfaces. Generally, an excess of β subunits exists in the cell, and the amount of α subunit determines the amount of receptor that will go to the cell surface (Santala and Heino 1991). Free α and β subunits do thus not exist at the cell surface.
Integrin β subunit
The β subunit contains a plexin-sempahorin-integrin (PSI) domain, a hybrid domain, a βI domain (Lee et al. 1995), and four cysteine-rich epidermal growth factor (EGF) repeats. The βI domain contains an Mg2+ coordinating MIDAS and a site adjacent to MIDAS (ADMIDAS) binding an inhibitory Ca2+ ion. This ADMIDAS site binds the Mn2+ ion leading to a conformational change resulting in an active form of the integrin (Humphries et al. 2003).
The β integrin chains share homology in the cytoplasmic tail, with NPX/Y motifs able to bind proteins containing PTB domains. In recent years, several proteins have been found to interact with the β subunit (Legate and Fassler 2009).
Some key proteins seem to be essential for binding and, in doing so, break salt bridges formed with the α subunit that keeps the integrin in the inactive conformation. A detailed study has recently clarified the interacting regions in the αIIb and β3 subunit transmembrane domains, suggesting a model for conformation-mediated changes over the membrane (Lau et al. 2009). Talin 1–2 and kindlins 1–3 seem to act synergistically to activate integrins by binding to integrin β subunit tails (Larjava et al. 2008; Senetar et al. 2007; Tadokoro et al. 2003), whereas filamin A negatively regulates activation (Kiema et al. 2006). Migfilin is another molecular switch that, by blocking the integrin-binding region of filamins (Ithychanda et al. 2009), can regulate integrin activation. More recently, the integrin-linked kinases (ILK) (Honda et al. 2009) and focal adhesion kinase (FAK) (Michael et al. 2009) enzymes taking part in outside-in signaling have also been shown to affect integrin activation via inside-out signaling.
Conformational changes in integrins
The crystallization of a soluble integrin heteodimer has made clear that integrins can exist in a compact bent conformation (Xiong et al. 2002). Later research has shown that this conformation represents an inactive conformation (Nishida et al. 2006). Some data suggest that integrins “breathe” and change between different conformations with individual variations as to their degree of bending. Furthermore, the bent conformation does not always seem to be inactive, especially with regard to small ligands (Askari et al. 2009). More recently, mechanical tension has been demonstrated to consolidate integrin contact points by further stretching the conformation to stabilize the active conformation, thereby increasing affinity (Askari et al. 2009; Astrof et al. 2006; Friedland et al. 2009). For integrin α5β1, the mechanical tension induces α5β1 engagement with the synergy site in fibronectin, in turn leading to FAK phosphorylation (Friedland et al. 2009). A recent synergy site in nephronectin has been suggested mainly to exert its action by bringing the RGD site into a best-fit conformation with regard to high-affinity integrin binding (Sato et al. 2009).
In addition to the affinity modulation that occurs in various activation states, integrin clustering by multivalent ligands, and possibly also changes in membrane fluidity, cause avidity changes of integrin contacts (Carman and Springer 2003).
For integrins that require a tight control of activity, such as platelet integrin αIIβ3 and β2 integrins, precise activating mechanisms must exist (Luo et al. 2007). When kindlin-3 is mutated, integrin activation in leukocytes and platelets fails (Moser et al. 2008; Svensson et al. 2009), demonstrating the central role of kindlin-3 for integrin activation on these cell types.
Ligand recognition
The ligand-binding site forms in a region at the intersection of the integrin α-chain β-propeller and the βI domain, with the α chain being central in determining ligand specificity. Integrins with an αI domain bind ligands via the αI domain, but since this ligand-binding causes distinct conformational changes in the I domain, this in turn affects the conformation of the β subunit (Luo et al. 2007). For some αI domains, interactions with the β chain are even needed for the proper folding of the αI domain (Valdramidou et al. 2008). The list of integrin ligands is long (Humphries et al. 2006; Johnson et al. 2009) and includes the major constituents of the ECM.
Prototypic integrin ligands and recognition sequences
The prototypic integrin ligand, fibronectin, contains the amino acid sequence RGDS at the apex of a flexible loop connecting two β-strands in the 10th fibronectin type III repeat (Dickinson et al. 1994). The RGD sequence is also present in vitronectin, fibrinogen, and the LAP complex part of inactive transforming growth factor-β (TGF-β) and many other ECM proteins (Humphries et al. 2006). Elegant studies have shown that the epithelial αvβ6 and αvβ8, by binding RGD in LAP, are the major integrins that activate TGF-β in vivo either by allosteric changes in TGF-β-LAP (αvβ6) or by inducing matrix metalloproteinase-14 and causing proteolytic release of TGF-β (αvβ8; Aluwihare et al. 2009; Mu et al. 2002; Munger et al. 1999). More recently, a mechanical-strain-dependent contribution of myofibroblast β1 integrins to TGF-β activation has been demonstrated (Wipff et al. 2007).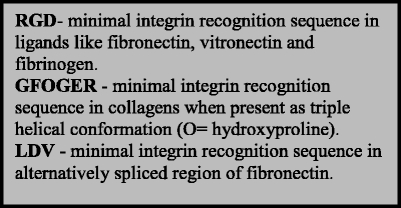
In many biochemistry and cell biology books, the RGD sequence is presented as the major integrin-binding sequence, but only a third of the integrins are known to bind this sequence (Table 1). RGD peptides can be used to assess the involvement of RGD-binding integrins in a certain event, but lack of effect of RGD peptides does not exclude integrin involvement in a process since two-thirds of integrins bind ligands RGD-independently.
Some isoforms of the basement membrane protein laminin contain an RGD sequence, but the sequence is not evolutionary conserved in these isoforms and is not recognized by the laminin-binding integrins. In laminin-111, the RGD site is masked in the native molecule (Aumailley et al. 1990; Schulze et al. 1996), whereas in laminin isoforms containing the α5 chain, it is exposed in the native molecule (Domogatskaya et al. 2008; Forsberg et al. 1994; Genersch et al. 2003; Sasaki and Timpl 2001). The full physiological importance of the αvβ1- and αvβ3-mediated binding to laminin-α5-containing laminins however remains unclear. As pointed out, the EHS-produced laminin-111 is a poor ligand for the laminin-binding integrins. Accumulating data suggest that a major integrin-binding site is present in laminin LG1-3 domains in the C-terminal part of the laminin heterotrimer (Ido et al. 2004; Kunneken et al. 2004). In addition to laminin α-chain-specifc sequences, C-terminal residues of both β- and γ-chains contribute/facilitate integrin binding (Ido et al. 2004; Kunneken et al. 2004). Splicing in the extracellular domain of integrin α7 affects it preference for certain laminin isoforms (Nishiuchi et al. 2003; Taniguchi et al. 2009; von der Mark et al. 2002; Table 1).
RGD sequences present in triple helical fibrillar collagen sequences are normally not available for fibronectin receptors in native fibrillar collagen (Davis 1992; Gullberg et al. 1990), only becoming available in denatured collagen I. Instead, the collagen-binding integrins recognize the triple-helical GFOGER sequence (Knight et al. 1998), or variants thereof, in native collagens. An assembled database on collagen-I-binding sites and mutations has enabled the construction of a model in which the GFOGER site is present in a cell-interactive domain that is suggested to be exposed once per microfibril unit (each microfibrillar unit is composed of five collagen triple helix monomers), allowing the clustering of integrins (Sweeney et al. 2008). However, a number of components in the fibrillar matrix might influence the availability of these sites. Interestingly, collagen IX, which is present in cartilage, has been shown to be a good ligand for all of the collagen-binding integrins (Kapyla et al. 2004), but in vivo, not all of the collagen-binding integrins are expressed in chondrocytes. In the case of α11β1 integrin, if cartilaginous tissue is not dissected free of perichondrium, a signal from α11 will be derived from α11 expression in the perichondrium (Tiger et al. 2001), which lacks collagen IX. The identity of the integrins that might physiologically be relevant as collagen IX receptors in various regions of cartilage thus remains to be defined.
Collagen IX lacks the prototypic collagen-binding integrin recognition sequence GFOGER, and the exact sequence awaits mapping. Certain variants of the GFOGER sequence might show specificity for the different collagen-binding integrins, as indicated by a recent finding showing that the bacterial collagen-like peptide, sscl, with the active binding site GLPGER, is preferred by α11β1-expressing cells over α2β1-expressing cells (Caswell et al. 2008).
Other integrin-binding motifs include the α4-integrin-binding sequence LDV (Clements et al. 1994; Komoriya et al. 1991), variants of which are also present in the adhesion molecules ICAMs and MadCAM recognized by β2 integrins and the α4β7 heterodimer, respectively (Wang and Springer 1998).
In addition to physiological ligands, integrin ligands generated by proteolysis are receiving increasing recognition. Endostatin (derived from collagen XVIII), endorepellin (derived from perlecan), and tumstatin derived from collagen α3 (IV) are the best-known examples (Bix and Iozzo 2005; Suhr et al. 2009; Table 1). In addition, integrins can bind snake toxins called disintegrins (Calvete et al. 2005; Swenson et al. 2007) and certain viruses (Stewart and Nemerow 2007) and bacteria (Hauck et al. 2006; Palumbo and Wang 2006). Some of these interactions occur outside the regular ligand-binding sites in the integrins and display distinct binding characteristics compared with the binding of physiological ligands.
Integrins as mechanical links
The first function established for integrins was their function as links between the ECM and the cytoskeleton. For a majority of integrins, the linkage is to the actin cytoskeleton (Geiger et al. 2009), whereas α6β4 connects to the intermediate filament system (Nievers et al. 1999). Recently, the intermediate filament protein vimentin has been shown to be dependent on β3 integrins for its recruitment to the cell surface (Bhattacharya et al. 2009), indicating that an intimate relationship exists between the various cytoskeletal networks and integrins.
Some of the components in this mechanical linkage, such as talin, play a dual role and also take part in activating integrins in an inside-out signaling mechanism (Tadokoro et al. 2003). A new dimension of integrins as mechanical links has come to the fore with the realization that integrins can act as mechanosensors and generate signals that affect cell physiology via complex intracellular signaling mechanisms including autocrine and paracrine mechanisms (Chen and O’Connor 2005; Linton et al. 2007; Millward-Sadler and Salter 2004; Millward-Sadler et al. 1999; Zhu et al. 2007). As mentioned above, mechanical tension can also increase integrin affinity.
Integrins as signaling receptors
Integrins are bi-directional signaling receptors involved in outside-in and inside-out signaling. The inside-out signaling mainly acts to bring the integrin into the active conformation. As previously mentioned, talins, kindlins, filamins, migfilin, FAK, but also ILK (Honda et al. 2009) can regulate integrin activation.
Upon ligand binding, integrins undergo conformation changes leading to outside-in signaling. This activates signaling events that are complex and cell-specific, depending on what other signaling receptors and signaling systems are available in the cell. The scope of this review does not include these events, but readers are referred to recent excellent reviews on this subject (Askari et al. 2009; Gahmberg et al. 2009; Larjava et al. 2008; Legate et al. 2009; Luo and Springer 2006).
Integrin expression
Integrins are widely expressed, and every nucleated cell in the body possesses a specific integrin signature. Importantly, the regulation of integrins is dynamic and quickly changes once cells are taken out of their normal environment. A few integrins are more restricted than others to certain cell lineages, but the expression if often developmentally regulated.
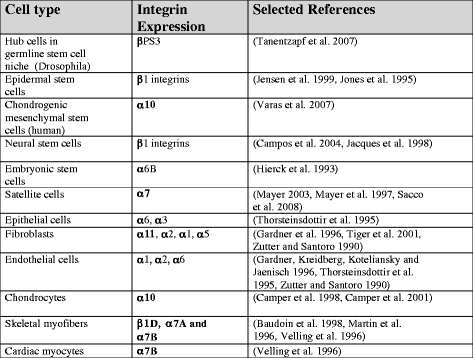
Most cells in the body express β1 integrin, and the use of β1 as part of a stem cell signature should thus be used with some caution. However, β1 integrin expression levels have been successfully used as a marker for epidermal stem cells, which express high levels of β1 integrins (Jensen et al. 1999). In Drosophila, the finding that βPS3 is needed to maintain the germ-line stem cell niche has shown the importance of integrin-mediated events for early stem cell function (Tanentzapf et al. 2007). In vitro studies have revealed that cell adhesive interactions can provide signals that keep stem cells undifferentiated on certain ECM molecules. The signals emanating from cell adhesion mediated via α6β1 and αvβ1 integrins to specific laminin isoforms can influence intracellular signaling to maintain the undifferentiated state (Domogatskaya et al. 2008). As stem cells differentiate, they change their integrin expression. In chondrogenic mesenchymal stem, the α10/α11 ratio reflects chondrogenic differentiation (Varas et al. 2007). Table 3 lists some integrins expressed on various characteristic cell types. The list is not complete and does not consider the dynamic changes in integrin expression that take place during growth and regeneration phases.
Table 3.
Integrin signatures of some selected cell types. Each cell in an organism contains a specific integrin signature under certain conditions. The integrin repertoire is dynamic, changes with developmental age, and is strongly responsive to microenvironmental conditions. Sometimes, a specific integrin isoform is indicative of the cell type or the differentiation status (integrins given in bold are the predominating integrins present)
Integrin function
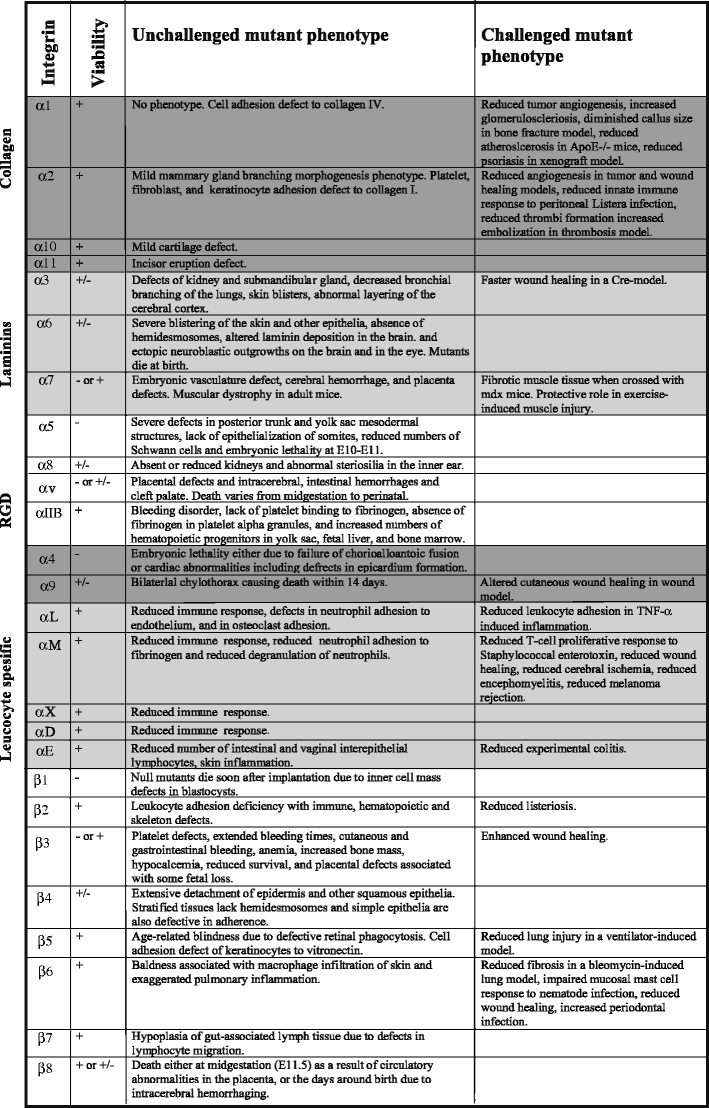
Integrins are essential cell adhesion receptors, and individual integrins have become specialized for certain functions. Although knockout animal models have provided essential information about integrin function (Table 4), several integrin functions remain to be clarified. Two examples come from the conditional deletion of β1 integrins in cartilage and skeletal muscle, respectively, and indicate important roles for β1 integrins during myogenesis and chondrogenesis (Aszodi et al. 2003; Schwander et al. 2003). The specific β1 integrin heterodimers involved in skeletal muscle and cartilage have not as yet been characterized. A combination of conditional deletions of multiple integrin α-chains might be needed to resolve these issues.
Table 4.
Phenotypes of unchallenged and challenged integrin mutant mice. The knockout data were collected from the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/) and, whenever needed, updated with relevant Pubmed (http://www.ncbi.nlm.nih.gov/pubmed/) retrieved original references
We have just started to comprehend the way that integrins work in complex biological systems. A new age with more refined assays, methods, and tools is likely to meet the challenge of understanding the integrated role of integrins in these biological systems.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aluwihare P, Mu Z, Zhao Z, Yu D, Weinreb PH, Horan GS, Violette SM, Munger JS. Mice that lack activity of {alpha}v{beta}6- and {alpha}v{beta}8-integrins reproduce the abnormalities of Tgfb1- and Tgfb3-null mice. J Cell Sci. 2009;122:227–232. doi: 10.1242/jcs.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askari JA, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. J Cell Sci. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrof NS, Salas A, Shimaoka M, Chen J, Springer TA. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszodi A, Hunziker EB, Brakebusch C, Fassler R. β1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 2003;17:2465–2479. doi: 10.1101/gad.277003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumailley M, Gerl M, Sonnenberg A, Deutzmann R, Timpl R. Identification of the Arg-Gly-Asp sequence in laminin a chain as a latent cell-binding site being exposed in fragment P1. FEBS Lett. 1990;262:82–86. doi: 10.1016/0014-5793(90)80159-g. [DOI] [PubMed] [Google Scholar]
- Baudoin C, Goumans M-J, Mummery C, Sonnenberg A. Knockout and knockin of the b1 exon D define distinct roles for integrin splice variants in heart function and embryonic development. Genes Dev. 1998;12:1202–1206. doi: 10.1101/gad.12.8.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R, Gonzalez AM, Debiase PJ, Trejo HE, Goldman RD, Flitney FW, Jones JC. Recruitment of vimentin to the cell surface by {beta}3 integrin and plectin mediates adhesion strength. J Cell Sci. 2009;122:1390–1400. doi: 10.1242/jcs.043042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bix G, Iozzo RV. Matrix revolutions: “tails” of basement-membrane components with angiostatic functions. Trends Cell Biol. 2005;15:52–60. doi: 10.1016/j.tcb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, Juarez P, Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Camper L, Hellman U, Lundgren-Akerlund E. Isolation, cloning, and sequence analysis of the integrin subunit a10, a b1-associated collagen binding integrin expressed on chondrocytes. J Biol Chem. 1998;273:20383–20389. doi: 10.1074/jbc.273.32.20383. [DOI] [PubMed] [Google Scholar]
- Camper L, Holmvall K, Wangnerud C, Aszodi A, Lundgren-Akerlund E. Distribution of the collagen-binding integrin alpha10beta1 during mouse development. Cell Tissue Res. 2001;306:107–116. doi: 10.1007/s004410100385. [DOI] [PubMed] [Google Scholar]
- Campos LS, Leone DP, Relvas JB, Brakebusch C, Fassler R, Suter U, ffrench-Constant C. Beta1 integrins activate a MAPK signalling pathway in neural stem cells that contributes to their maintenance. Development. 2004;131:3433–3444. doi: 10.1242/dev.01199. [DOI] [PubMed] [Google Scholar]
- Carman CV, Springer TA. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr Opin Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Barczyk M, Keene DR, Lukomska E, Gullberg DE, Lukomski S. Identification of the first prokaryotic collagen-sequence motif that mediates binding to human collagen receptors, integrins alpha 2beta 1 and alpha 11beta 1. J Biol Chem. 2008;283:36168–36175. doi: 10.1074/jbc.M806865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, O’Connor KL. Integrin alpha6beta4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene. 2005;24:5125–5130. doi: 10.1038/sj.onc.1208729. [DOI] [PubMed] [Google Scholar]
- Clements JM, Newham P, Shepherd M, Gilbert R, Dudgeon TJ, Needham LA, Edwards RM, Berry L, Brass A, Humphries MJ. Identification of a key integrin-binding sequence in VCAM-1 homologous to the LDV active site in fibronectin. J Cell Sci. 1994;107:2127–2135. doi: 10.1242/jcs.107.8.2127. [DOI] [PubMed] [Google Scholar]
- Davis GE. Affinity of integrins for damaged extracellular matrix: alpha v beta 3 binds to denatured collagen type I through RGD sites. Biochem Biophys Res Commun. 1992;182:1025–1031. doi: 10.1016/0006-291x(92)91834-d. [DOI] [PubMed] [Google Scholar]
- Dickinson CD, Veerapandian B, Dai XP, Hamlin RC, Xuong NH, Ruoslahti E, Ely KR. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J Mol Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Domogatskaya A, Rodin S, Boutaud A, Tryggvason K. Laminin-511 but not -332, -111, or -411 enables mouse embryonic stem cell self-renewal in vitro. Stem Cells. 2008;26:2800–2809. doi: 10.1634/stemcells.2007-0389. [DOI] [PubMed] [Google Scholar]
- Emsley J, King SL, Bergelson JM, Liddington RC. Crystal structure of the I domain from integrin α2β1. J Biol Chem. 1997;272:28512–28517. doi: 10.1074/jbc.272.45.28512. [DOI] [PubMed] [Google Scholar]
- Forsberg E, Lindblom A, Paulsson M, Johansson S. Laminin isoforms promote attachment of hepatocytes via different integrins. Exp Cell Res. 1994;215:33–39. doi: 10.1006/excr.1994.1311. [DOI] [PubMed] [Google Scholar]
- Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- Gahmberg CG, Fagerholm SC, Nurmi SM, Chavakis T, Marchesan S, Grönholm M. Regulation of integrin activity and signalling. Biochim Biophys Acta. 2009;1790:431–444. doi: 10.1016/j.bbagen.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailit J, Ruoslahti E. Regulation of the fibronectin receptor affinity by divalent cations. J Biol Chem. 1988;263:12927–12932. [PubMed] [Google Scholar]
- Gardner H, Kreidberg J, Koteliansky V, Jaenisch R. Deletion of integrin alpha 1 by homologous recombination permits normal murine development but gives rise to a specific deficit in cell adhesion. Dev Biol. 1996;175:301–312. doi: 10.1006/dbio.1996.0116. [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Genersch E, Ferletta M, Virtanen I, Haller H, Ekblom P. Integrin alphavbeta3 binding to human alpha5-laminins facilitates FGF-2- and VEGF-induced proliferation of human ECV304 carcinoma cells. Eur J Cell Biol. 2003;82:105–117. doi: 10.1078/0171-9335-00297. [DOI] [PubMed] [Google Scholar]
- Gullberg D, Turner DC, Borg TK, Terracio L, Rubin K. Different β1-integrin collagen receptors on rat hepatocytes and cardiac fibroblasts. Exp Cell Res. 1990;190:254–264. doi: 10.1016/0014-4827(90)90194-f. [DOI] [PubMed] [Google Scholar]
- Hauck CR, Agerer F, Muenzner P, Schmitter T. Cellular adhesion molecules as targets for bacterial infection. Eur J Cell Biol. 2006;85:235–242. doi: 10.1016/j.ejcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Hemler ME, Jacobson JG, Strominger JL. Biochemical characterization of VLA-1 and VLA-2. Cell surface heterodimers on activated T cells. J Biol Chem. 1985;260:15246–15252. [PubMed] [Google Scholar]
- Hierck BP, Thorsteinsdottir S, Niessen CM, Freund E, Iperen LV, Feyen A, Hogervorst F, Poelmann RE, Mummery CL, Sonnenberg A. Variants of the alpha 6 beta 1 laminin receptor in early murine development: distribution, molecular cloning and chromosomal localization of the mouse integrin alpha 6 subunit. Cell Adhes Commun. 1993;1:33–53. doi: 10.3109/15419069309095680. [DOI] [PubMed] [Google Scholar]
- Honda S, Shirotani-Ikejima H, Tadokoro S, Maeda Y, Kinoshita T, Tomiyama Y, Miyata T. Integrin-linked kinase associated with integrin activation. Blood. 2009;113:5304–5313. doi: 10.1182/blood-2008-07-169136. [DOI] [PubMed] [Google Scholar]
- Horwitz A, Duggan K, Greggs R, Decker C, Buck C. The cell substrate attachment (CSAT) antigen has properties of a receptor for laminin and fibronectin. J Cell Biol. 1985;101:2134–2144. doi: 10.1083/jcb.101.6.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MJ. Integrin structure. Biochem Soc Trans. 2000;28:311–339. [PubMed] [Google Scholar]
- Humphries MJ, Symonds EJ, Mould AP. Mapping functional residues onto integrin crystal structures. Curr Opin Struct Biol. 2003;13:236–243. doi: 10.1016/s0959-440x(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Hynes RO. The emergence of integrins: a personal and historical perspective. Matrix Biol. 2004;23:333–340. doi: 10.1016/j.matbio.2004.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido H, Harada K, Futaki S, Hayashi Y, Nishiuchi R, Natsuka Y, Li S, Wada Y, Combs AC, Ervasti JM, Sekiguchi K. Molecular dissection of the alpha-dystroglycan- and integrin-binding sites within the globular domain of human laminin-10. J Biol Chem. 2004;279:10946–10954. doi: 10.1074/jbc.M313626200. [DOI] [PubMed] [Google Scholar]
- Ithychanda SS, Das M, Ma YQ, Ding K, Wang X, Gupta S, Wu C, Plow EF, Qin J. Migfilin, a molecular switch in regulation of integrin activation. J Biol Chem. 2009;284:4713–4722. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques TS, Relvas JB, Nishimura S, Pytela R, Edwards GM, Streuli CH, ffrench-Constant C. Neural precursor cell chain migration and division are regulated through different beta1 integrins. Development. 1998;125:3167–3177. doi: 10.1242/dev.125.16.3167. [DOI] [PubMed] [Google Scholar]
- Jensen UB, Lowell S, Watt FM. The spatial relationship between stem cells and their progeny in the basal layer of human epidermis: a new view based on whole-mount labelling and lineage analysis. Development. 1999;126:2409–2418. doi: 10.1242/dev.126.11.2409. [DOI] [PubMed] [Google Scholar]
- Johnson MS, Lu N, Denessiouk K, Heino J, Gullberg D. Integrins during evolution: evolutionary trees and model organisms. Biochim Biophys Acta. 2009;1788:779–789. doi: 10.1016/j.bbamem.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Jones PH, Harper S, Watt FM. Stem cell patterning and fate in human epidermis. Cell. 1995;80:83–93. doi: 10.1016/0092-8674(95)90453-0. [DOI] [PubMed] [Google Scholar]
- Kapyla J, Jaalinoja J, Tulla M, Ylostalo J, Nissinen L, Viitasalo T, Vehvilainen P, Marjomaki V, Nykvist P, Saamanen AM, Farndale RW, Birk DE, Ala-Kokko L, Heino J. The fibril-associated collagen IX provides a novel mechanism for cell adhesion to cartilaginous matrix. J Biol Chem. 2004;279:51677–51687. doi: 10.1074/jbc.M409412200. [DOI] [PubMed] [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Knight CG, Morton LF, Onley DJ, Peachey AR, Messent AJ, Smethurst PA, Tuckwell DS, Farndale RW, Barnes MJ. Identification in collagen type I of an integrin α2β1-binding site containing an essential GER sequence. J Biol Chem. 1998;273:33287–33294. doi: 10.1074/jbc.273.50.33287. [DOI] [PubMed] [Google Scholar]
- Knudsen KA, Horwitz AF, Buck CA. A monoclonal antibody identifies a glycoprotein complex involved in cell-substratum adhesion. Exp Cell Res. 1985;157:218–226. doi: 10.1016/0014-4827(85)90164-8. [DOI] [PubMed] [Google Scholar]
- Komoriya A, Green LJ, Mervic M, Yamada SS, Yamada KM, Humphries MJ. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J Biol Chem. 1991;266:15075–15079. [PubMed] [Google Scholar]
- Kunneken K, Pohlentz G, Schmidt-Hederich A, Odenthal U, Smyth N, Peter-Katalinic J, Bruckner P, Eble JA. Recombinant human laminin-5 domains. Effects of heterotrimerization, proteolytic processing, and N-glycosylation on alpha3beta1 integrin binding. J Biol Chem. 2004;279:5184–5193. doi: 10.1074/jbc.M310424200. [DOI] [PubMed] [Google Scholar]
- Larjava H, Plow EF, Wu C. Kindlins: essential regulators of integrin signalling and cell-matrix adhesion. EMBO Rep. 2008;9:1203–1208. doi: 10.1038/embor.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RS, Corbi AL, Berman L, Springer T. Primary structure of the leukocyte function-associated molecule-1 alpha subunit: an integrin with an embedded domain defining a protein superfamily. J Cell Biol. 1989;108:703–712. doi: 10.1083/jcb.108.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Bankston LA, Arnaout MA, Liddington RC. Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure. 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Legate KR, Fassler R. Mechanisms that regulate adaptor binding to beta-integrin cytoplasmic tails. J Cell Sci. 2009;122:187–198. doi: 10.1242/jcs.041624. [DOI] [PubMed] [Google Scholar]
- Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- Leptin M, Aebersold R, Wilcox M. Drosophila position-specific antigens resemble the vertebrate fibronectin-receptor family. EMBO J. 1987;6:1037–1043. doi: 10.1002/j.1460-2075.1987.tb04856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton JM, Martin GR, Reichardt LF. The ECM protein nephronectin promotes kidney development via integrin alpha8beta1-mediated stimulation of Gdnf expression. Development. 2007;134:2501–2509. doi: 10.1242/dev.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Springer TA. Integrin structures and conformational signaling. Curr Opin Cell Biol. 2006;18:579–586. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Mark H, Williams I, Wendler O, Sorokin L, von der Mark K, Poschl E. Alternative splice variants of alpha 7 beta 1 integrin selectively recognize different laminin isoforms. J Biol Chem. 2002;277:6012–6016. doi: 10.1074/jbc.M102188200. [DOI] [PubMed] [Google Scholar]
- Martin PT, Kaufman SJ, Kramer RH, Sanes JR. Synaptic integrins in developing, adult, and mutant muscle: selective association of a1, a7A and a7B integrins with the neuromuscular junction. Dev Biol. 1996;174:125–139. doi: 10.1006/dbio.1996.0057. [DOI] [PubMed] [Google Scholar]
- Mayer U. Integrins: redundant or important players in skeletal muscle? J Biol Chem. 2003;278:14587–14590. doi: 10.1074/jbc.R200022200. [DOI] [PubMed] [Google Scholar]
- Mayer U, Saher G, Fassler R, Bornemann A, Echtermeyer F, von der Mark H, Miosge N, Poschl E, von der Mark K. Absence of integrin a7 causes a novel form of muscular dystrophy. Nat Genet. 1997;17:318–323. doi: 10.1038/ng1197-318. [DOI] [PubMed] [Google Scholar]
- Michael KE, Dumbauld DW, Burns KL, Hanks SK, García AJ. FAK modulates cell adhesion strengthening via integrin activation. Mol Biol Cell. 2009;20:2508–2519. doi: 10.1091/mbc.E08-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward-Sadler SJ, Salter DM. Integrin-dependent signal cascades in chondrocyte mechanotransduction. Ann Biomed Eng. 2004;32:435–446. doi: 10.1023/b:abme.0000017538.72511.48. [DOI] [PubMed] [Google Scholar]
- Millward-Sadler SJ, Wright MO, Lee H, Nishida K, Caldwell H, Nuki G, Salter DM. Integrin-regulated secretion of interleukin 4: a novel pathway of mechanotransduction in human articular chondrocytes. J Cell Biol. 1999;145:183–189. doi: 10.1083/jcb.145.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, Rifkin DB, Sheppard D. The integrin αvβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- Nievers MG, Schaapveld RQ, Sonnenberg A. Biology and function of hemidesmosomes. Matrix Biol. 1999;18:5–17. doi: 10.1016/s0945-053x(98)00003-1. [DOI] [PubMed] [Google Scholar]
- Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. Activation of leukocyte beta2 integrins by conversion from bent to extended conformations. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Nishiuchi R, Murayama O, Fujiwara H, Gu J, Kawakami T, Aimoto S, Wada Y, Sekiguchi K. Characterization of the ligand-binding specificities of integrin alpha3beta1 and alpha6beta1 using a panel of purified laminin isoforms containing distinct alpha chains. J Biochem. 2003;134:497–504. doi: 10.1093/jb/mvg185. [DOI] [PubMed] [Google Scholar]
- Oxvig C, Springer TA. Experimental support for a b-propeller domain in integrin a-subunits and a calcium binding site on its lower surface. Proc Natl Acad Sci USA. 1998;95:4870–4875. doi: 10.1073/pnas.95.9.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo RN, Wang C. Bacterial invasin: structure, function, and implication for targeted oral gene delivery. Current Drug Deliv. 2006;3:47–53. doi: 10.2174/156720106775197475. [DOI] [PubMed] [Google Scholar]
- Parise LV, Phillips DR. Reconstitution of the purified platelet fibrinogen receptor. Fibrinogen binding properties of the glycoprotein IIb-IIIa complex. J Biol Chem. 1985;260:10698–10707. [PubMed] [Google Scholar]
- Parise LV, Phillips DR. Fibronectin-binding properties of the purified platelet glycoprotein IIb-IIIa complex. J Biol Chem. 1986;261:14011–14017. [PubMed] [Google Scholar]
- Pierschbacher M, Hayman EG, Ruoslahti E. Synthetic peptide with cell attachment activity of fibronectin. Proc Natl Acad Sci USA. 1983;80:1224–1227. doi: 10.1073/pnas.80.5.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santala P, Heino J. Regulation of integrin-type cell adhesion receptors by cytokines. J Biol Chem. 1991;266:23505–23509. [PubMed] [Google Scholar]
- Sasaki T, Timpl R. Domain IVa of laminin alpha5 chain is cell-adhesive and binds beta1 and alphaVbeta3 integrins through Arg-Gly-Asp. FEBS Lett. 2001;509:181–185. doi: 10.1016/s0014-5793(01)03167-2. [DOI] [PubMed] [Google Scholar]
- Sato Y, Uemura T, Morimitsu K, Sato-Nishiuchi R, Manabe RI, Takagi J, Yamada M, Sekiguchi K (2009) Molecular basis of the recognition of nephronectin by integrin alpha 8beta 1. J Biol Chem (in press) [DOI] [PMC free article] [PubMed]
- Schulze B, Mann K, Poschl E, Yamada Y, Timpl R. Structural and functional analysis of the globular domain IVa of the laminin alpha 1 chain and its impact on an adjacent RGD site. Biochem J. 1996;314:847–851. doi: 10.1042/bj3140847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M, Leu M, Stumm M, Dorchies OM, Ruegg UT, Schittny J, Muller U. β1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell. 2003;4:673–685. doi: 10.1016/s1534-5807(03)00118-7. [DOI] [PubMed] [Google Scholar]
- Senetar MA, Moncman CL, McCann RO. Talin2 is induced during striated muscle differentiation and is targeted to stable adhesion complexes in mature muscle. Cell Motil Cytoskeleton. 2007;64:157–173. doi: 10.1002/cm.20173. [DOI] [PubMed] [Google Scholar]
- Springer TA, Miller LJ, Anderson DC. p150, 95, the third member of the Mac-1, LFA-1 human leukocyte adhesion glycoprotein family. J Immunol. 1986;136:240–245. [PubMed] [Google Scholar]
- Stewart PL, Nemerow GR. Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 2007;15:500–507. doi: 10.1016/j.tim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Suhr F, Brixius K, Bloch W. Angiogenic and vascular modulation by extracellular matrix cleavage products. Curr Pharma Des. 2009;15:389–410. doi: 10.2174/138161209787315756. [DOI] [PubMed] [Google Scholar]
- Svensson L, Howarth K, McDowall A, Patzak I, Evans R, Ussar S, Moser M, Metin A, Fried M, Tomlinson I, Hogg N. Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat Med. 2009;15:306–312. doi: 10.1038/nm.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney SM, Orgel JP, Fertala A, McAuliffe JD, Turner KR, Di Lullo GA, Chen S, Antipova O, Perumal S, Ala-Kokko L, Forlino A, Cabral WA, Barnes AM, Marini JC, San Antonio JD. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J Biol Chem. 2008;283:21187–21197. doi: 10.1074/jbc.M709319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson S, Ramu S, Markland FS. Anti-angiogenesis and RGD-containing snake venom disintegrins. Curr Pharma Des. 2007;13:2860–2871. doi: 10.2174/138161207782023793. [DOI] [PubMed] [Google Scholar]
- Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- Takada Y, Ye X, Simon S. The integrins. Genome Biol. 2007;8:215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, DeSimone DW, Fonda D, Patel RS, Buck C, Horwitz AF, Hynes RO. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell. 1986;46:271–282. doi: 10.1016/0092-8674(86)90744-0. [DOI] [PubMed] [Google Scholar]
- Tanentzapf G, Devenport D, Godt D, Brown NH. Integrin-dependent anchoring of a stem-cell niche. Nat Cell Biol. 2007;9:1413–1418. doi: 10.1038/ncb1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Ido H, Sanzen N, Hayashi M, Sato-Nishiuchi R, Futaki S, Sekiguchi K. The C-terminal region of laminin beta chains modulates the integrin binding affinities of laminins. J Biol Chem. 2009;284:7820–7831. doi: 10.1074/jbc.M809332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir S, Roelen BA, Freund E, Gaspar AC, Sonnenberg A, Mummery CL. Expression patterns of laminin receptor splice variants a6Ab1 and a6Bb1 suggest different roles in mouse development. Dev Dyn. 1995;204:240–258. doi: 10.1002/aja.1002040304. [DOI] [PubMed] [Google Scholar]
- Tiger CF, Fougerousse F, Grundstrom G, Velling T, Gullberg D. Alpha11beta1 integrin is a receptor for interstitial collagens involved in cell migration and collagen reorganization on mesenchymal nonmuscle cells. Dev Biol. 2001;237:116–129. doi: 10.1006/dbio.2001.0363. [DOI] [PubMed] [Google Scholar]
- Valdramidou D, Humphries MJ, Mould AP. Distinct roles of beta 1 MIDAS, ADMIDAS and LIMBS cation-binding sites in ligand recognition by integrin alpha 2beta 1. J Biol Chem. 2008;283:32704–32714. doi: 10.1074/jbc.M802066200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas L, Ohlsson LB, Honeth G, Olsson A, Bengtsson T, Wiberg C, Bockermann R, Jarnum S, Richter J, Pennington D, Johnstone B, Lundgren-Akerlund E, Kjellman C. Alpha10 integrin expression is up-regulated on fibroblast growth factor-2-treated mesenchymal stem cells with improved chondrogenic differentiation potential. Stem Cells Dev. 2007;16:965–978. doi: 10.1089/scd.2007.0049. [DOI] [PubMed] [Google Scholar]
- Velling T, Collo GA, Sorokin L, Durbeej MA, Zhang H, Gullberg D. Distinct alpha 7A beta 1 and alpha 7B beta 1 integrin expression patterns during mouse development: alpha 7A is restricted to skeletal muscle but alpha 7B is expressed in striated muscle, vasculature, and nervous system. Dev Dyn. 1996;207:335–371. doi: 10.1002/(SICI)1097-0177(199612)207:4<355::AID-AJA1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Wang J, Springer TA. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol Rev. 1998;163:197–215. doi: 10.1111/j.1600-065x.1998.tb01198.x. [DOI] [PubMed] [Google Scholar]
- Wilcox M, Brown N, Piovant M, Smith RJ, White RA. The Drosophila position-specific antigens are a family of cell surface glycoprotein complexes. EMBO J. 1984;3:2307–2313. doi: 10.1002/j.1460-2075.1984.tb02131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipff PJ, Rifkin DB, Meister JJ, Hinz B. Myofibroblast contraction activates latent TGF-1 from the extracellular matrix. J Cell Biol. 2007;179:1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Zhu CQ, Popova SN, Brown ER, Barsyte-Lovejoy D, Navab R, Shih W, Li M, Lu M, Jurisica I, Penn LZ, Gullberg D, Tsao MS. Integrin alpha 11 regulates IGF2 expression in fibroblasts to enhance tumorigenicity of human non-small-cell lung cancer cells. Proc Natl Acad Sci USA. 2007;104:11754–11759. doi: 10.1073/pnas.0703040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zutter MM, Santoro SA. Widespread histologic distribution of the a2b1 integrin cell-surface collagen receptor. m J Pathol. 1990;137:113–120. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.