Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 21.
Summary
In cancer cells, genetic alterations can activate proto-oncogenes, thereby contributing to tumorigenesis. However, the protein products of oncogenes are sometimes over-expressed without alteration of the proto-oncogene. Helping to explain this phenomenon, we found that when compared to similarly proliferating non-transformed cell lines, cancer cell lines often expressed substantial amounts of mRNA isoforms with shorter 3′ untranslated regions (UTRs). These shorter isoforms usually resulted from alternative cleavage and polyadenylation (APA). The APA had functional consequences, with the shorter mRNA isoforms exhibiting increased stability and typically producing ten-fold more protein, in part through the loss of microRNA-mediated repression. Moreover, expression of the shorter mRNA isoform of the proto-oncogene IGF2BP1/IMP-1 led to far more oncogenic transformation than did expression of the full-length, annotated mRNA. The high incidence of APA in cancer cells, with consequent loss of 3′UTR repressive elements, suggests a pervasive role for APA in oncogene activation without genetic alteration.
Introduction
Oncogene activation plays a central role in the pathogenesis of cancer. Mammalian genomes contain proto-oncogenes that function to regulate normal cell proliferation and differentiation. In cancer cells, these genes frequently have been activated—sometimes by a mutation that changes the encoded proteins and other times by a mutation that increases the expression of the gene. The increased expression can occur through several mechanisms, including gene amplifications that increase copy number and chromosomal translocations that replace the original promoter with a much more active one (Bishop, 1991; Rowley, 2001).
A recently recognized mechanism of oncogene activation is the loss of microRNA (miRNA) complementary sites (Mayr et al., 2007; Lee and Dutta, 2007). miRNAs specify posttranscriptional repression by pairing to short sites in mRNA targets, usually in 3′UTRs (Bartel, 2009). In the case of HMGA2 activation, chromosomal translocations swap the HMGA2 3′UTR for that of another gene, which results in loss of complementary sites for the let-7 miRNA and escape from repression (Mayr et al., 2007; Lee and Dutta, 2007). In principle, point substitutions or other lesions that disrupt individual target sites or that shorten the 3′UTR through premature cleavage and polyadenylation could have the same effect (Majoros and Ohler, 2007). Hinting at this possibility are events associated with a more aggressive subgroup of mantle cell lymphomas. Mantle cell lymphomas are characterized by the t(11;14)(q13;q32) translocation, which brings Cyclin D1 near the highly active immunoglobulin heavy chain gene promoter, leading to Cyclin D1 overexpression. In a subset of mantle cell lymphomas a second alteration of Cyclin D1, the shortening of its 3′UTR, which might relieve miRNA-directed repression, leads to an additional 1.6-fold increase in Cyclin D1 expression and correlates with both increased proliferation of the lymphoma cells and decreased overall survival of patients (Rosenwald et al., 2003). The reasons for 3′UTR shortening are unknown in a third of cases, but in the remaining cases either genomic deletions encompass much of the 3′UTR or point substitutions create premature cleavage and polyadenylation signals (Wiestner et al., 2007). For some cases in which DNA lesions are not found, the Cyclin D1 3′UTR might be shortened through alternative cleavage and polyadenylation (APA).
Cleavage and polyadenylation is required for the maturation of most mRNA transcripts (Proudfoot, 1991; Colgan and Manley, 1997). The pre-mRNA is cleaved 10–30 nt after the polyadenylation signal (AAUAAA and variants thereof) and an untemplated poly(A) tract is added. Although a strong polyadenylation signal usually is located at the 3′ end of the 3′UTR, nearly all genes have additional polyadenylation signals in their 3′UTRs, with about half of human genes possessing conserved APA signals with usage supported by expressed sequence tags (ESTs) (Tian et al., 2005). Use of APA signals often eliminates large parts of the 3′UTR, enabling escape from the stronger regulatory potential of longer 3′UTRs. Besides miRNA regulation, the lost regulatory sequences in the 3′UTR can influence mRNA nuclear export and cytoplasmic localization, as well as non-miRNA-mediated changes in mRNA stability and translational efficiency (Moore, 2005). Alternative mRNAs that differ in their 3′UTRs can exist in different tissues or developmental stages, and studies have shown that these mRNA isoforms can have different stability or translational activity (Miyamoto et al., 1996; Takagaki et al., 1996; Edwalds-Gilbert et al., 1997; Lutz, 2008). Over 30 years ago, stimulation of lymphocytes was shown to increase both RNA polyadenylation and the rate of protein synthesis, without a change in the rate of transcription (Coleman et al., 1974; Hauser et al., 1978). Recently, a genome-wide study identified shorter 3′UTRs in activated T cells compared to resting T cells, and found that in general shorter 3′UTRs were associated with cell proliferation (Sandberg et al., 2008).
We set out to explore the possibility that APA might be a mechanism by which genes can escape miRNA-mediated repression in cancer. We found that shorter 3′UTRs were indeed associated with tumor cells, over and above the association expected from their proliferative state. Reporter assays and immunoblots revealed the functional consequences of APA, showing that the shorter transcripts produced substantially more protein than did their full-length counterparts, in part through escape of miRNA-mediated targeting. Moreover, expressing the shorter but not the full-length isoform of the proto-oncogene Insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1/IMP-1/CRD-BP/ZBP-1) led to oncogenic transformation, illustrating that loss of repressive 3′UTR elements through APA can promote the oncogenic phenotype.
Results
Shorter 3′UTRs are associated with transformation
To find altered mRNA length and abundance, we used Northern blots probing RNA from 27 cancer cell lines from different tissues. The cell lines were derived from sarcomas and breast, lung and colon cancers, and compared to immortalized non-transformed epithelial cell lines and normal corresponding tissues. We chose to analyze 23 genes. Gene function was not considered when selecting these genes. One criterion for selecting a gene was that there were potential APA signals in the 3′UTR. AAUAAA is the canonical polyadenylation signal, but variants of this signal, including AUUAAA, AAGAAA, UAUAAA, AGUAAA are also used. These variants are used less often as the polyadenylation signal for the longest isoform but more often as proximal polyadenylation signals (Tian et al., 2005). When including these variants, nearly every gene had potential APA signals, and thus this criterion imposed only a minor constraint. The other selection criterion was that the mRNA have strong potential for miRNA regulation, as predicted by TargetScan (version 4.2, (Lewis et al., 2005; Grimson et al., 2007). With one exception (IMP-3), mRNAs with at least two sites to a single miRNA were chosen (Table 1). Reasoning that the miRNAs would be relevant only if expressed in the cancer cell lines, we considered only widely expressed miRNAs (e.g., let-7, miR-15/16 and miR-103/107) and confirmed their expression in the cell lines by Northern blots (Figure S1). For example, we investigated the top ten predicted targets of let-7.
Table 1.
Candidate genes
| Gene | Isoformsa | miRNA | Sitesb | Function |
|---|---|---|---|---|
| HMGA2 | >1 | let-7 | 7 | Architectural transcription factor; cancer, stem cell biology |
| C12orf28 | 0 | let-7 | 4 | Unknown |
| LIN28B | 1 | let-7 | 5 | Homolog of Lin28, reprogramming of human fibroblasts |
| TRIM71 | 0 | let-7 | 2 | Development |
| IMP-1 | 3 | let-7 | 5 | RNA-binding protein; mRNA stability, translational control, mRNA transport |
| ARID3B | 1 | let-7 | 5 | DNA-binding protein; development, oncogene |
| FIGN | 0 | let-7 | 3 | Development |
| PUNC | 0 | let-7 | 3 | Development |
| SMARCAD1 | 0 | let-7 | 2 | Helicase; chromatin remodeling |
| YOD1 | 1 | let-7 | 4 | Deubiquitinating enzyme |
| BACH1 | 1 | let-7 | 2 | Transcription factor |
| CPEB2 | 1 | let-7 | 3 | RNA-binding protein; development, cell division, senescence, synaptic plasticity |
| IMP-3 | 2 | let-7 | 1 | RNA-binding protein |
| IMP-2 | 2 | let-7 | 2 | RNA-binding protein |
| FGF2 | 3 | miR-15/16 | 5 | Mitogen; development, wound healing, tumor growth |
| PLAG1 | 1 | miR-15/16 | 2 | Zink finger protein; rearranged in pleomorphic adenoma |
| MYB | 1 | miR-15/16 | 2 | Transcription factor; oncogene |
| TACC1 | 0 | miR-15/16 | 2 | Oncogene |
| CCND2 | 2 | miR-15/16 | 3 | Cyclin; oncogene |
| CCND1 | 2 | miR-15/16 | 2 | Cyclin; oncogene |
| DICER1 | 2–3 | miR-103/107 | 6 | miRNA biogenesis |
| RAB10 | 2 | miR-103/107 | 3 | Ras-related GTP-binding protein |
| RUNX1 | 0 | miR-27 | 2 | Subunit of transcription factor; hematopoiesis |
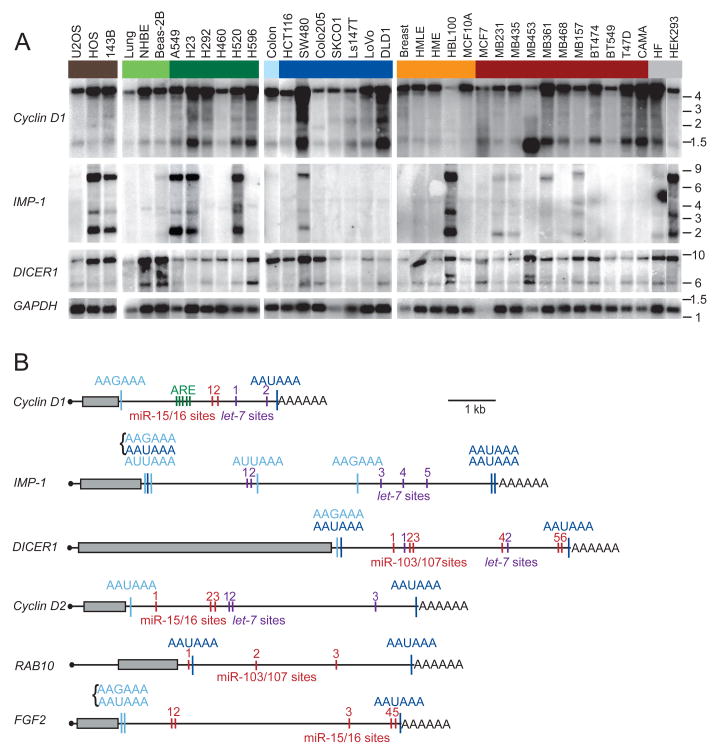
Sixteen of the 23 candidate genes were expressed in the cell lines. In nine of these 16, more than one mRNA isoform was detected (Figures 1A and S2). To investigate if the difference in mRNA length was due to APA, 3′ RACE (rapid amplification of cDNA ends) was performed. For three of the nine genes with multiple isoforms, the differences in mRNA lengths were due to alternative splicing (data not shown), but for the other six, the differences were due to APA (Figure 1B). For the shorter mRNA isoforms of these six genes, the canonical AAUAAA signal was used in three cases, the variant AAGAAA in three cases, the variant AUUAAA in two cases, and in the other two cases the signals were clustered (AAGAAAAAUAAA), making it difficult to assign which one was used (Figure 1B). Thirty percent of these signals (all AAUAAA) were conserved at the orthologous position in all four mammals examined (human, mouse, rat and dog), and in many of the other cases signals were present at non-orthologous positions. The most 3′ located polyadenylation signal was always AAUAAA and always conserved at the orthologous position.
Figure 1. APA leads to shorter 3′UTRs.
(A) Northern blots of human cell lines and tissues, successively stripped and reprobed for the indicated mRNAs. Groupings include sarcoma cell lines (brown), normal lung tissue, cultured human bronchial epithelial cells (NHBE) and immortalized lung epithelial cell line (Beas-2B, light green), lung cancer cell lines (dark green), normal colon tissue (light blue), colon cancer cell lines (dark blue), normal breast tissue and immortalized breast epithelial cell lines (orange), breast cancer cell lines (red), and other cell lines (grey), which were immortalized fibroblasts (HF) and an embryonic kidney cell line (HEK293). GAPDH expression served as loading control. Migration of markers with lengths indicated (kb) is shown at the right.
(B) Schematic illustration of mRNAs with alternative isoforms due to APA. Grey boxes show protein coding region; black line represents untranslated regions, and AAAAAA represents the poly(A) tail. Shown are functional APA signals identified by 3′ RACE, with signals conserved at the orthologous position in four genomes (human, mouse, rat, dog) in dark blue, and more poorly conserved signals in light blue. Brackets indicate closely spaced signals. Also shown are negative regulatory elements identified in the 3′UTRs, including AU-rich elements (ARE, green), let-7 sites (purple), and sites to the other widely expressed miRNA with the most sites in that 3′UTR (red).
GU- or U-rich sequences are often found downstream of functional cleavage and polyadenylation signals and are important for signal recognition (Proudfoot, 1991; Colgan and Manley, 1997). Indeed, examining composition of the 200 nucleotides surrounding the functional proximal polyadenylation signals and comparing it with that of the nucleotides surrounding the apparently nonfunctional polyadenylation signals in the genes for which only the full-length mRNA isoform was detected revealed an over-representation of U-rich sequences within 50 nt downstream of the functional poly(A) signals (p = 0.02; Figure S3).
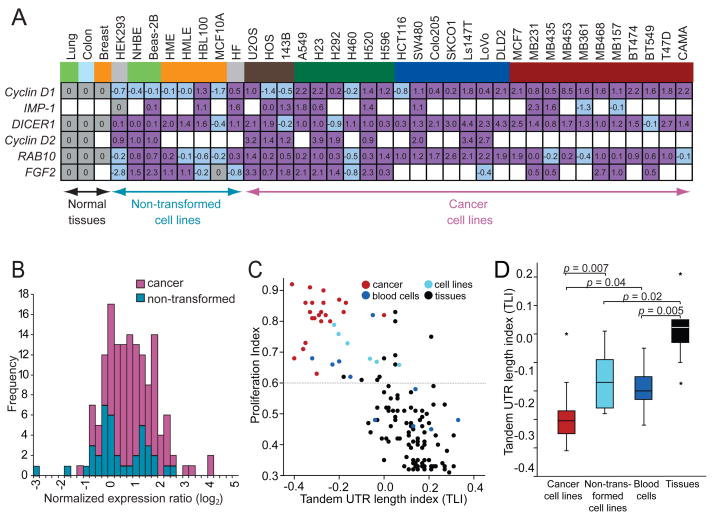
The genes with alternative isoforms included Cyclin D1, DICER1 and RAB10, which were expressed in all samples, and IMP-1, Cyclin D2 and FGF2, which were expressed in at least a third of the samples. The shorter mRNAs usually were detectable in normal tissues but more prominent in cancer lines (Figures 1A and S2). The bands of the longest and the shortest mRNA isoform were quantified to determine the expression ratio. Comparing the ratios between the cell lines and their normal corresponding tissues revealed that a majority of the cell lines expressed a higher fraction of the shorter mRNA isoform (p <10−3) (Figure 2A, purple). These results concurred with those of Sandberg et al. (2008), who associated shorter 3′UTRs with a proliferative state. However, the previous study did not distinguish between cancer cell lines and non-transformed proliferating cells. When making this distinction for the lines used in our study, higher amounts of shorter mRNAs were detected significantly more often in cancer cell lines than in non-transformed cell lines (p <10−4), indicating that shorter mRNAs are associated with transformation even more than they are associated with proliferation (Figure 2B). Indeed, no significant difference was found between the non-transformed cell lines compared with normal tissues (p = 0.35; Figure 2B).
Figure 2. APA leads to shorter 3′UTRs preferentially in cancer cells.
(A) Quantification of alternative mRNA isoforms. Expression ratio of shortest to longest mRNA isoform from Figures 1A and S2 was normalized to that of the corresponding normal tissue and shown as log2 values. For the sarcoma cell lines no normal tissue was available, therefore the values were normalized to the median of expression of all normal tissues. IMP-1 was not expressed in any of the normal tissues, and thus IMP-1 ratios were normalized to that of HEK293, because for the other genes examined the expression ratios of the mRNA isoforms in HEK293 cells were similar to those of the normal tissues. FGF2 was not expressed in normal breast tissue; therefore FGF2 ratios of the breast cell lines were normalized to that MCF10A, a non-transformed breast epithelial line, whose expression ratio of short and long mRNA isoforms resembled that of normal breast tissue for the other genes analyzed. Ratios used for normalization are shaded grey; those with a higher fraction of shorter isoform are purple; those with a lower fraction of shorter isoform are shaded light blue, and cases without detectable mRNA are blank. Cells and tissues are color-coded as in Figure 1A.
(B) Distributions of alternative mRNAs for cancer cells and non-transformed cells. Plotted are the values from Figure 2A, which are the expression ratios of shorter to longer mRNA isoforms, normalized to the corresponding normal tissue. The value for Cyclin D1 in the breast cancer cell line MB453, which was 8.5 (log2), is not included because a deletion encompassing the 3′UTR is responsible for the shorter mRNA (Lebwohl et al., 1994). For non-transformed cell lines, the distribution is not significantly displaced from zero (p = 0.35), indicating similar ratios of shorter to longer mRNA isoforms as the normal tissues. For cancer cell lines, the ratio is significantly displaced from both zero (p <10−4) and the distribution for non-transformed cell lines (p <10−4), indicating increased ratios of the shorter isoforms.
(C) Genome-wide analyses of 3′UTR length and cellular proliferation. The figure is redrawn from Sandberg et al. (2008), except data points are colored to indicate different types of samples, with cancer samples red (including 22 cancer cell lines and 1 cancer sample), non-transformed cell lines light blue, blood cells (including resting and stimulated T cells, B cells and monocytes) dark blue and normal tissues black. The tandem UTR length index (TLI) is a relative measure of 3′UTR length derived from array data(Sandberg et al., 2008).
(D) The 3′UTR length as measured by TLI for different sample types, focusing only on proliferating cells. TLI values from samples of panel (C) above a proliferation threshold of proliferation index = 0.6 [dotted line in panel (C)] were plotted (median, horizontal line; 25th through 75th percentile, box; range, error bars; outliers, *).
One possibility raised by our findings was that APA-mediated shortening of 3′UTRs is a cancer-associated phenomenon, without any association with proliferation after accounting for the association with cellular transformation. Alternatively, a more global analysis might still reveal an association with proliferation. To address this issue, we revisited the dataset of Sandberg et al. (2008), who analyzed array data across a broad panel of mammalian cell lines and tissues for differences in both proliferation and 3′UTR length. They created a tandem UTR length index (TLI) to assess aggregate expression of extended 3′UTR regions relative to overall gene-expression levels, and correlated this UTR-length measurement with a gene-signature-based measure of cellular proliferation, called the proliferation index, across a panel of 135 samples. These samples included 22 cancer cell lines, one cancer sample, five non-transformed cell lines, ten samples derived from blood cells and 97 tissues. After subgrouping their data based on these sample types (Figure 2C), we confirmed that even after excluding the cancer lines, proliferation influenced 3′UTR length: Stimulation of blood cells, including T cells, B cells and monocytes, significantly reduced 3′UTR length when compared to their unstimulated counterparts (p = 0.04) or normal tissues (p = 0.005), and non-transformed cell lines also had shorter 3′UTRs than normal tissues (p = 0.02; Figure 2D). But most strikingly, the cancer cell lines had the shortest 3′UTRs (Figures 2C and 2D, red), which were significantly shorter than the 3′UTRs of non-transformed cell lines (p = 0.007) despite comparable proliferation indices (p = 0.6). This analysis extending our Northern blot results from a limited number of genes to a genome-wide scale confirmed that shorter 3′UTRs are associated with transformation over and above the association expected from their proliferative state.
Shorter mRNAs have greater stability and produce more protein
What might be the functional consequence of shorter 3′UTRs? In all cases investigated, predicted miRNA target sites were lost in the shorter isoform (Figure 1B). Because miRNAs often destabilize their target mRNAs (Bartel, 2009), the stabilities of the longer and the shorter mRNAs were examined in nine cell lines from diverse tissues for three genes with the most robust mRNA Northern signals (IMP-1, Cyclin D1 and Cyclin D2). Cells were treated with Actinomycin D to inhibit transcription, total RNA was collected, and the decay of the mRNA isoforms was investigated by Northern blot analysis. Across different tissues and different genes, the shorter mRNA was on average 2.6-times more stable than was the longer mRNA (Table 2).
Table 2.
mRNA half-life (t1/2).
| Cancer tissue | Cell line | Long mRNA t1/2 (h)* | Short mRNA t1/2 (h)* | Short/long |
|---|---|---|---|---|
| IMP-1 | ||||
| Lung | 4.2 ± 1.4 | 9.0 ± 2.5 | 2.1 | |
| Colon | SW480 | 6.2 ± 0.4 | 12.3 ± 4.3 | 2.0 |
| Breast | HBL100 | 2.3 ± 0.9 | 6.4 ± 1.0 | 2.8 |
| Breast | MDA-MB231 | 4.1 ± 0.5 | 6.7 ± 4.0 | 1.6 |
| Breast | MDA-MB361 | 1.8 ± 0.3 | 4.5 ± 1.6 | 2.5 |
| Cyclin D1 | ||||
| Breast | MCF-7 | 1.9 ± 0.2 | 3.8 ± 0.7 | 2.0 |
| Breast | MDA-MB361 | 8.0 ± 0.1 | 22.4 ± 6.3 | 2.8 |
| Breast | MDA-MB453 | 1.5 ± 0.5 | 6.3 ± 2.7 | 4.2 |
| Cyclin D2 | ||||
| Sarcoma | U2OS | 2.3 ± 0.2 | 3.5 ± 1.1 | 1.5 |
| Colon | SW480 | 3.1 ± 0.9 | 20.6 ± 12.4 | 6.6 |
| Colon | LoVo | 13.8 ± 1.4 | 9.3 ± 1.8 | 0.7 |
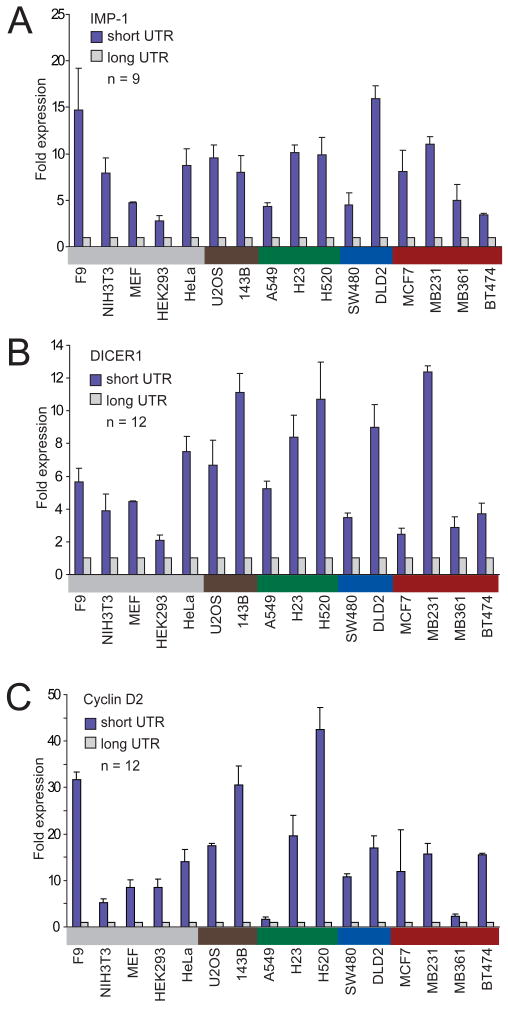
In addition to mRNA destabilization, miRNAs can also direct translational repression of their target mRNAs, which adds to their impact on protein output (Lee et al., 1993; Wightman et al., 1993; Filipowicz et al., 2008). To investigate if different amounts of protein were produced from the shorter and the longer isoforms, we used reporter assays. Both the short and the long 3′UTR isoforms of IMP-1, Cyclin D2, and DICER1 were each cloned downstream of a luciferase reporter gene. To ensure that each long isoform was expressed, for these reporters all the proximal polyadenylation signals that were functional (as detected by 3′ RACE) were mutated (Figure 1B). A strong poly(A) signal from SV40 virus was added downstream of the endogenous polyadenylation signal of the short isoform, as well as that of the long isoform, to ensure both isoforms were expressed as intended. Luciferase reporter gene expression was measured for all three genes in 16 cell lines from different tissues. For all genes in all the cell lines tested, the shorter mRNA isoform produced more protein (Figure 3). The protein expression derived from the shorter mRNA was 1.6–42-fold higher than that from the longer mRNA (IMP-1 median, 8.0 fold; DICER1 median, 5.5 fold; Cyclin D2 median, 14.8 fold; average difference overall, 10 fold).
Figure 3. The shortest mRNA isoform leads to higher protein expression than does the full-length isoform.
(A) Luciferase expression from a reporter containing the 3′UTR of the shortest IMP-1 isoform, compared to that from the reporter containing the 3′UTR of the full-length IMP-1 isoform. Assays were performed in 16 cell lines from various tissues, color-coded as in Figure 1A. The number of transfections is shown (n = 9, performed in three independent experiments, each with three transfections of each reporter), with error bars indicating the standard deviation for these transfections.
(B) As in (A), but using reporters with the DICER1 3′UTRs.
(C) As in (A), but using reporters with the Cyclin D2 3′UTRs.
Contribution of miRNA regulation on differential protein expression levels
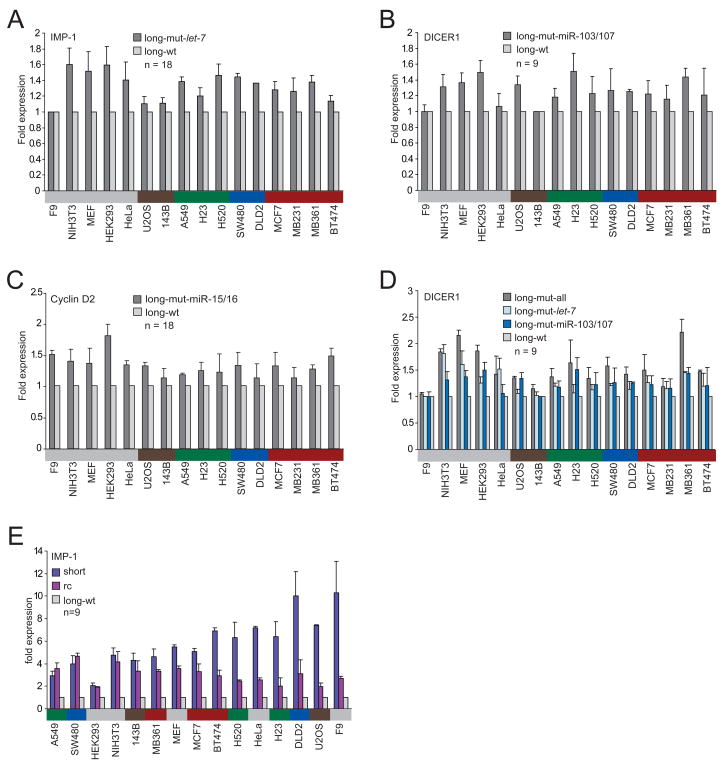
Although the greater mRNA destabilization and translational repression observed for the longer isoforms was consistent with regulation by the miRNAs expressed endogenously within the cell lines, other types of posttranscriptional repression lost in the shorter isoforms also might have explained some or all of the observed differences. To begin to determine the impact of miRNA regulation on protein expression, we mutated the miRNA complementary sites for the miRNAs with the highest number of sites. The five let-7 sites in the IMP-1 3′UTR, the six sites for miR-103/107 in the DICER1 3′UTR and the three sites for miR-15/16 in the Cyclin D2 3′UTR were mutated in the context of the long isoform (Figure 1B), and luciferase activity was examined, comparing activity from reporters with intact sites with that from those with mutant sites. Loss of these miRNA sites led to expression increases of 1.0–1.8 fold across 16 cell lines from various tissues (IMP-1 median, 1.4, range, 1.0–1.6; Cyclin D2 median, 1.3, range, 1.1–1.8; DICER1 median, 1.2, range, 1.0–1.5)—rather modest effects when considering the number of sites mutated (Figures 4A–4C). When the endogenous miRNAs were supplemented by transfecting more of the cognate miRNAs, the reporter gene expression differences increased up to 3.3 fold (data not shown), implying regulatory potential beyond that mediated by the miRNAs at their endogenous levels and suggesting that the variability in response to the endogenous miRNAs might depend on expression levels of the cognate miRNAs. Quantitative Northern blots of miRNAs across the cell lines showed that, overall, let-7 levels were the highest, miR-15/16 levels were intermediate, and miR-103/107 levels were about 5 times lower than those of let-7 (Figure S1). In addition to its six miR-103/107 sites, the DICER1 3′UTR has two let-7 sites. Mutating these sites led to a median expression increase of 1.3 (range 1.0–1.8 fold), which was higher than that of mutating the six miR-103/107 sites, consistent with the idea that let-7, based on its higher expression, might have been more effective in these cells (Figure 4D).
Figure 4. Contributions of miRNA regulation and 3′UTR length to the difference in protein expression observed between the long and the short mRNA isoforms.
(A) Repression of the IMP-1 3′UTR by the let-7 miRNA endogenously expressed within the cells. Luciferase expression of a reporter possessing the full-length IMP-1 3′UTR with five mutant let-7 sites (long-mut-let-7) is compared with that of a reporter with five intact let-7 sites (long-wt, Figure 1B). The number of transfections is shown (n = 18, performed in six independent experiments, each with three transfections of each reporter), with error bars indicating standard deviation. Cells are color-coded as in Figure 1A.
(B) Repression of the DICER1 3′UTR by the miR-103/107 endogenously expressed within the cells. As in (A) but examining the contribution of the six miR-103/107 sites in the full-length DICER1 3′UTR.
(C) Repression of the Cyclin D2 3′UTR by the miR-15/16 endogenously expressed within the cells. As in (A) but examining the contribution of the three miR-15/16 sites in the full-length Cyclin D2 3′UTR.
(D) Repression of the DICER1 3′UTR by the let-7 miRNA endogenously expressed within the cells. As in (B) but examining the contribution of the two let-7 sites, disrupting either only the two let-7 sites (long-mut-let-7) or both the two let-7 sites and the six miR-103/107 sites (long-mut-all).
(E) Influence of the length of the IMP-1 3′UTR on protein expression. The reverse complement (rc) of the difference between the shortest and the full-length mRNA isoforms of IMP-1 was cloned downstream the shortest 3′UTR, while mutating the proximal poly(A) signal. Expression of the reporter possessing the shortest 3′UTR and of that also possessing the rc, each normalized to expression of the reporter containing the full-length isoform. The number of transfections is shown (n = 9, performed in three independent experiments, each with three transfections of each reporter), with error bars indicating standard deviation.
To find additional miRNAs endogenously expressed in these cell lines and to measure the relative expression of the miRNAs examined, we profiled the miRNAs by high-throughput sequencing. The let-7 miRNA family was either the highest or the second highest sequenced family in the investigated cell lines (with the exception of HEK293 cells where it ranked eighth highest) and corresponded to 3.3%–34% (median, 16.8%) of all miRNA sequencing reads in the cells (Table S1). Consistent with the Northern blots, miR-103/107 was sequenced less frequently, ranking 3–16 of all miRNA families expressed in these cells and corresponding to 0.74%–5.8% of the reads (median, 2.4%). Considering the expression levels of the different miRNAs in the different cell types, variability in miRNA expression explained a significant fraction of the variability in reporter expression (_r_2 = 0.20, p = 0.001, Figure S4). By taking this correlation into account and extrapolating to the other miRNA sites (conserved and nonconserved) predicted by TargetScan to match coexpressed miRNAs, we estimated the maximum repression attributable to miRNAs to range from 1.9–4.1 fold for the three 3′UTRs in the 12 cell lines with profiled miRNAs (IMP-1 median, 2.8, range, 2.5–3.5; DICER1 median, 2.7, range, 1.9–3.2; Cyclin D2 median, 2.4, range, 1.9–4.1; Figure S4 and Table S2). These rough estimates were typically lower than the average increases in luciferase reporter gene expression for the short 3′UTR in these 12 lines (IMP-1, 7.7 fold, DICER1: 6.6 fold, Cyclin D2: 15.3 fold). Thus, loss of miRNA regulation appears to account for a quarter to two-thirds of the increase in protein-expression levels observed with the short 3′UTRs. In 20–40% of the cell lines (dependent on the gene investigated) loss of miRNA regulation might even explain all the difference in protein expression. However, for the majority of cell lines miRNA regulation explained only a part of the difference in protein expression between the short and the long mRNAs, indicating a net repressive effect of other regulatory phenomena lost in the shorter UTRs as a consequence of APA.
Impact of 3′UTR sequence versus 3′UTR length
One explanation for the repression not attributable to miRNA targeting might be the length of the mRNA itself. The difference in length between the shorter and the longer mRNA isoforms was substantial (IMP-1, 6.3 kb, or 71% of the mRNA; DICER1, 4.3 kb, or 42% of the mRNA; Cyclin D2, 3.5 kb, or 54% of the mRNA). Shorter mRNAs might more readily form a closed-loop structure, which enhances translation, as shown recently in yeast (Amrani et al., 2008). To find out if the length or specific sequences within the longer mRNA were responsible for the difference in luciferase activity, the reverse complement (rc) of the UTR segment present in the long but not the short IMP-1 mRNA isoform was cloned downstream the short 3′UTR, while disrupting the proximal poly(A) signal. In four of the 15 cell lines tested, the short and rc reporters had comparable protein production, which averaged 4-times higher than that from a reporter with the long 3′UTR, suggesting that repressive sequences recognized by _trans_-acting factors present preferentially in these cell lines specify translational repression or mRNA destabilization (Figure 4E). In the other cell lines, however, expression of the rc construct led to intermediate gene expression. Although 3′UTR length might have some influence on gene expression in these cell lines, our results for the other four lines speak against this as a general phenomenon in human cells.
The shorter Cyclin D2 mRNA leads to more cells in S phase
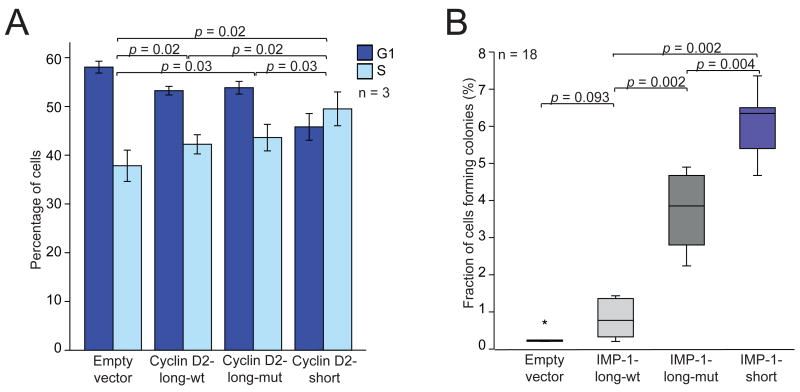
Having found that the shorter mRNA isoforms were more stable and led to higher protein expression, we wanted to test if there might be a phenotypic consequence of expressing the shorter rather than the longer isoform. For Cyclin D2, we expressed either the full-length mRNA or its shorter isoform in breast cancer cell lines. Cyclin D2 is not expressed endogenously in these cell lines because of promoter hypermethylation(Evron et al., 2001). In MCF7 cells, we observed an effect on the cell-cycle profile, with a lower fraction of cells in G1 accompanied by a higher fraction of cells that had entered S phase (Figure 5A), which was concordant with the function of Cyclin D2 in overcoming the G1/S checkpoint (Lukas et al., 1995). This effect was significantly greater in cells expressing the short mRNA of Cyclin D2 compared with those expressing the long mRNA, and correlated with Cyclin D2 mRNA and protein expression in these cells (Figure S5).
Figure 5. Functional consequences of expressing short or long mRNA isoforms.
(A) The short mRNA isoform of Cyclin D2 increases the fraction of cells in S phase. For MCF7 cells stably transduced with retroviral vectors, the percentage of cells that were in G1 and S phase were measured using fluorescence-activated cell sorting (FACS) analysis. Plotted is the mean (± s.d.) of three independent experiments.
(B) The short mRNA isoform of IMP-1 promotes oncogenic transformation. For NIH3T3 cells stably transduced with retroviral vectors, the percentage that yielded colonies after 28 days was plotted (median, horizontal line; 25th through 75th percentile, box; range, error bars; outlier, *; n = 18 from six independent experiments, each in triplicate). The vectors expressed either the long mRNA isoform of IMP-1 with mutations designed to prevent APA but retaining intact let-7 sites (IMP-1-long-wt) or the analogous long mRNA isoform with mutant let-7 sites (IMP-1-long-mut) or the shortest mRNA isoform of IMP-1 (IMP-1-short).
The shorter IMP-1 mRNA is oncogenic
IMP-1 is an RNA-binding protein that is overexpressed in a variety of human cancers, including those of the breast, lung and colon, where it potentially plays a role in stabilizating several target mRNAs, including c-myc, β-catenin and β-TrCP1 (Ross et al., 1997; Doyle et al., 1998; Nielsen et al., 1999; Noubissi et al., 2006). To learn the relative effects of the IMP-1 isoforms, we assayed colony formation in soft-agar after transducing cells with retroviral vectors expressing either the short or long mRNA isoform. Expressing the full-length IMP-1 isoform (with mutations designed to prevent APA) produced a number of colonies comparable to that of expressing the empty vector (p = 0.09), whereas expressing the short isoform greatly promoted cell transformation (p = 0.002, Figure 5B). Much of this transformation was attributable to loss of miRNA targeting, in that mutating the let-7 sites significantly enhanced transformation from the long isoform (p = 0.002) and correlated with IMP-1 mRNA and protein expression in these cells (Figure S6). The shorter isoform was not only able to transform fibroblasts but also human breast epithelial cell lines (Figure S7). Taken together, our results indicate that APA associated with cancer cells can activate an oncogene, thereby potentially contributing to the pathogenesis of cancer.
Discussion
Recent reports propose that a change in 3′UTR length by means of APA is a coordinated mechanism for altering expression of many genes during T cell activation (Sandberg et al., 2008), neuronal activation (Flavell et al., 2008) or embryonic development (Ji et al., 2009). For a better understanding of gene regulation, it will be important to identify additional cellular, developmental and disease states that lead to increased use of proximal APA signals. We observed shorter 3′UTRs in cancer cell lines compared with non-transformed cell lines, despite similar proliferation rates of the transformed and non-transformed lines, thereby linking 3′UTR shortening with oncogenic transformation even more than with cellular proliferation.
Many proto-oncogenes play roles in proliferation and differentiation of normal cells and must be highly regulated to prevent malignant transformation. In normal cells, the full-length, annotated transcripts were expressed and presumably responsible for normal proto-oncogene function, but in cancer cells, substantially more shorter isoforms were also expressed, which typically differed from the full-length mRNAs only in the length of their 3′UTRs. Our reporter assays and immunoblots revealed that the APA-mediated 3′UTR shortening can have striking functional consequences in the cancer cell lines, with the shorter mRNA isoforms typically producing ten-times more protein. These results suggested that the cancer-associated shortening of 3′UTRs could activate oncogenes, thereby reinforcing the transformed state. In support of this hypothesis, we found that the shorter IMP-1 isoform promoted oncogenic transformation far more efficiently than did the longer one.
The oncogenic potential of IMP-1 was previously demonstrated using a transgenic mouse model with IMP-1 under the promoter of whey acidic protein, which is induced in mammary epithelial cells in pregnant and lactating female mice (Tessier et al., 2004). The construct overexpressing IMP-1 in these mice contained only the first 288 nt of the 6.3 kb annotated 3′UTR. Our results, which showed both that cancer cells endogenously express a shorter isoform with a 369-nt 3′UTR (with the orthologous mouse isoform estimated to have a 336-nt 3′UTR) and that this shorter isoform is the one that was oncogenic, support the in vivo relevance of the previous transgenic model.
Part of the IMP-1 oncogene activation observed in our soft-agar assays was explained by escape of the shorter isoform from miRNA-mediated repression. Since the discovery of miRNAs in mammalian cells, much has been learned about their roles in tumorigenesis. Some miRNAs, including the miR-17~92 cluster are amplified in human cancers and act as oncogenes when overexpressed in mice (Ota et al., 2004; He et al., 2005). Others, including miR-15/16, miR-34 and the let-7 miRNAs, are deleted or downregulated in cancers and are reported to act as tumor-suppressor genes (Calin et al., 2002; Johnson et al., 2005; Mayr et al., 2007; Yu et al., 2007; He et al., 2007). These miRNAs playing tumor-suppressor roles can do so only when their targets retain the cognate regulatory sites. Thus, another important mechanism for oncogenic transformation is the loss of miRNA sites in the mRNAs of oncogenes. This loss can occur through genetic aberrations, such as the translocations that abrogate let-7 repression of the HMGA2 oncogene (Mayr et al., 2007; Lee and Dutta, 2007). Our results uncovered an epigenetic mechanism (using the broader sense of the word “epigenetic”) that achieved the same effect: oncogenes can escape miRNA-mediated repression through 3′UTR shortening due to the APA prevalent in cancer cells. Our data suggests that this epigenetic mechanism is more prevalent than is the genetic one because cancer cells had the shortest UTRs as indicated by their low TLI (a transcriptome-wide index), and APA generated the different transcripts in over a third of the expressed genes examined in detail.
Escape from miRNA-mediated repression explained only part of the upregulation conferred by APA. In addition to miRNAs, AU-rich or GU-rich elements can specify repression (Chen and Shyu, 1995; Vlasova et al., 2008), but a search of the tested 3′UTRs did not reveal these elements. Thus, our results implicated additional, as-yet-undefined regulatory elements in these 3′UTRs. Interestingly, the net effect of these elements was never activating—whenever an effect was observed, it was repressive. Although activating regulatory elements undoubtedly occur in some 3′UTRs and the net effect of all RNA elements might be activating in some 3′UTRs or cell types, our results support the idea that regulatory phenomena acting on 3′UTRs are generally repressive. This conclusion holds despite the bias in selecting for study 3′UTRs with multiple sites to coexpressed miRNAs, because even after all those sites were mutated, we still observed only repression. The overall repressive character of 3′UTRs could help explain why nearly all successful transgenic mouse models of tumorigenesis overexpress oncogenes missing large segments of their annotated 3′UTRs (Ruther et al., 1987; Fedele et al., 2002; Primrose and Twyman, 2006). As with IMP-1, our results indicate that these models expressing shorter versions of the mRNA are more biologically relevant than might have been anticipated when they were first generated.
One of the most interesting open questions regarding APA in cancer cells is, what mechanism underlies the recognition and increased utilization of proximal polyadenylation signals? In the sequence surrounding the proximal polyadenylation sites we never found point mutations that would have changed the strength of the polyadenylation signal, with the caveat that by 3′ RACE we investigated only the sequences upstream of the cleavage sites. Although mutations downstream of the cleavage sites cannot be excluded, we hypothesize that differential expression of _trans_-acting factors explains the use of proximal polyadenylation sites in cancer cells. Factors that might influence the choice of polyadenylation signal include limiting components of the polyadenylation machinery, RNA-binding proteins that bind in the vicinity of the proximal signal and influence recognition by the polyadenylation machinery (Takagaki et al., 1996; Martincic et al., 1998; Veraldi et al., 2001; Lutz, 2008; Wang et al., 2008), and perhaps factors that influence transcriptional elongation rate(Kornblihtt, 2005). To begin to identify such factors, we examined published array data comparing breast cancer cells with a breast epithelial cell line, MCF10A (Hoeflich et al., 2009). A survey of the constitutive components of the polyadenylation machinery and other candidates from the literature revealed several that were significantly upregulated in the cancer lines (Figure S8). These included the mRNAs of cleavage and polyadenylation specificity factor 1 (CPSF1) and cleavage stimulation factor 2 (CSTF2), which recognize the poly(A) signal and accessory sequences including the downstream G/U-rich sequence, respectively, raising the intriguing possibility that an increase of these factors might help increase utilization of sub-optimal proximal poly(A) signals in cancer cells.
We imagine a complex scenario in which some _trans_-acting factors act globally, some act tissue specifically, and some act gene specifically, with the combinatorial expression of all the different _trans_-acting factors determining the probability of using each proximal polyadenylation signal. The observation of higher amounts of shorter mRNAs in cancer cells compared with normal cells, with no examples of the reverse for any of the genes and cell types studied, suggested a role for globally acting factors. That some cell lines showed high amounts of shorter transcripts for all genes investigated further implicated the role of globally acting factors. Nonetheless, the differences observed for different genes in different cell types suggested a role for additional factors acting more specifically. Such complexity could explain the differential impact of different oncogenes in different tissues. Indeed, some of the genes for which we did not observe alternative mRNAs are known oncogenes in tissues not included in our panel of cell lines (ARID3B in neuroblastomas, MYB and PLAG1 in hematological malignancies). Perhaps shorter mRNA transcripts might be found in the tissues where these genes have oncogenic effects. Moreover, the prospect of some factors acting more specifically opens the possibility for exceptions to the trend of shorter isoforms expressed preferentially in cancer cells. Combinatorial use of tissue-specific and gene-specific _trans_-acting factors could for some genes (most intriguingly, for tumor-suppressor genes) lead to higher amounts of shorter mRNAs in normal cells rather than in cancer cells.
The prevalence of APA in cancer lines brought to the fore the question of what influence it might have on oncogenes, and our results for IMP-1 supported the idea that APA was activating. However, APA creates shorter, less repressed isoforms of more than just known oncogenes, as illustrated for DICER1. Because some of these genes likely act in opposition to oncogenes, the net functional significance of APA in cancer cells is unknown, and in principle could even be tumor suppressive. During both normal development and cancer development there is often a dichotomy of cell proliferation and differentiation (Derynck and Wagner, 1995). The association of APA with cell proliferation suggests that it might represent a coordinated gene-expression program that antagonizes differentiation during normal development. Accordingly, we propose that cancer cells coopt and exaggerate this proliferation/de-differentiation program with the net effect of enhanced tumorigenesis.
Our observations that the shorter mRNAs were found in transformed cells and that expression of the shorter mRNA of IMP-1 (and presumably other oncogenes) can lead to transformation suggests a APA-mediated feed-forward loop in cancer that might lead to a more aggressive phenotype. This proposal is in agreement with the report on mantle cell lymphomas, in which the patients that have shorter Cyclin D1 3′UTRs have the worst prognosis (Rosenwald et al., 2003; Wiestner et al., 2007). However, when this process of generating shorter mRNAs would start and whether APA might play a role in early tumorigenesis is unclear. In general, shorter mRNAs were not observed in non-transformed cell lines, which usually already have one hit towards cancer but are not yet fully transformed. Perhaps if an early lesion activates a signaling pathway that is able to change the expression of a key _trans_-acting factor, then APA would contribute to early steps in oncogenesis.
Oncogenes are reported to be overexpressed in human tumors much more frequently than genetic lesions are detected at these loci. Our results could explain some of this discrepancy, because overexpression of oncogenes due to APA-mediated shortening of mRNA transcripts was a widespread phenomenon in the cancer cells investigated. The epigenetic nature of this mechanism for oncogene activation suggests that it can be reversed, perhaps providing a new strategy for cancer treatment.
Experimental procedures
Northern blots
Total RNA was isolated using Tri-reagent (Ambion). miRNAs were detected as described, loading 20 μg per lane (Mayr et al., 2007). mRNA was detected using a protocol adapted from Cold Spring Harbour Protocols (Sambrook et al., 2006), loading in each lane 1.5–2 μg polyadenylated RNA purified from total RNA with Oligotex (Qiagen).
mRNA stability
Cell lines were treated with Actinomycin D (10 μg/ml), and after 0, 2, 4, 6 and 8 hours total RNA was isolated and analyzed on Northern blots.
3′ RACE
This protocol was adapted from Cold Spring Harbour Protocols (Sambrook et al., 2006). 1 μg total RNA was used to generate cDNA with Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions using TAP as a primer. The first PCR was done with a gene-specific forward primer and AP as reverse primer. Nested PCR was done with a nested gene-specific forward primer and MAP as reverse primer. The PCR product from the nested PCR was separated on an agarose gel, cloned and sequenced. If only full-length mRNAs were found by 3′ RACE, Northern blots were reprobed with different probes in the ORF to find alternatively spliced isoforms.
Luciferase assays
Luciferase assays were performed as described (Mayr et al., 2007). To account for differences in plasmid preparations, values for each wild-type reporter were normalized to those for its cognate mutant-site reporter by using values from cells that lacked or expressed the relevant miRNA at low levels (F9 cells for the let-7 miRNA, DLD2 cells for miR-15/16 and 143B for miR-103/107).
Small RNA cloning
Small RNAs were cloned and analyzed as described previously (Grimson et al., 2008). The miRNA cloning data have been deposited at the NCBI Gene Expression Omnibus (GEO) repository (GSE16579).
Cell cycle analysis by FACS
Cells were plated at comparable densities, harvested after 48h, fixed with ethanol and stained with propidium iodide (50μg/ml) and RNase A (40U/ml) and DNA content was measured on a FACS Calibur HTS (Becton Dickinson). The percentage of diploid cells in G1, S and G2 were analyzed by ModFitLT V3.1.
Soft-agar assay
Soft-agar assays were performed as described (Mayr et al., 2007).
Statistics
The Kruskal-Wallis test was used to analyze the difference between several independent subgroups. Mann Whitney test was applied to analyze the difference between two independent supgroups. The Wilcoxon test was used to make pairwise comparisons using SPSS 14.0.
Supplementary Material
01
Acknowledgments
We thank Calvin Jan (CJ) and Andrew Grimson (AG) for very helpful advice throughout the project, Wendy Johnston for preparing the small RNA libraries for the cell lines, Gina Lafkas for help with the soft-agar assay, AG and Rosaria Chang for analyzing the Solexa reads, and Olivia Rissland, AG, CJ and Alena Shkumatava for helpful comments on the manuscript. Supported by grants from the Deutsche Forschungsgemeinschaft (C.M.) and the NIH (D.B.). D.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We have eight supplemental figures, two supplemental tables and supplemental text, including experimental procedures and references.
References
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JM. Molecular themes in oncogenesis. Cell. 1991;64:235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Coleman MS, Hutton JJ, Bollum FJ. Terminal riboadenylate transferase in human lymphocytes. Nature. 1974;248:407–409. doi: 10.1038/248407a0. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Derynck R, Wagner EF. Cell differentiation. Curr Opin Cell Biol. 1995;7:843–844. doi: 10.1016/0955-0674(95)80068-9. [DOI] [PubMed] [Google Scholar]
- Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res. 1998;26:5036–5044. doi: 10.1093/nar/26.22.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B, Buluwela L, Weitzman SA, Marks J, Sukumar S. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001;61:2782–2787. [PubMed] [Google Scholar]
- Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R, Pierantoni GM, Outwater E, et al. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene. 2002;21:3190–3198. doi: 10.1038/sj.onc.1205428. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser H, Knippers R, Schafer KP. Increased rate of RNA-polyadenylation. An early response in Concanavalin A activated lymphocytes. Exp Cell Res. 1978;111:175–184. doi: 10.1016/0014-4827(78)90247-1. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, O'Brien C, Boyd Z, Cavet G, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Lebwohl DE, Muise-Helmericks R, Sepp-Lorenzino L, Serve S, Timaul M, Bol R, Borgen P, Rosen N. A truncated cyclin D1 gene encodes a stable mRNA in a human breast cancer cell line. Oncogene. 1994;9:1925–1929. [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lukas J, Bartkova J, Welcker M, Petersen OW, Peters G, Strauss M, Bartek J. Cyclin D2 is a moderately oscillating nucleoprotein required for G1 phase progression in specific cell types. Oncogene. 1995;10:2125–2134. [PubMed] [Google Scholar]
- Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Majoros WH, Ohler U. Spatial preferences of microRNA targets in 3′ untranslated regions. BMC Genomics. 2007;8:152. doi: 10.1186/1471-2164-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze MT, Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc Natl Acad Sci USA. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Chiorini JA, Urcelay E, Safer B. Regulation of gene expression for translation initiation factor eIF-2 alpha: importance of the 3′ untranslated region. Biochem J . 1996;315(Pt 3):791–798. doi: 10.1042/bj3150791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi FK, Elcheva I, Bhatia N, Shakoori A, Ougolkov A, Liu J, Minamoto T, Ross J, Fuchs SY, Spiegelman VS. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- Primrose SB, Twyman RM. Principles of Gene Manipulation and Genomics. Blackwell Publishing; 2006. [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- Ruther U, Garber C, Komitowski D, Muller R, Wagner EF. Deregulated c-fos expression interferes with normal bone development in transgenic mice. Nature. 1987;325:412–416. doi: 10.1038/325412a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Separation of RNA according to size: Electrophoresis of glyoxylated RNA through agarose gels. Cold Spring Harbor Protocols:pdb.prot4057. 2006 doi: 10.1101/pdb.prot4057. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 2004;64:209–214. doi: 10.1158/0008-5472.can-03-2927. [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, Liu H, Rosenwald A, Muller-Hermelink HK, Ott G, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01