Regulation of axonal trafficking of cytochrome c oxidase IV mRNA (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 1.
Published in final edited form as: Mol Cell Neurosci. 2010 Feb 6;43(4):422–430. doi: 10.1016/j.mcn.2010.01.009
Abstract
Trafficking and local translation of axonal mRNAs play a critical role in the development and function of this neuronal subcellular structural domain. In this report, we studied cytochrome c oxidase subunit IV (COXIV) mRNA trafficking into distal axons of primary superior cervical ganglia (SCG) neurons, and provide evidence that axonal trafficking and mitochondrial association of the mRNA is mediated by an element located in a 38 bp-long, hairpin-loop forming region within the 3’UTR of the transcript. Our results also suggest that suppression of local translation of COXIV mRNA results in significant attenuation of axonal elongation. Taken together, the results provide the first evidence for the existence of a _cis_-acting axonal transport element within a nuclear-encoding mitochondrial gene, and demonstrate the importance of the axonal trafficking and local translation of nuclear-encoded mitochondrial mRNAs in axonal growth.
Keywords: Sympathetic neurons, superior cervical ganglion, intra-axonal translation, mRNA localization, 3’UTR, axonal growth, mitochondria, ATP synthesis
Introduction
Mitochondria in the axons and presynaptic nerve terminals fulfill distinct functions and have been shown to be closely associated with synapses and tethered to vesicle release sites (Zenisek and Matthews, 2000). To date, little is known about the half-life and biogenesis of synaptically localized mitochondria. Evidence that neuronal mitochondrial biogenesis does occur in the axon at a significant distance from the cell body has been recently provided (Amiri and Hollenbeck, 2008). Over the past few years, it has also become widely accepted that a distinct subset of nuclear-encoding mitochondrial mRNAs are selectively transported to the distal structural/functional domains of the neuron, including the axon and presynaptic nerve terminal (Gioio et al., 2001; Gioio et al., 2004; Willis et al., 2007; Taylor et al., 2009). Local proteins synthesized from these mRNAs play a key role, not only for mitochondrial function, but also in the development of the neuron and the function of the axon and nerve terminal. In addition, these studies called attention to the importance of local translation of COXIV mRNA, and its regulation by a brain-specific microRNA, miR-338, that regulates COXIV synthesis locally with distinct consequences on mitochondrial function, such as ATP generation, and axonal function as monitored by neurotransmitter uptake (Aschrafi et al., 2008).
The molecular mechanisms responsible for the transport of COXIV or other nuclear-encoded mitochondrial mRNAs into the axon are unknown. Subcellular mRNA localization and local translation within dendrites and axons are posttranscriptional mechanisms that generally require _cis_-acting sequences for their localization. These gene sequences are usually found in the 3’ untranslated region (3’UTR) of the transcript, and it was suggested that RNA-binding proteins recognize specific secondary structures in the 3’UTR, forming messenger ribonucleoprotein (mRNP) particles that are subsequently transported along microtubules to specific sites (Bassell and Singer, 1997; Kohrmann et al., 1999).
In this work, we used superior cervical ganglia (SCG) neurons cultured in compartmentalized Campenot cultures to examine the axonal transport and translation of COXIV mRNA. Using green fluorescent protein (GFP)-tagged constructs, we identified a 38 bp-long fragment within the COXIV 3’UTR that was required for mitochondrial targeting and axonal localization of the mRNA. Secondary RNA structure analysis of the 3’UTR suggests that these _cis_-acting regulating sequences are situated in a hairpin-loop forming region. In addition, our investigations identified local COXIV synthesis as an important determinant of axonal growth, as over-expression or knockdown of axonal COXIV levels significantly increased or attenuated neurite elongation, respectively.
Results
The COXIV 3’UTR is sufficient for axonal mRNA transport
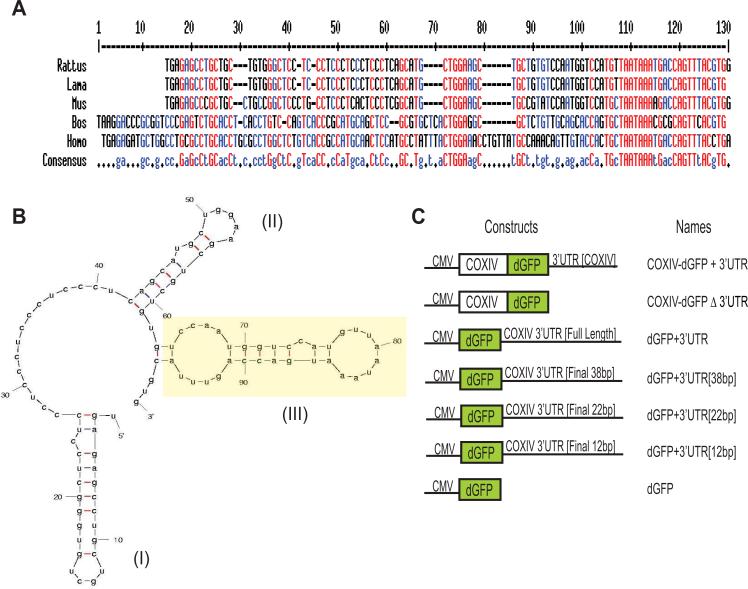
Most of the _cis_-acting sequences that have been identified as being involved in dendritic or axonal localization are situated in the 3' UTR of the mRNA (Martin and Zukin, 2006). Initial comparative sequence analysis revealed that the 3'UTR of the mammalian COXIV gene is highly conserved (Fig. 1A). In addition, an RNA secondary structure prediction analysis using Mfold (Zuker, 2003) indicated that this 3’UTR consisted of three well-conserved stem-loop structures, that might have functional significance (Fig. 1B). Previous investigation of the COXIV 3’UTR confirmed that the target site for the brain-specific miR-338 was positioned on the second hairpin-loop structure, in an exposed position facilitating microRNA accessibility and subsequent regulation of COXIV expression (Aschrafi et al., 2008).
Fig. 1.
The 3'UTR of mammalian COXIV mRNA is highly conserved. (A) Sequence alignment of the 3'UTR of COXIV mRNAs from five mammalian species shows a high degree of sequence conservation. The red and the blue shading represent nucleotides conserved among all five and in four out of the five species, respectively. Each 3’UTR sequence begins with the stop codon. (B) Structure of the COXIV 3’UTR, as determined by secondary structure prediction analysis (Mfold). Yellow highlighted hairpin-loop segment (III) facilitates mRNA transport into distal axons. (C) Overview of constructs used in this study. Plasmid constructs consisting of the COXIV ORF followed by the destabilized GFP (dGFP) cDNA that contain the COXIV 3’UTR [+3’UTR] or the SV40 3′UTR [Δ3’UTR], are diagrammed. In addition, schematic representation of reporter gene plasmids carrying the dGFP cDNA followed by either the full length 3’UTR of rat COXIV, the final 38 bp, 22 bp or 12 bp of the COXIV 3’UTR, respectively, is shown. Expression of all fusion gene products was driven by the human cytomegalovirus promoter (CMV).
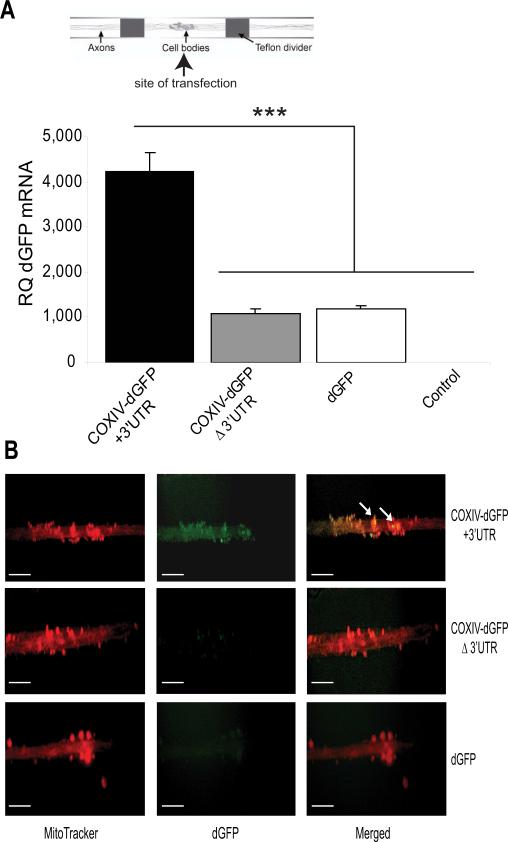
Our previous studies also suggested that the nuclear-encoded COXIV mRNA is localized constitutively in relatively high abundance in the distal axons and presynaptic terminals of sympathetic neurons (Hillefors et al., 2007; Aschrafi et al., 2008). To determine whether the 3’UTR of COXIV mRNA contained functional transport signals for directing mRNAs into distal axons, we employed an experimental strategy that encompassed the transfection of chimeric gene expression plasmids into primary SCG neurons. These chimeric gene constructs consisted of the COXIV ORF followed by a destabilized GFP cDNA (dGFP; with a known protein half-life of approximately 2 h) that contained the COXIV 3’UTR [COXIV-dGFP+3’UTR], or the SV40 3′UTR [COXIV-dGFPΔ3’UTR] (Fig. 1C). These vectors expressing chimeric reporter mRNAs were introduced into SCG neurons by transfecting the soma and proximal axons located in the center compartment of Campenot chambers (see scheme Fig. 2A). One to two days after transfection, we used GFP-specific primers and qRT-PCR to quantify COXIV-dGFP mRNA levels in the distal axons of SCG neurons that were located in the lateral compartments of the Campenot cultures. It has been previously shown that somatically introduced GFP transcripts remain restricted to the somata of sympathetic neurons (Muslimov et al., 1997). As shown in Figure 2A, COXIV-dGFP mRNAs lacking the 3’UTR of COXIV were detected at similar levels as the dGFP mRNAs in total RNA prepared from distal axons, while levels of the chimeric mRNAs carrying the 3’UTR of COXIV were present at significantly higher levels in the axonal compartment. This observation suggested that the COXIV 3’UTR was both necessary and sufficient for targeting COXIV mRNAs to the distal axons. Following the transfection protocols described above, we also tested whether the COXIV mRNAs targeted to the distal axons were translated locally. In previous studies, the local translation of functionally important mRNAs in distal parts of neurons has been demonstrated utilizing destabilized GFP reporter constructs (Macchi et al., 2003). Fluorescence microscopy was employed to visualize dGFP expression in the distal axons of SCG neurons. The experiments revealed that the 3’UTR of COXIV significantly increased axonal expression of the COXIV-dGFP chimera, as compared to COXIV-dGFP lacking the COXIV 3’UTR, and dGFP, respectively (Fig. 2B). In addition, a significant amount of the COXIV-dGFP fluorescence was localized to small, punctuate structures. Next, mitochondrial association of the fusion proteins was assessed using co-localization studies in axons expressing the chimeric mRNAs followed by axonal staining with MitoTracker Red, a mitochondrion-specific fluorescent dye (Fig. 2B). This analysis revealed that the N-terminally located COXIV amino acid sequence was able to target the COXIV-dGFP fusion protein to the mitochondria, as shown in yellow in the merged images in Figure 2B. This finding indicates that the translated COXIV-dGFP is targeted to the mitochondria.
Fig. 2.
COXIV mRNA 3’UTR contains an axonal targeting element. (A_)_ SCG cultures were transfected with the chimeric COXIV-dGFP constructs (2 μg) in the center compartment of the Campenot chambers, as shown. Using GFP-specific primers, quantitation of COXIV-dGFP mRNA levels in the distal axons of SCG neurons were determined by qRT-PCR 24 h after plasmid DNA or sham-transfection (control). GFP mRNA levels are expressed relative to β-actin mRNA. Error bars represent the SEM for n = 3 samples. ***, p < 0.0001. (B) COXIV-dGFP+3’UTR puncta colocalize with the mitochondrial marker MitoTracker in the distal SCG axons, indicating local translation of the COXIV-dGFP mRNA and mitochondrial association of the translation product. Arrowheads depict examples of areas of co-localization. The scale bars represent 100 μm.
The 3’UTR of COXIV mRNA contains an axonal targeting element
To identify the _cis_-acting axonal targeting sequences in the 3’UTR of the rat COXIV mRNA, we created vectors expressing chimeric reporter mRNAs. In the control vector, a transcript encoding dGFP is synthesized under the control of the CMV promoter and contains a SV40 3’UTR. It has been shown previously that the mRNA derived from such a construct is restricted to cell bodies of primary neurons (Goetze et al., 2003). To investigate whether discrete regions of the COXIV 3’UTR are capable of facilitating axonal localization of the dGFP reporter transcripts, various cDNA subfragments from the 3’UTR were inserted into the CMV-driven plasmid at the 3’-end of the dGFP cDNA. These vectors expressing chimeric reporter mRNAs were introduced into primary SCG neurons by employing the transfection protocol outlined in the studies described in Figure 2; i.e. introducing the vectors into the soma and proximal axons located in the center compartment of Campenot cultures by lipofection, followed by qRT-PCR quantification of GFP mRNAs in the distal axons located in the lateral side compartments. These experiments revealed that the chimeric mRNA carrying the entire COXIV 3’UTR was present in the distal axons at significantly higher levels than the control mRNAs, carrying the dGFP followed by the SV40 3’UTR (Fig. 3A). The investigation of the subfragments of this 3’UTR further identified a highly conserved 38 bp fragment, which is located at the terminal end of the COXIV 3’UTR (highlighted in Fig. 1B), and has the potential to form a hairpin-loop structure, that acted as the minimal transport element for the trafficking of dGFP mRNAs into distal axons. As shown in Figure 3A, the insertion of this cDNA subfragment into a CMV promoter-driven dGFP plasmid at the 3’-end of the GFP cDNA resulted in a significant increase in mRNA abundance in the distal axons, an increase that was similar to that observed for the full 3’UTR of COXIV. To assess whether the increased axonal dGFP mRNA abundance was due to axonal transport, or based on an increased molecular stability conferred by the addition of COXIV 3’UTR, we performed a qRT-PCR analysis of the total RNA isolated from transfected somas located in the center compartment of Campenot cultures. Relative mRNA quantity in the soma of SCG axons was assessed by total RNA isolation, followed by subsequent treatment with DNaseI and DpnI to remove the methylated, DpnI sensitive plasmid DNA that was introduced by transfection. The subsequent qRT-PCR confirmed that, unlike the different mRNA quantities obtained in the distal axons, nearly equal amounts of the various chimeric dGFP transcripts were present in the cell bodies after transfection (data not shown), indicating that the increase in dGFP mRNA seen by the fusion of the full 3’UTR or the final 38 bp fragment is due to axonal transport of the mRNAs.
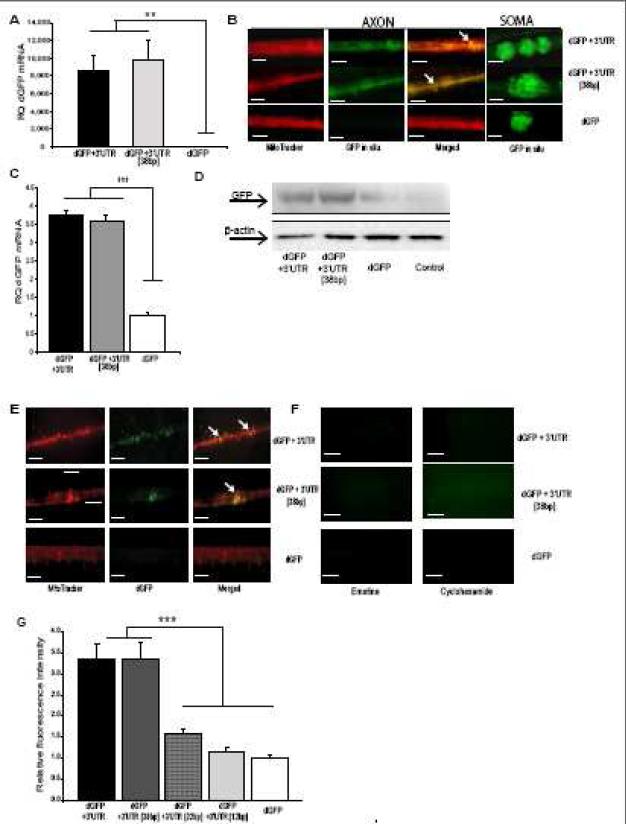
Fig. 3.
The final 38 bp within the COXIV mRNA 3’UTR defines axonal localization, mitochondrial targeting, and dGFP local translation. (A) SCG cultures (6 DIV) were transfected with the dGFP constructs (2 μg) in the center compartment as shown in Fig. 2. Quantitation of GFP mRNA levels in the distal axons of SCG neurons was determined by qRT-PCR 24 h after DNA transfection. GFP mRNA levels are expressed relative to β-actin mRNA. Error bars represent the SEM for n = 3 samples (dGFP constructs versus sham-transfected controls). Student's t test, **p < 0.001. (B) SCG neuronal cell bodies were transfected as in (A). Distal axons and cell bodies were processed for in situ hybridization with a FITC-labeled LNA probe specific for GFP. Higher-magnification images illustrate co-localization of dGFP mRNAs carrying the entire COXIV 3’UTR or the final 38bp of this 3’UTR with axonal mitochondria, but complete absence of FITC fluorescence in axons of dGFP-control transfected neurons. Representative examples from one of four independent experiments are shown. FITC-labeled, scramble, LNA probe served as negative control, and produced only faint fluorescence under identical hybridization and wash conditions (not shown). Scale bar, 100 μm (C) SCG cultures (6 DIV) were transfected with the dGFP constructs as described in (A). Total RNA was prepared in the presence of 3 mM MgCl2 from an enriched mitochondrial fraction in distal axons following the protocols described in (Russo et al., 2006). Quantitation of dGFP mRNA levels from total mitochondrial RNA was determined by qRT-PCR 24 h after DNA transfection. dGFP mRNA levels are expressed relative to COXII mRNA. Error bars represent the SEM for n = 3 samples (dGFP constructs versus sham-transfected controls). Student's t test, ***p < 0.0001. (D) Protein lysates (5 μg) were prepared from distal axons of SCG neurons that were transfected as in (A), and analyzed by western blot using a monoclonal GFP antibody. Levels of β-actin protein served as an internal control. (E) dGFP encoded by transcripts containing either the entire COXIV 3’UTR or the final 38 bp segment are visualized using fluorescence microscopy as puncta that colocalize with the mitochondrial marker MitoTracker in the distal SCG axons, indicating mitochondrial association of the dGFP translation product. In contrast, dGFP encoded by transcripts devoid of the COXIV 3’UTR sequence were not detected in axons. Arrowheads depict areas of colocalization. The scale bars represent 100 μm. (F) Fluorescent image analysis of distal axons treated with protein synthesis inhibitors. Application of emetine or cyclohexamide for 3-4 h caused complete disappearance of the dGFP fluorescent signal in the axon. The scale bars represent 100 μm. (G) A 38 bp fragment within the COXIV 3’UTR represents the minimal axonal transport element of the mRNA. SCG cultures (6 DIV) were transfected with dGFP vectors (2 μg) that contained the full 3’UTR (103 bp), or final sub-fragments in length of 38 bp, 22 bp, and 12 bp of the COXIV 3’UTR, respectively, in the center compartment. Axonal GFP levels were visualized by fluorescence microscopy, and quantified using Image J. Fluorescence levels were normalized to the fluorescence intensity of axons from non-transfected neurons. Data are averages ±SEM from the measurement of 35-45 axons. The experiment was repeated three times with similar results. ANOVA, ***, p < 0.0001.
Initially, we assumed that the observed co-localization of the reporter gene and the mitochondria (Fig. 2B) was attributable to amino acid sequences present in the N-terminal flanking COXIV protein. However, previous studies in yeast and HeLa cells suggested that translocation of specific nuclear-encoded mRNAs to the vicinity of mitochondria required _cis_-acting signals present in the 3’UTR of the RNA molecule (Corral-Debrinski et al., 1999; Corral-Debrinski et al., 2000). To investigate whether the full 3’UTR or the final 38 bp region could facilitate mitochondrial association of the message, we transfected SCG neurons with either the dGFP, dGFP+3’UTR, or dGFP+3’UTR [38 bp] vectors, as described above. Following transfection, the subcellular localization of the mRNA was monitored by fluorescent in situ hybridization with a LNA-modified oligonucleotide probe directed against the GFP coding region. dGFP+3’UTR, or dGFP+3’UTR [38 bp] mRNAs were readily detected in neuronal cell bodies and distal axons (Fig. 3B). In contrast, the control mRNA, dGFP, was undetectable in distal axons and was found almost exclusively in the cell bodies. Using MitoTracker Red to stain axonal mitochondria, we found that the full 3’UTR, or the hairpin-loop forming 38 bp fragment were sufficient for mitochondrial co-localization of the reporter gene mRNA, as compared to control dGFP mRNA carrying the SV40 3’UTR (Fig. 3_B_). To further examine the importance of the 38 bp region in localizing the COXIV mRNA to the vicinity of mitochondria, we prepared an enriched mitochondrial fraction from distal axons in the presence of magnesium (Reinhart et al., 1982). The presence of dGFP mRNAs in total RNA purified from mitochondrial fractions was then determined by qRT-PCR, using the COXII gene, a mitochondrial DNA-encoded gene, to verify fractionation efficiency and as an internal control. These experiments revealed that the final 38 bp segment of COXIV 3’UTR significantly enhanced mitochondrial co-localization of dGFP reporter mRNA in distal axons (Fig. 3C), suggesting that the COXIV 3’UTR is sufficient for mitochondrial association of the message (Fig. 3C).
Local translation of COXIV in the axons
To monitor the spatial distribution of protein synthesis that might be facilitated by the COXIV 3’UTR-mediated mRNA transport, we investigated local protein synthesis derived from the actively transported mRNAs by Western blot analysis and fluorescent microscopy. Total protein lysates (5 μg) from the distal axons that were transfected in their cell bodies with either the dGFP, dGFP+3’UTR, or dGFP+3’UTR [38 bp] vectors, were analyzed by Western blot assay using monoclonal antibodies against GFP protein. This study revealed that higher levels of GFP protein were detected in axons that were expressing the dGFP-3’UTR [full-length], or [38 bp], indicating that the identified 38 bp segment within the 3’UTR is enhancing axonal levels of the dGFP protein (Fig. 3D). We also employed fluorescence microscopy to visualize dGFP local translation in the distal axons of SCG neurons. Using MitoTracker to fluorescently label axonal mitochondria, the experiments revealed that the full 3’UTR, or the 38 bp fragment were sufficient for mitochondrial co-localization of the newly synthesized reporter gene proteins (Fig. 3E). To confirm the local translation of COXIV protein, emetine and cyclohexamide, protein synthesis inhibitors which act by different mechanisms, were added to the lateral compartment of Campenot cultures harboring the distal axons that were transfected in their cell bodies with the dGFP reporter constructs described above. Addition of emetine or cyclohexamide to the culture media for 3-4 h resulted in the disappearance of the GFP fluorescent signal further supporting the local translation of GFP in distal axons (Fig. 3F).
To also assess whether the identified 38 bp fragment represents the minimal functional element for axonal trafficking of the message, we created dGFP vectors that contained the final 22 bp, or 12 bp segments of COXIV 3’UTR, by inserting these cDNA subfragments into CMV promoter-driven dGFP plasmids. Subsequently, we transfected the SCG soma with these vectors, and determined whether these sub-fragments had the capacity to transport dGFP mRNA and promote local protein synthesis in distal axons. As shown in Figure 3G, little fluorescence above background levels was detected in the axons that were transfected in their soma with these vectors, indicating that the terminal 38 bp segment of the 3’UTR represents the minimal axonal transport element of the mRNA. This finding raises the possibility that the putative hairpin-loop formed by this fragment may be crucial for recognition of trans-acting RNA-binding proteins requisite for the axonal trafficking of the COXIV mRNA.
Axonal synthesis of COXIV is important for mitochondrial function and neurite elongation
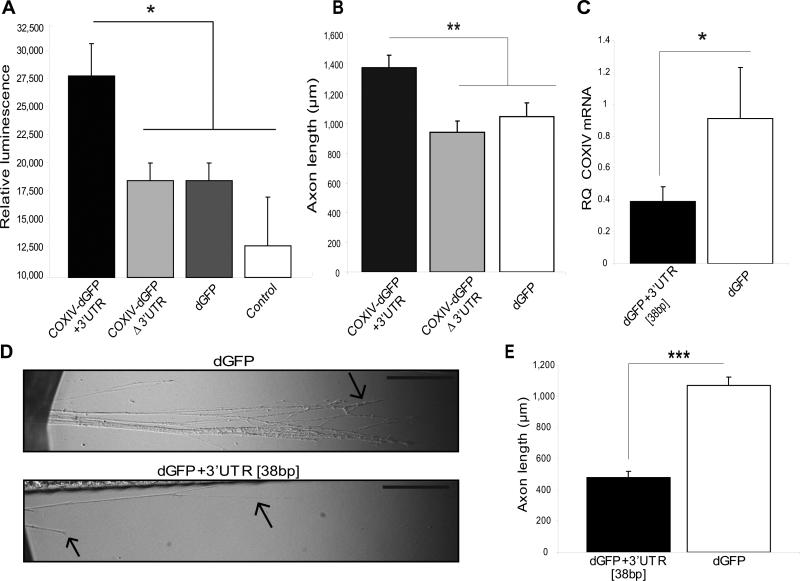
To examine the functional significance of the axonal trafficking of COXIV mRNA, we evaluated the effects of the chimeric COXIV-dGFP mRNAs transport into the distal axons on local ATP levels and neurite elongation. In previous studies, we provided evidence that axonal translation of COXIV had functional importance for the axons by employing siRNAs that silenced local rat COXIV expression, and demonstrated a significant decrease in basal oxygen consumption, and ATP levels in the distal axons (Aschrafi et al., 2008). In the present study, we measured ATP levels in the distal axons, 48 h after transfection of soma with COXIV-dGFP [+3’UTR], COXIV-dGFP [Δ3’UTR], or dGFP constructs. Using a luminometric ATP assay, a marked increase in axonal ATP levels was observed in SCG neurons overexpressing the COXIV-dGFP [+3’UTR] mRNA, as compared to neurons overexpressing COXIV-dGFP [Δ3’UTR] mRNA, and control neurons overexpressing dGFP mRNA, respectively (Fig. 4A). These findings suggested that local overexpression of COXIV-dGFP, due to the presence of functional elements within the COXIV mRNA that enhance axonal localization of the message, significantly enhances mitochondrial function. These data also raise the possibility that the COXIV-dGFP fusion protein escapes rapid, dGFP-mediated degradation upon mitochondrial integration, and retains its biological function.
Fig. 4.
Axonal trafficking and protein synthesis of COXIV mRNA is important for axonal respiration and elongation. (A) A marked increase in axonal ATP levels was observed upon local translation of COXIV-dGFP carrying the COXIV 3’UTR but not the SV40 3’UTR. ATP levels were assessed using CellTiter-Glo luminescent cell viability assay from Promega as previously described (Aschrafi et al., 2007). Values were plotted in arbitrary luminescence units. Data represent mean ±SEM; One-Way ANOVA, *, p<0.01. (B) Axons locally translating COXIV-dGFP mRNA containing the 3’UTR elongate more rapidly when grown in compartmented cultures than axons translating the construct lacking the 3’UTR or dGFP mRNA alone. Axon length is shown for primary SCG neurons that were transfected with the COXIV-dGFP constructs or dGFP 24 h after initial plating in compartmented dishes. Length of distal axons located in the side compartments was measured 2-3 d after the transfection of neurons located in the central compartment. Data are averages ±SEM from measurement of 35-45 axons. ANOVA, **, p < 0.001. (C) Over-expression of dGFP mRNA containing the final 38 bp segment of the COXIV 3’UTR reduces abundance of endogenous COXIV mRNA levels in distal axons. SCG cultures were transfected with dGFP-3’UTR [38 bp], or dGFP (1 μg each) in the center compartment as shown in Fig. 2. Quantitation levels of endogenous COXIV mRNA in the distal axons of SCG neurons were determined by qRT-PCR 24 h after DNA transfection. COXIV mRNA levels are expressed relative to β-actin mRNA. Error bars represent the SEM for n = 3 samples. *, p < 0.05. (D) Overexpression of dGFP mRNA containing the 38bp 3’UTR attenuates neurite outgrowth. SCG neurons were cultured for 1 d and were subsequently transfected with dGFP-3’UTR [38 bp], or dGFP constructs (2 μg each). Arrows indicate location of axonal distal or terminal regions. Scale bars: 10 μm. (E) Neurite extension was measured after 2 d in the lateral sides of compartmentalized culture. Data are averages ±SEM from measurement of 30-40 axons. The experiment was repeated three times with similar results. Student's t test, ***, p < 0.0001.
To examine if the local translation of COXIV mRNA plays an important role in axonal growth, we investigated the requirements of local synthesis of COXIV for normal axonal elongation in rat sympathetic neurons. Using the Campenot compartmentalized cultures, we transfected 3-d-old neuronal cultures in the center compartment with the COXIV-dGFP [+3’UTR], COXIV-dGFP [Δ3’UTR], or dGFP constructs and the axons were allowed to grow into the lateral side compartments. The extension of all neurites entering the lateral side compartments was subsequently measured after 2-4 d, and quantified using Image J. This analysis revealed that cultures which carried out local translation of COXIV-dGFP in their distal axons exhibited a 30-40% increase in length as compared to cultures expressing COXIV-dGFP [Δ3’UTR] mRNAs, and dGFP mRNAs, respectively (Fig. 4B).
To further determine the functional importance of the identified 38 bp 3’ UTR as a putative _cis_-acting element responsible for axonal trafficking of the message, we investigated whether overexpression of the dGFP-3’UTR [38 bp] affected the relative abundance and transport of the endogenous COXIV mRNA in the distal axons. In this experiment, SCG neurons were transfected with the dGFP vectors containing the 38 bp segment, and COXIV mRNA levels in the distal axons were assessed by qRT-PCR 24 h after transfection. As shown in Figure 4C, endogenous levels of COXIV mRNA in distal axons were reduced by approximately 60%, as compared to the axonal COXIV mRNA levels in neurons transfected with the control dGFP vector. Moreover, the reduction of endogenous axonal COXIV mRNA levels by the transfection and overexpression of dGFP-3’UTR [38 bp] resulted in an attenuation of neurite elongation (~50%), suggesting that competitive inhibition of endogenous COXIV mRNA trafficking and a subsequent reduction in local translation from this mRNA affected axonal growth (Fig. 4D, E). This observation further underscores the importance of local translation of COXIV for mitochondrial function and axonal elongation.
Silencing of local COXIV expression reduces axonal elongation rate
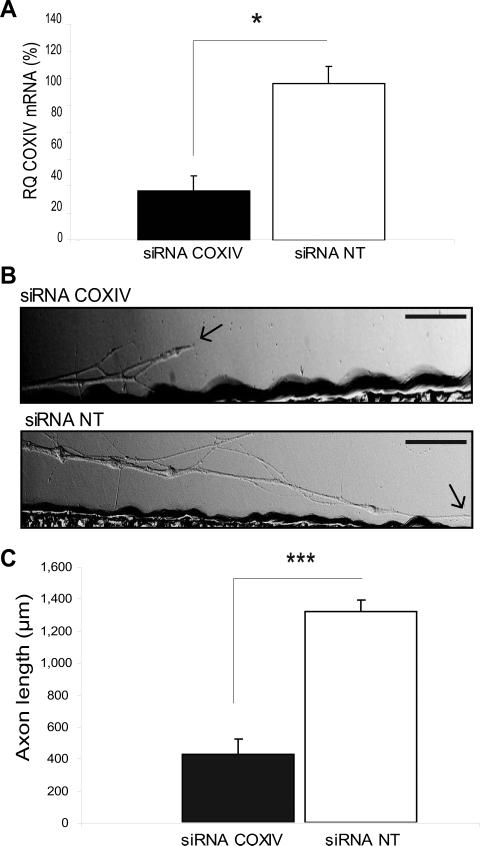
Since overexpression of COXIV levels resulted in a significant increase in axonal elongation rate (Fig. 4B), we investigated the effects of the knock-down of local COXIV expression on axonal growth. Toward this end, siRNAs targeted against COXIV mRNA were lipofected into the 3-d-old axons grown in the lateral compartments of Campenot cultures. qRT-PCR analysis revealed that COXIV mRNA levels were reduced by 80% within 24 h following siRNA transfection, as compared to axons transfected with a non-targeted siRNA (Fig. 5A). In contrast to these findings, the introduction of siRNA into the distal axons had no effect on somal levels of COXIV mRNA and protein 24 h post-transfection (Aschrafi et al., 2008). Additionally, knock-down of the COXIV levels by siRNA resulted in a 75 – 80% reduction in rates of axonal elongation as compared to control axons (Fig. 5B, C). These data suggest that the local translation of COXIV mRNA and its subsequent effects on mitochondrial activity play an important role in facilitating elongation of the axon.
Fig. 5.
siRNA-mediated knockdown of local COXIV levels attenuates neurite elongation in sympathetic neurons. (A) siRNA oligonucleotides (25 nM) targeted against rat COXIV mRNA were introduced into distal axons of SCG neurons by lipofection and COXIV mRNA quantitated 24 h later by qRT-PCR. Quantitation showed a significant reduction in COXIV mRNA levels after the introduction of siRNA (*, p < 0.05, Student's t test). (B) SCGs were cultured for 3 days and neurites located in the lateral compartments of compartmentalized cultures were transfected with siRNA (25 nM) targeted against rat COXIV mRNA, or transfected with non-targeted control siRNA (25 nM). Arrows indicate location of growth cones. Scale bars: 100 μm. (C) Neurite extension was measured 24 h after transfection. Data are averages ±SEM from measurement of 12-24 axons. The experiment was repeated three times with similar results. Student's t test, ***, p < 0.0001.
Discussion
Asymmetric subcellular distribution of RNA is critical for proper embryonic development, establishing differences in cell fate, and for neuronal function (Chartrand et al., 2001). Recent studies suggested a distinct role of local translation of COXIV mRNA in the modulation of mitochondrial activity and axonal function (Aschrafi et al., 2008). In this report, we provide data on the axonal targeting of COXIV mRNA, and explore the functional importance of the trafficking and local translation of this mRNA to the distal axons. The relatively short size of the COXIV 3’UTR, and the high degree of conservation among the genes of mammalian COXIV homologues suggest that this gene segment could provide an attractive model to characterize the _cis_-acting elements that contain information for the targeted delivery of the nuclear-encoded mitochondrial mRNAs to distal axons, as well as the regulation of their local translation.
Previously, on the basis of a sequence analysis (Mfold), the existence of three structural / functional motifs in the 3’UTR of the rat COXIV mRNA was predicted, and subsequent investigations raised the possibility that the enhanced accessibility of the COXIV 3’UTR as a miR-338 binding site was facilitated by the specific folding of the second hairpin-loop within this 3’UTR (see Fig. 1) (Kaplan et al., 2009). The studies presented here suggest that an evolutionarily conserved hairpin-loop structure located in the 3’UTR of the mRNA functioned as the minimal element (38 bp) for mitochondrial association and axonal transport in SCG neurons. When the entire COXIV 3’UTR, or only the final 38 bp segment of this 3’UTR were attached to a dGFP reporter and introduced into the cell bodies of SCG neurons, it elevated GFP levels in the distal segments of axons located in the lateral compartment of Campenot chambers. These results established a RNA-based mechanism to direct the expression of a nuclear-encoded mitochondrial gene transcript to the vicinity of axonal mitochondria in the distal axons. Previously, it was generally accepted that nuclear-encoded proteins destined for mitochondria are translated in the cytoplasm and subsequently transported into the organelles located in the distal parts of cells by complex translocation machinery. Our results, and data provided by others suggest that nuclear mRNAs are targeted to the organelle surface and raise the possibility that the messages are transported via these organelles to the distal axons of SCG neurons (Sylvestre et al., 2003). The local synthesis for mitochondrial protein in distal axons therefore establishes an intimate link between translation and translocation to ensure the viability of the organelle located in these neuronal domains (Gioio et al., 2001; Margeot et al., 2002).
Although previous studies have demonstrated a role of 3’UTR structural motifs in the regulation of mRNA stability (Bevilacqua and Blose, 2008), our results suggest that the identified gene segment within the COXIV 3’UTR elevated reporter gene levels via alteration in mRNA transport rather than molecular stability. The investigations further suggested that the hairpin-loop formation of the identified 38 bp-long transport element may be important for the axonal trafficking and mitochondrial association of the reporter gene, since reducing the length of the gene segment into 22 bp or 12 bp segments resulted in marked reduction in the mRNA transport of the reporter gene. A number of studies have proposed the model that _trans_-acting factors recognize a specific secondary structure in the RNA, often a hairpin–loop structure (Chartrand et al., 1999). This model has proven to be correct for a number of _cis_-acting elements in a variety of organisms (reviewed in Jambhekar et al., 2007). This may also prove the case in the axonal trafficking of nuclear-encoded mitochondrial mRNAs. For example, despite bioinformatics investigation, we were unable to identify a conserved consensus sequence similar to that found in the COXIV 3’UTR in other nuclear-encoded mitochondrial mRNAs. This could be explained by the fact that these “zipcodes” might be recognized by multiple transport complexes. Alternatively, the absence of a consensus transport sequence could be explained by the specific secondary structure of the hairpin–loop present in the COXIV 3’UTR, which may be recognized by RNA binding proteins mediating the axonal transport of multiple nuclear-encoded mitochondrial mRNAs. The analysis of the components of the RNP complexes mediating mitochondrial association and axonal localization of COXIV mRNA is therefore an important future approach to assess the existence of a distinct transport mechanism for the axonal transport of nuclear-encoded mitochondrial mRNAs.
Earlier studies conducted in differentiated P19 cells identified a fragment containing 240 bp of the tau 3’UTR, which was specifically required for axonal targeting (Aronov et al., 2001). The exchange between the 3’UTR of tau mRNA and the 3’UTR of MAP2 mRNA, a dendritically localized message, demonstrated that the subcellular localization of these mRNAs depended on the specific 3’UTR targeting signal. The significant distances of axonal mitochondria from the parental cell bodies might augment the dependence of mitochondria on local translation in distal axons (Mattson et al., 2008). Consistent with this postulate, our studies indicate that local translation of mitochondrial mRNAs may regulate the activity of mitochondria in the axons, and may provide axons with a mechanism to alter the metabolic activity in response to neuronal activity, or altered local environmental conditions. In addition, the local translation of nuclear-encoded mitochondrial mRNAs in distal axons could play a key role in mitochondrial biogenesis to facilitate the response to chronic stress or recovery after injury (Amiri and Hollenbeck, 2008). This hypothesis is supported by our findings that regulation of local translation of COXIV mRNA in distal axons of SCG neurons has marked effects on the rate of axonal elongation.
The intracellular mechanisms underlying the regulation of axonal growth are not well understood, although it has been demonstrated that extracellular factors, such as NGF, can modulate axon growth rates (Campenot, 1994). Recently, Jaffrey and associates have shown that NGF-stimulated outgrowth is protein synthesis dependent (Hengst, et al. 2009). Our findings are in agreement with the aforementioned studies, which demonstrate that in the presence of NGF, overexpression of COXIV in distal axons enhanced axon growth, while its downregulation resulted in an attenuation of axon growth. Previous findings indicated that local regulation of COXIV levels resulted in changes in mitochondrial activity, as judged by alterations in mitochondrial membrane potential and ATP synthesis, and consequently had significant effects of axonal respiration and function. Also, the importance of ATP provided by the mitochondria in axonal growth has been demonstrated earlier: the ATP is required for a number of pivotal processes during outgrowth, including growth cone motility, organelle transport, and cytoskeletal assembly (Bernstein and Bamburg, 2003). Moreover, mitochondria have been shown to be particularly abundant in the region of an active growth cone (Ruthel and Hollenbeck, 2003), and this organelle accumulation was shown to be important for sustained axonal outgrowth. In another line of research, the regulation of mitochondrial gene expression by mitochondrial cyclic AMP response element-binding protein (CREB) was provided (Lee et al., 2005). This finding raises the possibility that decreased mitochondrial CREB activity contributes to the mitochondrial dysfunction and neuronal loss associated with neurodegenerative disorders. In addition, it was shown that actin polymerization and depolymerization at growth cones, consume a significant fraction the total ATP of the neuron (Bernstein and Bamburg, 2003). Our studies suggest that regulation of mitochondrial activity by local translation of nuclear-encoded mRNAs contribute to the mitochondrial effects on axonal elongation, and further underscores the importance of local translation in axonal growth.
Recent studies have proposed a connection between mitochondrial dysfunction and a number of neurological and psychological disorders, including schizophrenia, bipolar disorder, and neurodegenerative disorders, such as amyotrophic lateral sclerosis, Parkinson's and Alzheimer's disease (reviewed in Konradi et al., 2004; Shao et al., 2008). The outcomes of this investigation support the notion of the significance of trafficking and local translation of nuclear-encoded mitochondrial mRNAs particularly in neurons with long axons, and propose a novel mechanism for the maintenance of axonal function and viability, dysregulation of which could ultimately underlie neurodegenerative and / or neurodevelopmental disorders.
In conclusion, in this report we present the first evidence for the existence of a _cis_-acting transport element within a nuclear-encoding mitochondrial gene that is responsible for directing axonal mRNA transport and the association of the mRNA with its target organelle. The studies further suggest that suppression of local COXIV translation in distal axons results in significant attenuation in axon growth. Future work will be designed to delineate the molecular mechanisms underlying the subcellular trafficking and organelle targeting of nuclear-encoded mitochondrial mRNAs.
Experimental Methods
Neuronal cell cultures
SCG were obtained from 3-d-old Harlan Sprague–Dawley rats, and dissociated neurons plated in the center compartment of compartmentalized Campenot culture dishes as previously described (Hillefors et al., 2007). Cells were cultured in serum-free medium containing 20 mM KCl, NGF (50 ng/ml), and 20 U/ml Penicillin/20 μg/ml Streptomycin (Hyclone) for 3-10 days prior to use, with media changes every 3-4 days. The complete culture media, including NGF, was present in both the central and side compartments throughout the culture period and during all experimental procedures. After 2 days in culture, 5-fluoro-2’-deoxyuridine (50 μM) was added to the culture medium to inhibit the growth of non-neuronal cells, and remained in the media for the duration of the experiment. The side compartments, which contained the distal axons used in these experiments, contained no neuronal soma or non-neuronal cells, as judged by phase-contrast microscopy, as well as ethidium bromide and acridine orange staining. To further ensure the purity of the axonal sample, qRT-PCR of axonal RNA were performed to assess for the absence of glial (GFAP) and cell body-specific mRNAs.
dGFP constructs
All constructs used in this study were cloned in frame into pd2eGFP-N1 expression vector (Clontech) and verified by sequence analysis and restriction enzyme digest analysis. The schematic representation of the constructs used in this study is shown in Figure 1_C_. The sense and antisense strands of oligonucleotides coding for the full-length rat COXIV 3’UTR (Refseq ID BC084719) or truncated fragments of the 3’UTR were synthesized (Invitrogen). Oligonucleotides were annealed and ligated into the NotI and XbaI sites following the destabilized eGFP (dGFP) open-reading-frame. In addition, expression constructs encoding fusion proteins of COXIV with dGFP were created by cloning the full length COXIV coding sequence (amplified from rat brain) in frame into the SacI and AgeI sites of the pd2eGFP-N1 vector containing a human cytomegalovirus promoter (Clontech).
Staining of axonal mitochondria
To evaluate the mitochondrial localization of the COXIV fusion proteins, mitochondria in axons or cell bodies of SCG neurons were stained using 25 nM (or 400 nM in in situ hybridization co-staining experiments) reduced MitoTracker red CM-H2XRos (Invitrogen). The reduced probe does not fluoresce until it enters an actively respiring cell, in which case it is oxidized to a fluorescent probe that becomes selectively sequestered in the mitochondria. Following MitoTracker staining for 10 min (20 min for in situ hybridization co-staining experiments), axons were washed twice with PBS followed by fixation using 4% paraformaldehyde in PBS.
Axonal protein synthesis inhibition and image acquisition and analysis
The axons in both side compartments were pre-incubated with the protein synthesis inhibitors emetine or cycloheximide, as described in Hillefors et al. (2007). Images were acquired on a fluorescent microscope (Nikon Eclipse TE300 with Super High Pressure Mercury Lamp power supply) to excite the MitoTracker-stained mitochondria (red fluorescence with excitation at 535 nm and emission at 590 nm), and GFP green fluorescence (excitation at 485 nm and emission at 525 nm). Photomicrographs of the stained axons were collected and saved as RBG images. Co-localization of COXIV-dGFP fusion proteins with the mitochondria were analyzed using Image J.
Analyses of dGFP and COXIV mRNA
COXIV and dGFP mRNA levels were determined by quantitative real-time RT-PCR (qRT-PCR), in total RNA samples prepared from SCG axons using the Cells-to-Signal lysis buffer (Ambion) and gene-specific primers for COXIV and GFP (Qiagen). Total RNA was incubated with RNase-free DNaseI to remove trace plasmid DNA contamination from the side compartments. qRT-PCR analyses were conducted essentially as previously described (Hillefors et al., 2007). The relative levels of each transcript were normalized to β-actin or ribosomal protein S18 mRNA to provide an internal control for reverse transcription and axonal density. RNA values are expressed relative to control by the comparative threshold (CT) method.
In situ hybridization
In situ hybridization for GFP was performed following the protocol described in Janes et al. (2004), with minor modifications. The neurons were washed in pre-warmed medium and in PBS for 5 min, fixed in pre-warmed 4% paraformaldehyde in complete medium for 25 min at 37° C, washed again in PBS and permeabilized in 0.01% Triton-X for 10 min at room temperature. After washing with PBS for 5 min, fluorescein isothiocyanate (FITC)-conjugated locked nucleic acid (LNA) probes (sequence: 5’ TGAAMCTTGTGGCCGTTTACGTC 3’, Exiqon) were hybridized to GFP mRNA overnight at 50° C. The hybridization solution contained 50% formamide, 10% dextran sulfate, 2 × saline sodium citrate (SSC), 10 mM EDTA, 25 mM NaH2PO4 (pH 7.4), 250 ng/μL sheared salmon sperm DNA and 40 nM FITC-conjugated LNA probe. The neurons were washed once in 2×SSC at room temperature, followed by one wash in 2×SSC and one in 0.5×SSC, each for 5 min at 40°C. A scrambled LNA probe was used as a negative control.
Western blot analysis
Distal axons in the side compartments of Campenot chambers or soma and proximal axons in the central compartment were harvested separately and lysed in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, and Complete protease inhibitor cocktail (Roche). Protein concentration was assessed using the micro BCA Assay kit (Pierce). Equal amounts of each lysate were separated on a SDS-polyacrylamide gel. For SDS-PAGE, axonal homogenates were boiled in NuPage sample buffer (Invitrogen) and resolved on 4-12% linear gradient gels or 12% mini-gels. Proteins were transferred to PVDF membranes that were then blocked, probed with antibodies and developed using the ECL Advance kit (GE Health Care). β-actin and GFP antibodies were purchased from Sigma, and Covance, respectively.
Transfection of neurons with siRNAs and dGFP plasmids
Two independent siRNAs targeting rat COXIV were used for silencing in SCG neurons. The siRNA sense and antisense strands were purchased from Dharmacon; the sequences are provided in Aschrafi et al. (2008). The introduction of siRNAs (25 nM final concentration) into the distal axons or the soma and proximal axons of young SCG neurons (3 DIV) was accomplished by lipofection using siPORT NeoFX (Ambion). The Campenot compartmented culture used in the present studies contained two side compartments that harbored the distal axons, which were transfected independently. The dGFP constructs were transfected into the center compartment of the culture dishes (1 DIV for the neurite outgrowth assays and 6-10 DIV for all other investigations) using FuGENE HD Transfection Reagent (Roche). At least three plates were transfected for each condition and all experiments were repeated three times.
Measurement of axonal outgrowth
Images of axons were obtained using an inverted phase-contrast microscope (Nikon). The length of distal axons present in the side compartments of the Campenot cultures were measured 2-3 d post-transfection with the use of an ocular micrometer and the neurite tracing and quantification tool Neuron J, from Image J software (NIH). All axons from each culture dish were measured and a total of at least 35 axons for each condition were used for statistical comparisons.
Bioinformatics and statistical analysis
Constructs and primers used in this report were designed using VectorNTI (Invitrogen). Multiple sequence alignment was performed using ClustalW, and the secondary structure prediction analysis of the COXIV 3’UTR was conducted using Mfold (Zuker, 2003). Quantitative data are presented as the mean ± SEM. Student's t-test was used to determine significant differences between two groups. One-Way ANOVA was employed to analyze differences among multiple groups; p values ≤ 0.05 were considered significant.
Acknowledgements
This work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amiri M, Hollenbeck PJ. Mitochondrial biogenesis in the axons of vertebrate peripheral neurons. Dev. Neurobiol. 2008;68:1348–1361. doi: 10.1002/dneu.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S, Aranda G, Behar L, Ginzburg I. Axonal tau mRNA localization coincides with tau protein in living neuronal cells and depends on axonal targeting signal. J. Neurosci. 2001;21:6577–6587. doi: 10.1523/JNEUROSCI.21-17-06577.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell G, Singer RH. mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J. Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, Blose JM. Structures, kinetics, thermodynamics, and biological functions of RNA hairpins. Annu. Rev. Phys. Chem. 2008;59:79–103. doi: 10.1146/annurev.physchem.59.032607.093743. [DOI] [PubMed] [Google Scholar]
- Campenot RB. NGF and the local control of nerve terminal growth. J. Neurobiol. 1994;25:599–611. doi: 10.1002/neu.480250603. [DOI] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Chartrand P, Singer RH, Long RM. RNP localization and transport in yeast. Annu. Rev. Cell. Dev. Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Belgareh N, Blugeon C, Claros MG, Doye V, Jacq C. Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo. Mol. Microbiol. 1999;31:1499–1511. doi: 10.1046/j.1365-2958.1999.01295.x. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Blugeon C, Jacq C. In yeast, the 3' untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 2000;20:7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J. Neurosci. Res. 2001;64:447–453. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Lavina ZS, Jurkovica D, Zhang H, Eyman M, Giuditta A, Kaplan BB. Nerve terminals of squid photoreceptor neurons contain a heterogeneous population of mRNAs and translate a transfected reporter mRNA. Eur. J. Neurosci. 2004;20:865–872. doi: 10.1111/j.1460-9568.2004.03538.x. [DOI] [PubMed] [Google Scholar]
- Goetze B, Grunewald B, Kiebler MA, Macchi P. Coupling the iron-responsive element to GFP--an inducible system to study translation in a single living cell. Sci. STKE. 2003;2003:L12. doi: 10.1126/stke.2003.204.pl12. [DOI] [PubMed] [Google Scholar]
- Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell. Mol. Neurobiol. 2007;27:701–716. doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes MS, Hanson BJ, Hill DM, Buller GM, Agnew JY, Sherwood SW, Cox WG, Yamagata K, Capaldi RA. Rapid analysis of mitochondrial DNA depletion by fluorescence in situ hybridization and immunocytochemistry, potential strategies for HIV therapeutic monitoring. J. Histochem. Cytochem. 2004;52:1011–1018. doi: 10.1369/jhc.3A6209.2004. [DOI] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. Axonal Protein Synthesis and the Regulation of Local Mitochondrial Function. In: Koenig E, editor. Cell biology of the axon. Springer; Berlin/Heidelberg: 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrmann M, Haubensak W, Hemraj I, Kaether C, Lessmann VJ, Kiebler MA. Fast, convenient, and effective method to transiently transfect primary hippocampal neurons. J. Neurosci. Res. 1999;58:831–835. [PubMed] [Google Scholar]
- Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch. Gen. Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim CH, Simon DK, Aminova LR, Andreyev AY, Kushnareva YE, Murphy AN, Lonze BE, Kim KS, Ginty DD, Ferrante RJ, Ryu H, Ratan RR. Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J. Biol. Chem. 2005;280:40398–40401. doi: 10.1074/jbc.C500140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Hemraj I, Goetze B, Grunewald B, Mallardo M, Kiebler MA. A GFP-based system to uncouple mRNA transport from translation in a single living neuron. Mol. Biol. Cell. 2003;14:1570–1582. doi: 10.1091/mbc.E02-08-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeot A, Blugeon C, Sylvestre J, Vialette S, Jacq C, Corral-Debrinski M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21:6893–6904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites, an overview. J. Neurosci. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites, a cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 1997;17:4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart PH, Taylor WM, Bygrave FL. A procedure for the rapid preparation of mitochondria from rat liver. Biochem J. 1982;204:731–735. doi: 10.1042/bj2040731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A, Russo G, Cuccurese M, Garbi C, Pietropaolo C. The 3'-untranslated region directs ribosomal protein-encoding mRNAs to specific cytoplasmic regions. Biochim. Biophys. Acta. 2006;1763:833–843. doi: 10.1016/j.bbamcr.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Ruthel G, Hollenbeck PJ. Response of mitochondrial traffic to axon determination and differential branch growth. J. Neurosci. 2003;23:8618–8624. doi: 10.1523/JNEUROSCI.23-24-08618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, Jones EG, Bunney WE, Vawter MP. Mitochondrial involvement in psychiatric disorders. Ann. Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre J, Margeot A, Jacq C, Dujardin G, Corral-Debrinski M. The role of the 3' untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol. Biol. Cell. 2003;14:3848–3856. doi: 10.1091/mbc.E03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Jeon NL, Cotman CW. Axonal mRNA in Uninjured and Regenerating Cortical Mammalian Axons. J. Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J. Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenisek D, Matthews G. The role of mitochondria in presynaptic calcium handling at a ribbon synapse. Neuron. 2000;25:229–237. doi: 10.1016/s0896-6273(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]