Respiratory Virus Pneumonia after Hematopoietic Cell Transplantation (HCT): Associations between Viral Load in Bronchoalveolar Lavage Samples, Viral RNA Detection in Serum Samples, and Clinical Outcomes of HCT (original) (raw)
Abstract
Background. Few data exist on respiratory virus quantitation in lower respiratory samples and detection in serum from hematopoietic cell transplant (HCT) recipients with respiratory virus-associated pneumonia.
Methods. We retrospectively identified HCT recipients with respiratory syncytial virus (RSV), parainfluenza virus, influenza virus, metapneumovirus (MPV), and coronavirus (CoV) detected in bronchoalveolar lavage (BAL) samples, and we tested stored BAL and/or serum samples by quantitative polymerase chain reaction.
Results. In 85 BAL samples from 82 patients, median viral loads were as follows: for RSV (_n_=35), 2.6×106 copies/mL; for parainfluenza virus (_n_=35), 4.9×107 copies/mL; for influenza virus (_n_=9), 6.8×105 copies/mL; for MPV (_n_=7), 3.9×107 copies/mL; and for CoV (_n_=4), 1.8×105 copies/mL. Quantitative viral load was not associated with mechanical ventilation or death. Viral RNA was detected in serum samples from 6 of 66 patients: 4 of 41 with RSV pneumonia, 1 with influenza B, and 1 with MPV/influenza A virus/CoV coinfection (influenza A virus and MPV RNA detected). RSV detection in serum was associated with high viral load in BAL samples (_P_=.05), and viral RNA detection in serum was significantly associated with death (adjusted rate ratio, 1.8; _P_=.02).
Conclusion. Quantitative polymerase chain reaction detects high viral loads in BAL samples from HCT recipients with respiratory virus pneumonia. Viral RNA is also detectable in the serum of patients with RSV, influenza, and MPV pneumonia and may correlate with the severity of disease.
Respiratory virus infections are associated with high morbidity and mortality after hematopoietic cell transplantation (HCT). The most common viruses that cause progression from upper to lower respiratory tract disease in HCT recipients are respiratory syncytial virus (RSV), parainfluenza virus (PIV), influenza virus, and human metapneumovirus (MPV), with mortality rates up to 25%–45% within 30 days after progression to pneumonia [1–10]. Recent studies suggest that PIV and RSV, in particular, are associated with the complication of fixed airflow obstruction after HCT, further contributing to transplant-related mortality [11, 12].
Higher viral loads for cytomegalovirus (CMV) and herpes simplex virus in bronchoalveolar lavage (BAL) specimens from immunosuppressed patients have been associated with clinical outcomes, such as severe respiratory illness and death [13, 14]. Quantitative polymerase chain reaction (PCR) analysis of BAL samples from predominantly immunosuppressed patients, including lung transplant recipients, showed that high MPV load was associated with severe pneumonia and complications requiring prolonged hospitalization [15]. Similar studies are lacking for HCT recipients.
In immunocompetent hosts, respiratory virus replication is generally limited to the respiratory epithelium. However, for viruses such as avian influenza A(H5N1) virus, seasonal influenza virus, and severe acute respiratory syndrome-associated coronavirus (CoV), detection of viral RNA by PCR and isolation (of influenza virus [16–21]) from plasma or serum samples has been described; these viruses may cause disseminated infection with replication outside the respiratory tract [16–27]. Notably, among individuals with avian influenza A(H5N1) infection, viral RNA in blood has been detected only in fatal cases and is associated with higher pharyngeal viral loads, indicating possible viral dissemination associated with poor prognosis [16, 17, 27]. Detection of other viruses, such as RSV and rhinovirus, in serum, whole blood, and peripheral blood mononuclear cell samples by PCR and culture has rarely been described in neonates and children, although there is no consistent correlation between virus detection and disease severity [28–32]. Serum DNA viral load in immunocompromised patients with disseminated adenovirus or CMV infection reflects disease activity and can be used to predict severity and monitor response to antiviral treatment [33–39]. The presence of respiratory virus RNA in serum has not been systemically evaluated among HCT recipients.
Methods
Patients and samples. We retrospectively identified a cohort of 104 HCT recipients who experienced 108 episodes of respiratory virus-associated pneumonia within 1 year after HCT. Four patients had 2 distinct episodes of pneumonia caused by different viruses, separated by ⩾1 month. All patients underwent HCT at the Fred Hutchinson Cancer Research Center between 1993 and 2007 and provided written informed consent allowing use of stored specimens and medical records. The study was approved by the center's Institutional Review Board.
All patients eligible for analysis had radiographic and clinical evidence of lower respiratory tract disease confirmed by respiratory virus detection in BAL. BAL samples were obtained from adults (⩾18 years old) by washing with 90–150 mL of sterile, isotonic saline via bronchoscopy; smaller volumes were used for children. Pneumonia was virologically confirmed by testing BAL samples with direct fluorescent antibody, culture, shell vial, or qualitative reverse-transcription PCR (RT-PCR) for RSV, PIV types 1–4, influenza A/B viruses, and rhinoviruses (or PCR for adenoviruses) and by qualitative RT-PCR alone for MPV and the non-severe acute respiratory syndrome human CoVs (OC43, 229E, HKU1, and NL63). A multiplexed qualitative PCR panel has been available for testing of clinical samples at our center since May 2006. BAL samples were also submitted for routine bacterial, fungal, and acid-fast bacilli cultures and for detection of CMV and herpes simplex virus.
All patients had serum and/or plasma samples prospectively collected weekly for laboratory monitoring during the first 100 days and at varying intervals for up to 1 year after HCT. Although both serum and plasma samples were tested, these will be referred to collectively as serum samples throughout this article. BAL and residual serum samples were stored frozen at -20°C or -70°C. For this analysis, we obtained 1–3 serum samples that corresponded most closely to the date of bronchoscopy. We included patients with either stored BAL or stored serum samples; concomitant stored BAL and serum samples were available from a subset of patients. Stored BAL samples that had previously tested positive for RSV, PIV, influenza A virus/B virus, MPV, or CoV were tested by quantitative RTPCR for the previously detected virus. Stored serum samples were also tested by quantitative RT-PCR for the virus that had been previously detected in BAL samples.
Quantitative RT-PCR assays. Total nucleic acids were isolated from 200 _µ_L of BAL specimens, and quantitative RT-PCR assays were performed using 10 _µ_L of specimen for each reaction [40–43]. Each quantitative assay was linear from 10 to 108 viral copies/reaction, with a 95% limit of detection of 10 copies/reaction (or 1000 copies/mL) [40–43]. A different nucleic acid extraction method (QIAamp Viral RNA Mini kit; Qiagen) was used for serum, which enabled processing of a larger volume (50 _µ_L), providing a sensitivity of 200 copies/mL for a cutoff of 10 copies/reaction. Serum samples were tested by quantitative RT-PCR using the same primers and method described for BAL specimens [40–43]. All PCR methods were performed according to College of American Pathologist standards, and the laboratories passed proficiency testing in viral diagnostics.
Statistical analysis. The Wilcoxon rank-sum test was used to compare quantitative viral loads in BAL samples among patients grouped by clinical characteristics. Cox proportional hazards regression was used to analyze overall survival at 1 and 6 months by quantitative viral loads in the first BAL sample. The model was adjusted for potential confounders, including age, stem cell source (peripheral blood stem cells vs bone marrow and cord blood), presence of lymphopenia (⩽100 lymphocytes/ _µ_L ⩽7 days before collection of BAL fluid), mechanical ventilation at or within 30 days after BAL, and the presence or absence of copathogens. Copathogens were defined as pathogenic bacteria, fungi, or opportunistic viruses from the same BAL sample or a concomitant lung biopsy sample. For these evaluations, each patient was represented once using the first viral load available from the first episode of pneumonia.
Characteristics of patients with or without viral RNA detected in serum were compared using the Wilcoxon rank-sum test for nonparametric continuous data and the Fisher exact test for categorical variables. Poisson regression with robust standard error estimates was used to calculate the prevalence rate ratio (RR) for clinical outcomes among HCT recipients with or without detection of viral RNA [44]. The models were adjusted for stem cell source, presence of lymphopenia, and interval between HCT and BAL sample [4–6]. Age, donor type (HLA-identical sibling vs alternate donors), sex, conditioning regimen (nonmyeloablative vs myeloablative), CMV risk group (donor and recipient seronegative vs other), underlying disease risk, presence of acute grade 2–4 graft-vs-host disease at or before diagnosis of pneumonia, presence or absence of copathogens, and year of transplant were also considered potential confounders. These confounders were included in the multivariate model if they altered the adjusted RR for the outcomes of interest by ⩾10%. For RSV pneumonia, the model was limited to 1 confounder.
Differences were considered statistically significant at _P_>.05 (2-sided). No adjustments were made for multiple comparisons.
Results
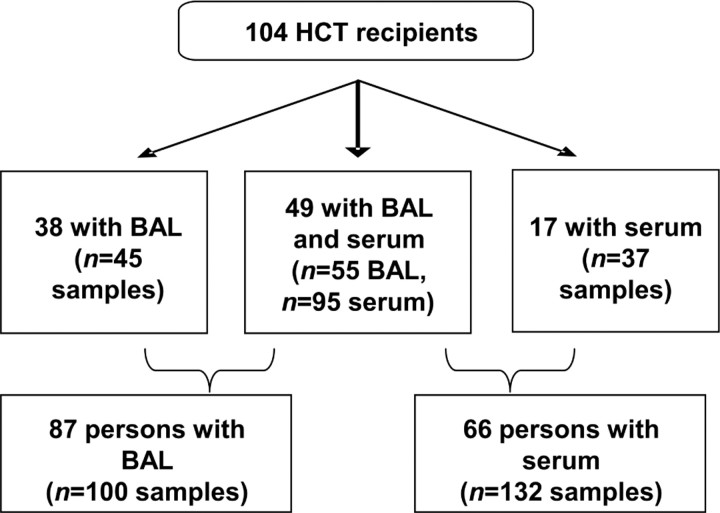
Of 104 HCT recipients, 87 had 100 stored BAL samples available for testing (Figure 1). Thirty-eight patients had 45 stored BAL samples without serum samples available, and 17 patients had 37 stored serum samples without BAL samples. Forty-nine patients had 55 concomitant BAL and 95 concomitant serum samples. Serum samples were collected a median of 1 day after BAL samples (interquartile range, 3 days before to 4 days after BAL collection).
Figure 1.
Specimens available for testing from 104 hematopoietic cell transplant (HCT) recipients. BAL, bronchoalveolar lavage.
Study cohort characteristics, grouped by type of samples available, are shown in Table 1. Most patients had lymphopenia in the week before the diagnosis of pneumonia. The groups differed with respect to the proportion of pneumonia episodes accompanied by copathogens. Of 27 patients with pulmonary copathogens, 11 had >1 type. Aspergillus was most common, with 9 infections confirmed as Aspergillus fumigatus. Twelve patients had CMV pneumonia, and 10 had coexisting bacterial pathogens (including Pseudomonas aeruginosa in 5 patients, Streptococcus pneumoniae in 3, Staphylococcus aureus in 2, coliforms in 3, acinetobacter in 1, and nocardia in 1). Other copathogens included Candida species in 2 patients, Mycobacterium fortuitum in 1, and Rhizopus species in 1.
Quantitative respiratory virus detection in BAL fluid. Five BAL samples from 5 HCT recipients who had previously tested positive were negative on retesting (RSV in 2 samples, influenza A virus in 2, and MPV in 1); that is, viral RNA was below the limit of detection by quantitative RT-PCR. These samples were collected in 1990, 1999, and 2006 (3 samples) and had previously been positive by culture (RSV [2 samples]), direct fluorescent antibody (influenza A virus [2 samples] and RSV), and RT-PCR (MPV). We analyzed quantitative viral load in association with the date of sample collection and found no correlation between viral load and timing within the study period to suggest that sample degradation may have consistently contributed to lack of RNA amplification (Pearson coefficient, 0.08). Further analyses were performed using the 95 amplifiable BAL samples from 82 HCT recipients.
Three patients had 2 separate episodes of pneumonia, and 9 patients had >1 BAL sample per infection. Using the BAL sample with maximum quantitative viral load for each respiratory virus per episode of pneumonia (85 BAL samples), the median respiratory virus copy number for each was as follows: for RSV (_n_=35), 2.6×106 copies/mL (range, 1.5×103 to 1.0×109); for PIV (_n_35), 4.9×107 copies/mL (range, 2.7×103 to 1.1×109); for influenza virus (_n_9), 6.8×105 copies/mL (range, 7.4×103 to 8.3×108); for MPV (_n_7), 3.9×107 copies/mL (range, 2.9×104 to 2.8×108); and for CoV (_n_4), 1.8×105 copies/mL (range, 2.5×103 to 2.0×107) (Figure 2A).
Figure 2.
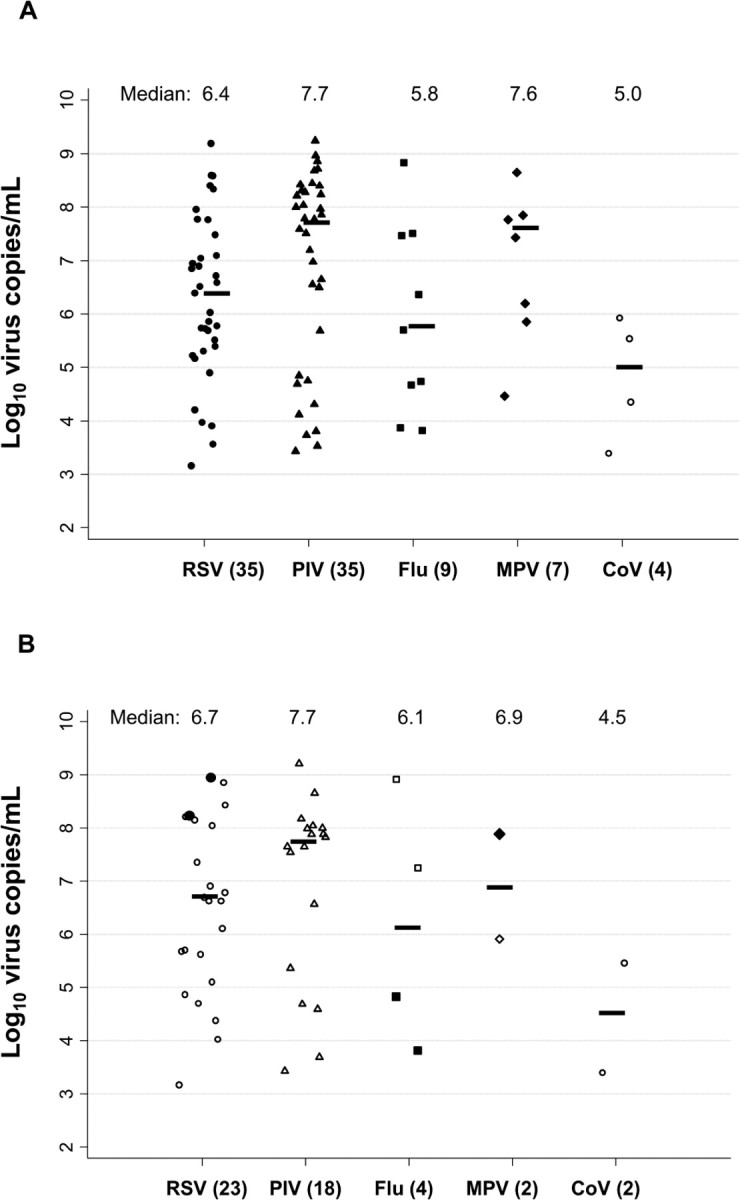
Respiratory virus-specific quantitative viral loads in bronchoalveolar lavage (BAL) fluid. The BAL sample with the maximum viral load per pneumonia episode is shown and was used to calculate the median value for each virus. A, Eighty-five BAL samples from 82 hematopoietic cell transplant (HCT) recipients (the total for viruses is 90 because 3 samples included multiple viruses). B, Forty-nine BAL samples from 44 HCT recipients with corresponding BAL and serum samples (the total for viruses is 49 because 2 samples included multiple viruses and 2 patients had 2 episodes of pneumonia). Black markers (2 for respiratory syncytial virus [RSV], 2 for influenza virus [Flu], and 1 for metapneumovirus [MPV]) indicate that viral RNA was detected in the corresponding serum sample; white markers indicate that the serum sample was negative for the same respiratory virus. Parenthetical values on the horizontal axis indicate the number of positive samples for each virus; medians are shown at the top of the graph. Horizontal bars represent the median value for each virus. PIV, parainfluenza virus; CoV, coronavirus.
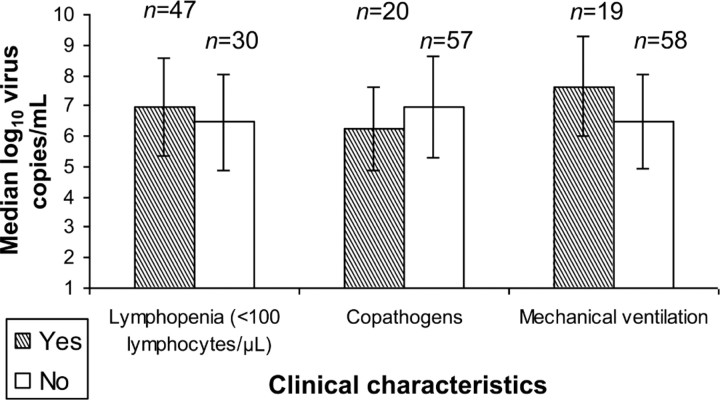
Quantitative respiratory virus detection in BAL samples and clinical outcomes. For 77 patients, we examined the association of quantitative viral load in BAL samples from first episodes of pneumonia (33 RSV, 29 PIV, 5 MPV, 8 influenza, and 2 CoV) with the presence of lymphopenia, the presence of copathogens, and the need for mechanical ventilation. We excluded 3 patients with multiple respiratory viruses detected in BAL samples and 2 patients for whom the first positive BAL sample was not available. There was no statistically significant difference in median quantitative viral load between patients with and those without lymphopenia (⩽100 lymphocytes/_µ_L ⩽7 days before collection of BAL fluid) or copathogens; there was a trend toward a higher viral load in patients who required mechanical ventilation (_P_=.06) (Figure 3). There was no association between quantitative viral load measured in the first BAL sample and overall survival at 1 or 6 months (data not shown).
Figure 3.
Median quantitative bronchoalveolar lavage (BAL) viral load from the first positive BAL samples associated with respiratory virus pneumonia in 77 patients, in the presence or absence of clinical characteristics.
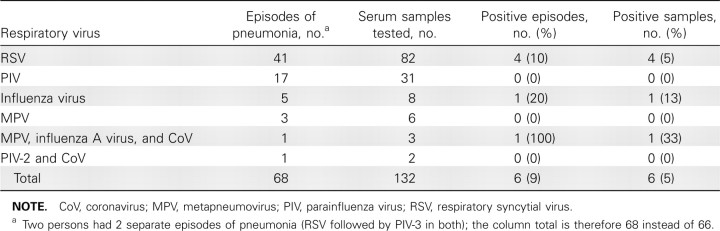
Quantitative respiratory virus detection in serum samples. RT-PCR was used to test 132 serum samples obtained from 66 HCT recipients near the date of diagnostic BAL samples, corresponding to 68 episodes of pneumonia (RSV in 41, PIV in 17, influenza virus in 5, MPV in 3, PIV-2/CoV coinfection in 1, and MPV/influenza A virus/CoV coinfection in 1) (Table 2). Forty-nine patients had concomitant BAL (_n_=55) and serum (_n_=95) samples. Respiratory viral RNA was detected in serum samples from 6 patients: 4 (10%) with RSV pneumonia, 1 with influenza B, and the 1 patient with MPV/influenza A virus/ CoV coinfection (influenza virus and MPV RNA detected).
Table 2.
Quantitative Reverse-Transcription Polymerase Chain Reaction Results for Serum Samples from 66 Hematopoietic Cell Transplant Recipients
The median serum RNA values were as follows: for RSV, 5.3×102 copies/mL (range, 3.0×102 to 1.2×104); for influenza B virus, 3.3×102 copies/mL; for MPV, 7.9×102; and for influenza A virus, 3.7×102 copies/mL. The 6 positive serum samples were collected a median of 1 day after the closest concomitant positive BAL sample (range, 11 days before to 6 days after), similar in timing to the 126 negative serum samples (median, 1 day after BAL sample; range, 16 days before to 19 days after) (_P_=.78).
Figure 2_B_ provides quantitative viral loads for BAL samples corresponding to negative and positive serum samples. For 2 patients with RSV RNA detected in serum, corresponding BAL samples with maximum viral load had 1.7×108 to 1.0×109 RNA copies/mL detected. One patient with influenza B virus RNA detected in serum had 6.7×104 copies/mL in the BAL sample, and the patient with MPV and influenza A virus detected in serum had 7.8×107 and 7.4×103 copies/mL, respectively, in the BAL sample. No viral RNA was detected in the serum samples of 18 patients with PIV pneumonia, even among patients with the highest BAL viral loads.
The association between quantitative RSV load in BAL samples and detection of RSV RNA in serum was analyzed for 23 HCT recipients with RSV pneumonia and quantitative viral loads from concurrent BAL and serum samples. The median maximum BAL RSV load was higher in 2 patients with RSV RNA detected in serum samples than in patients with no RSV RNA detected (5.9×108 vs 3.2×106 copies/mL; _P_=.05).
Detection of viral RNA in serum and clinical outcomes. All 6 patients with viral RNA detected in serum underwent allogeneic bone marrow transplantation with pneumonia and serum RNA detection in the first 120 days after HCT; additional characteristics are listed in Table 3. Serum viral RNA was detected in 3 patients after ⩾1 week of antiviral therapy, aerosolized ribavirin for RSV in 2 patients (patients 3 and 4), and oseltamivir in 1 (patient 6). Positive serum samples were collected within 1–12 days of positive BAL samples. Five of the 6 patients died within 1 week of a positive serum or BAL sample, and viral pneumonia was regarded as the final diagnosis and cause of death. Autopsies were performed for patients 5 and 6; both had diffuse alveolar damage and negative viral culture results, and focal bronchiolitis obliterans organizing pneumonia was also reported for patient 5. No patients with viral RNA detected in serum had bacterial, fungal, or viral copathogens in BAL fluid.
Characteristics from Table 1 were similar for patients with and those without viral RNA detected in serum (data not shown). When the entire population was analyzed, there was no difference between these subgroups in the timing of diagnoses of pneumonia after HCT. However, for the subset of 40 patients with RSV pneumonia after HCT, pneumonia was diagnosed sooner after transplantation in the 4 patients with RSV RNA detected in serum than in those without RSV RNA in serum (median interval, 10 [range, 4–27] vs 48 days [range, 8–237]; _P_=.02).
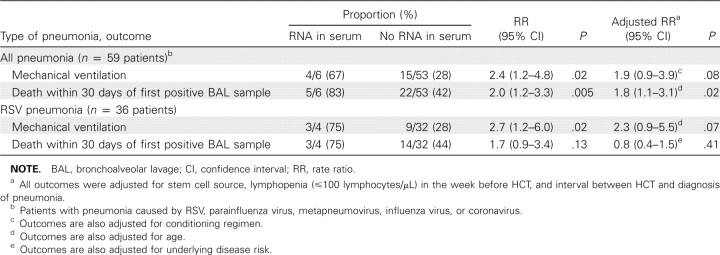
For first episodes of respiratory virus-associated pneumonia, detection of viral RNA in serum was assessed as a risk factor for use of mechanical ventilation or death within 30 days after the first positive BAL sample. This analysis was restricted to the 59 patients who underwent allogeneic transplantation. In univariate analysis, patients with viral RNA detected in serum had an increased risk of mechanical ventilation (RR, 2.4; _P_=.02) and death (RR, 2.0; _P_=.005) within 30 days after the first positive BAL sample; the association with death persisted in an adjusted model (RR, 1.8; _P_=.02) (Table 4).
Table 4.
Outcomes among Allogeneic Hematopoietic Cell Transplant (HCT) Recipients with Post-HCT Pneumonia, With and Without Viral RNA Detection in Serum, Shown Separately for Patients with Pneumonia Due to All Viral Infections and Patients with Respiratory Syncytial Virus (RSV) Pneumonia Alone
Discussion
We have presented the first description of quantitative viral load in BAL and detection of viral RNA in serum samples among HCT recipients with respiratory virus pneumonia. We found high viral loads in BAL samples for all virus types, findings similar to those of another study in which MPV was identified by quantitative RT-PCR in BAL and bronchial wash samples from predominantly immunocompromised patients [15]. We also detected viral RNA in serum among a subset of HCT recipients soon after transplantation with respiratory virus pneumonia caused by RSV, influenza virus, or MPV but not among patients with PIV or CoV pneumonia. Patients with viral RNA detected in serum samples had an increased risk of death, suggesting that RNA detection may correlate with disease severity and poor outcome. Among patients with RSV, RNA detection in serum samples was associated with earlier diagnosis after transplantation.
Detection of respiratory viral RNA in serum samples from these patients may indicate systemic viral dissemination associated with poor prognosis. We did not have access to other specimens to evaluate for the presence or replication of virus in extrapulmonary sites. Pathogenic viral dissemination is just one possible mechanism to explain our findings. Alternative explanations include (1) physical release of intact viral particles into the circulation resulting from high viral loads in the respiratory tract that leads to epithelial cell death and (2) detection of virus or viral RNA from antigen-presenting cells, including pulmonary macrophages and dendritic cells, which gain direct access to the bloodstream during severe infection. It may be that detection of viral RNA in serum is typical in respiratory virus infections among HCT recipients and that use of sensitive RT-PCR assays enabled us to detect this occurrence. Further studies are necessary to investigate whether patients with upper respiratory tract infection alone or with disease that progresses from the upper to the lower respiratory tract may also have respiratory virus RNA detected in serum samples.
Implications of viral RNA detection in serum probably vary for different viruses. Studies of avian influenza virus suggest that detection of viral RNA in serum or plasma is a marker of disease severity and poor outcome [16, 17, 27]. Indeed, in avian influenza virus infection a viremic phase may contribute greatly to pathogenesis. Although we do not have definitive evidence of detection of replication-competent virus, we did find that detection of RNA in serum was associated with an increased risk of death. Among patients with RSV infection, high levels of RNA were detected in concomitant BAL samples at the time of RNA detection in serum samples, suggesting that the amount of infecting virus may influence the likelihood of serum RNA detection. This may indicate that we are detecting RNA that has spilled over because of pulmonary tissue damage. However, for the 2 patients with influenza virus detected in serum, the corresponding BAL viral load was considerably lower, suggesting detection of a true viremic phase. Clinically, complications of influenza include myocarditis and encephalopathy, although it is not clear whether these represent direct viral invasion or an aberrant host immune response [45–48]. These are important questions for investigation, because RT-PCR testing of serum for respiratory viruses may offer an adjunctive method for diagnosis of severe viral respiratory pneumonia and may have important implications for therapy and monitoring in certain patients. Further study is needed to determine whether viruses such as influenza virus and RSV have a true viremic phase associated with the detection of viral RNA in serum; if so, this may indicate that aerosolized therapy has limitations for some HCT recipients with viral pneumonia. Treatment with systemic antivirals may be beneficial for these patients.
The major strength of this study is that it provides the first quantitative analysis of respiratory virus RNA in BAL samples and, to our knowledge, is the largest study conducted to date to evaluate respiratory viral load in BAL and serum samples from HCT recipients. Our stored repository of BAL and serum samples, collected prospectively during the study period, provided a valuable opportunity to evaluate a large quantity of specimens. Importantly, all patients received a standardized diagnostic work-up for pneumonia, including bronchoscopy at the first clinical or radiographic indication of lower respiratory disease. Although detection of viral RNA alone does not ensure that replicating virus is present, nucleic acid detection in serum has been associated with transmission and/or disease severity for both RNA and DNA viruses [24, 27, 35, 38, 49, 50]. PCR testing is often used for prompt, sensitive diagnosis of pneumonia in patients with possible lower respiratory tract disease, particularly in the immunocompromised population. Furthermore, many transplant centers are moving toward molecularbased laboratory techniques for diagnosis of respiratory viruses, so PCR testing of serum may soon be widely available. Our infrequent sampling of serum may have even underestimated the frequency of detection of circulating RNA. More frequent and regular sampling, as well as cell-based assays performed at the time of pneumonia, may increase the frequency of detection of respiratory viral RNA, because others have reported respiratory viral RNA detection in whole blood or peripheral blood mononuclear cell samples but not in serum samples [28, 30– 32].
The retrospective nature of this study is limiting. Varying collection volumes for BAL samples may have influenced quantitative viral results, although we would not expect BAL dilution factors of 1–2-fold to cause major differences in viral load. In a previous study of quantitative testing in nasal wash samples in which collection volumes were recorded, uncorrected viral loads were compared with viral loads corrected by sample volume, and differences in samples were within the range of assay reproducibility (<0.5 log10 viral load) [7]. In this study, we assumed that 1 copy of viral RNA equals 1 virus particle for all viruses studied-that is, that we are detecting genomic RNA. We do not know the proportion of viral genomes that are infectious. If not all genomic copies in a specimen are infectious in vivo, quantifying viral RNA by PCR may overestimate the number of infectious particles. In addition, a small number of BAL samples tested negative by RT-PCR. At times, these samples were stored at -20°C, a temperature not optimal for longterm RNA storage. Thus, it is not surprising that degradation may have occurred and that viral RNA was undetectable in a few samples. We did not see a trend of decreasing quantitative viral load over time to suggest that duration of storage affected RNA amplification. Because we identified only 6 patients with viral RNA detected in concomitant serum samples, our ability to perform more rigorous multivariate analysis of outcomes was limited by sample size.
In conclusion, our analysis found that respiratory viruses may be detected at high virus copy numbers in BAL samples and that detection of viral RNA in serum may be more frequent than previously appreciated among HCT recipients with virologically confirmed respiratory virus pneumonia soon after transplantation. This study provides evidence that detection of respiratory virus RNA in the bloodstream of severely immunocompromised patients may be associated with poor outcome. Circulating respiratory viral RNA may provide some explanation of viral pathogenesis, especially in RSV infection, for which RSV RNA was documented in serum samples of 10% of patients. Larger studies are needed to validate these findings and to determine whether detection of respiratory virus RNA in serum is associated with disease severity in HCT recipients and other severely immunocompromised populations.
Table 1.
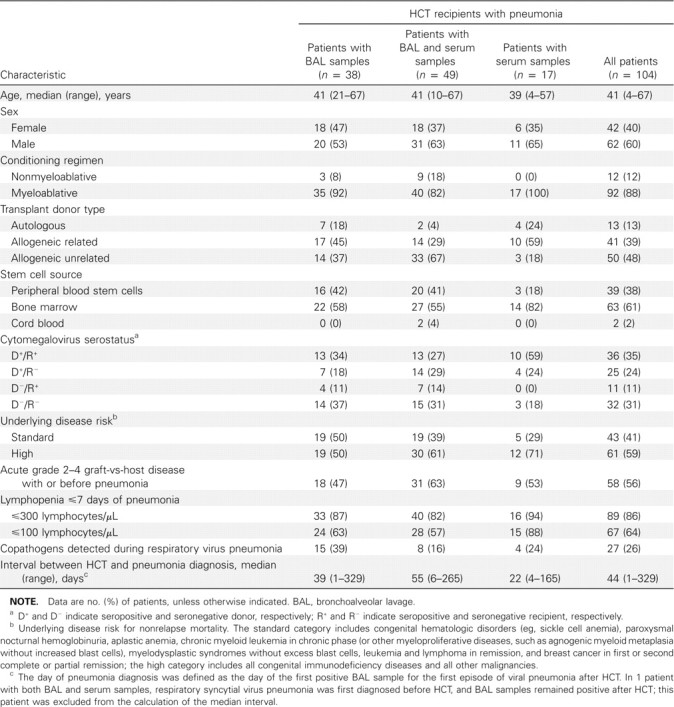
Characteristics of 104 Hematopoietic Cell Transplant (HCT) Recipients with Virologically Confirmed Respiratory Virus Pneumonia, Grouped by Stored Samples Available for Respiratory Virus Testing by Quantitative Reverse-Transcription Polymerase Chain Reaction
Table 3.
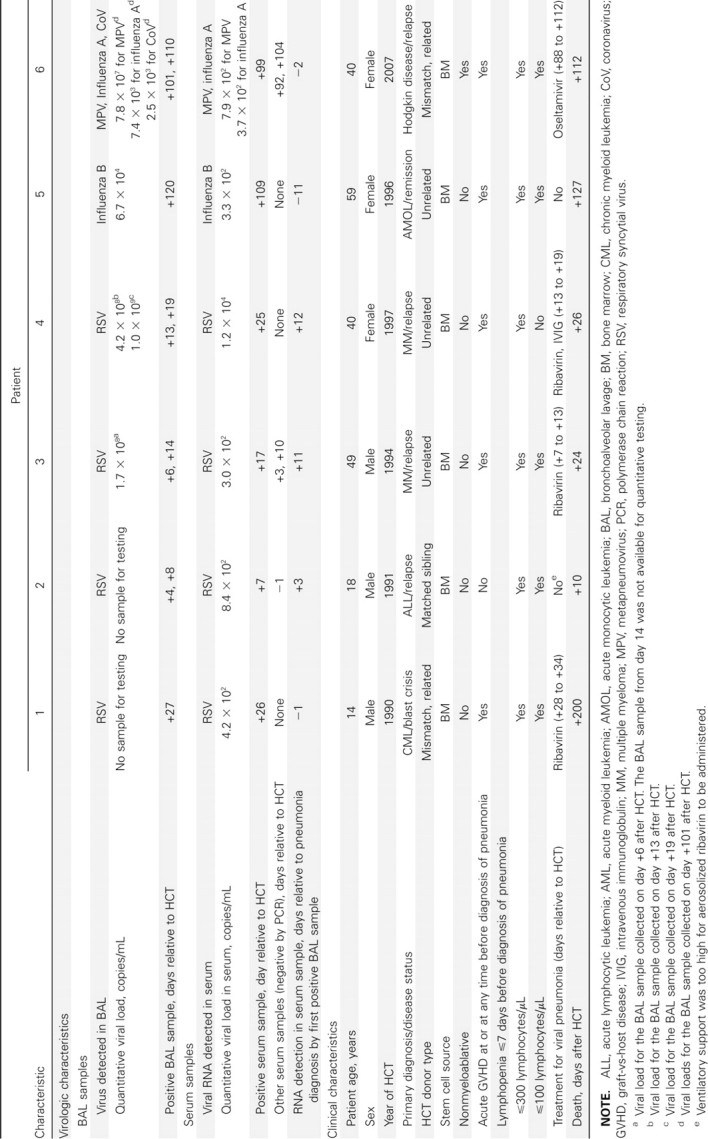
Characteristics of 6 Hematopoietic Cell Transplant (HCT) Recipients with Pneumonia and Respiratory Virus RNA Detected in Serum Samples
Acknowledgments
We thank Amalia Magaret for providing statistical expertise. We also thank Craig Silva, Sanam Hussein, Peter Choe, Nido Nguyen, and George Counts for database services and Anne Cent, Bruce Ulness, Nancy Wright, Terry Stevens-Ayers, Kristen White, Tera Matson, Sam Chatterton-Kirchmeier, Jason Daza, and Vikram Nayani for specimen processing, testing, and laboratory expertise.
Footnotes
Potential conflicts of interest: A.P.C. has received research support from MedImmune. J.A.E. has received research support from Sanofi Pasteur, MedImmune, ADMA Biologics, Adamas Pharmaceuticals, and Novartis. M.B. has received research support from ADMA Biologics, Adamas Pharmaceuticals, and Roche Pharmaceuticals and is a consultant for Roche Pharmaceuticals and Novartis. All other authors report no potential conflicts.
Presented in part: 45th Annual Meeting of the Infectious Diseases Society of America, San Diego, California, 4-7 October 2007 (abstract 764).
Financial support: National Institutes of Health (grants CA18029, HL081595, K23HL091059, L40AI071572, K24HL093294, and K24AI071113); MedImmune (Pediatric Fellowship Grant Award and Pediatric Infectious Diseases Society Fellowship Award to A.P.C.).
References
- 1.Boeckh M, Berrey MM, Bowden RA, Crawford SW, Balsley J, Corey L. Phase 1 evaluation of the respiratory syncytial virus-specific monoclonal antibody palivizumab in recipients of hematopoietic stem cell transplants. J Infect Dis. 2001;184:350–354. doi: 10.1086/322043. [DOI] [PubMed] [Google Scholar]
- 2.Champlin RE, Whimbey E. Community respiratory virus infections in bone marrow transplant recipients: the M.D. Anderson Cancer Center experience. Biol Blood Marrow Transplant. 2001;7(suppl):8S–10S. doi: 10.1053/bbmt.2001.v7.pm11777103. [DOI] [PubMed] [Google Scholar]
- 3.Ljungman P, Gleaves CA, Meyers JD. Respiratory virus infection in immunocompromised patients. Bone Marrow Transplant. 1989;4:35–40. [PubMed] [Google Scholar]
- 4.Martino R, Porras RP, Rabella N, et al. Prospective study of the incidence, clinical features, and outcome of symptomatic upper and lower respiratory tract infections by respiratory viruses in adult recipients of hematopoietic stem cell transplants for hematologic malignancies. Biol Blood Marrow Transplant. 2005;11:781–796. doi: 10.1016/j.bbmt.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. Parainfluenza virus infections after hematopoietic stem cell transplantation: risk factors, response to antiviral therapy, and effect on transplant outcome. Blood. 2001;98:573–578. doi: 10.1182/blood.v98.3.573. [DOI] [PubMed] [Google Scholar]
- 6.Nichols WG, Guthrie KA, Corey L, Boeckh M. Influenza infections after hematopoietic stem cell transplantation: risk factors, mortality, and the effect of antiviral therapy. Clin Infect Dis. 2004;39:1300–1306. doi: 10.1086/425004. [DOI] [PubMed] [Google Scholar]
- 7.Peck AJ, Englund JA, Kuypers J, et al. Respiratory virus infection among hematopoietic cell transplant recipients: evidence for asymptomatic parainfluenza virus infection. Blood. 2007;110:1681–1688. doi: 10.1182/blood-2006-12-060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raboni SM, Nogueira MB, Tsuchiya LR, et al. Respiratory tract viral infections in bone marrow transplant patients. Transplantation. 2003;76:142–146. doi: 10.1097/01.TP.0000072012.26176.58. [DOI] [PubMed] [Google Scholar]
- 9.Roghmann M, Ball K, Erdman D, Lovchik J, Anderson LJ, Edelman R. Active surveillance for respiratory virus infections in adults who have undergone bone marrow and peripheral blood stem cell transplantation. Bone Marrow Transplant. 2003;32:1085–1088. doi: 10.1038/sj.bmt.1704257. [DOI] [PubMed] [Google Scholar]
- 10.Wendt CH, Weisdorf DJ, Jordan MC, Balfour HH, Jr, Hertz MI. Parainfluenza virus respiratory infection after bone marrowtransplantation. N Engl J Med. 1992;326:921–926. doi: 10.1056/NEJM199204023261404. [DOI] [PubMed] [Google Scholar]
- 11.Chien JW, Martin PJ, Gooley TA, et al. Airflow obstruction after myeloablative allogeneic hematopoietic stem cell transplantation. Am J Respir Crit Care Med. 2003;168:208–214. doi: 10.1164/rccm.200212-1468OC. [DOI] [PubMed] [Google Scholar]
- 12.Erard V, Chien JW, Kim HW, et al. Airflow decline after myeloablative allogeneic hematopoietic cell transplantation: the role of community respiratory viruses. J Infect Dis. 2006;193:1619–1625. doi: 10.1086/504268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chemaly RF, Yen-Lieberman B, Chapman J, et al. Clinical utility of cytomegalovirus viral load in bronchoalveolar lavage in lung transplant recipients. Am J Transplant. 2005;5:544–548. doi: 10.1111/j.1600-6143.2005.00747.x. [DOI] [PubMed] [Google Scholar]
- 14.Gooskens J, Templeton KE, Claas EC, van Bussel MJ, Smit VT, Kroes AC. Quantitative detection of herpes simplex virus DNA in the lower respiratory tract. J Med Virol. 2007;79:597–604. doi: 10.1002/jmv.20861. [DOI] [PubMed] [Google Scholar]
- 15.Sumino KC, Agapov E, Pierce RA, et al. Detection of severe human metapneumovirus infection by real-time polymerase chain reaction and histopathological assessment. J Infect Dis. 2005;192:1052–1060. doi: 10.1086/432728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchy P, Mardy S, Vong S, et al. Influenza A/H5N1 virus infection in humans in Cambodia. J Clin Virol. 2007;39:164–168. doi: 10.1016/j.jcv.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Chutinimitkul S, Bhattarakosol P, risuratanon S, et al. H5N1 influenza A virus and infected human plasma. Emerg Infect Dis. 2006;12:1041–1043. doi: 10.3201/eid1206.060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong MD, Bach VC, Phan TQ, et al. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. N Engl J Med. 2005;352:686–691. doi: 10.1056/NEJMoa044307. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann NI, Gust ID. Viraemia in influenza: a report of two cases. Med J Aust. 1971;2:1166–1169. doi: 10.5694/j.1326-5377.1971.tb92768.x. [DOI] [PubMed] [Google Scholar]
- 20.Naficy K. Human influenza infection with proved viremia: report of a case. N Engl J Med. 1963;269:964–966. doi: 10.1056/NEJM196310312691807. [DOI] [PubMed] [Google Scholar]
- 21.Roberts GT, Roberts JT. Postsplenectomy sepsis due to influenzal viremia and pneumococcemia. Can Med Assoc J. 1976;115:435–437. [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WK, Fang CT, Chen HL, et al. Detection of severe acute respiratory syndrome coronavirus RNA in plasma during the course of infection. J Clin Microbiol. 2005;43:962–965. doi: 10.1128/JCM.43.2.962-965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steininger C, olzmann H, Zwiauer KF, Popow-Kraupp T. Influenza A virus infection and cardiac arrhythmia during the neonatal period. Scand J Infect Dis. 2002;34:782–784. doi: 10.1080/00365540260348653. [DOI] [PubMed] [Google Scholar]
- 24.Ng EK, Hui DS, Chan KC, et al. Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin Chem. 2003;49:1976–1980. doi: 10.1373/clinchem.2003.024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori I, Nagafuji H, Matsumoto K, Kimura Y. Use of the polymerase chain reaction for demonstration of influenza virus dissemination in children. Clin Infect Dis. 1997;24:736–737. [PubMed] [Google Scholar]
- 26.Likos AM, Kelvin DJ, Cameron CM, Rowe T, Kuehnert MJ, Norris PJ. Influenza viremia and the potential for blood-borne transmission. Transfusion. 2007;47:1080–1088. doi: 10.1111/j.1537-2995.2007.01264.x. [DOI] [PubMed] [Google Scholar]
- 27.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yui I, Hoshi A, Shigeta Y, akami T, Nakayama T. Detection of human respiratory syncytial virus sequences in peripheral blood mononuclear cells. J Med Virol. 2003;70:481–489. doi: 10.1002/jmv.10421. [DOI] [PubMed] [Google Scholar]
- 29.Rohwedder A, Keminer O, Forster J, Schneider K, Schneider E, Werchau H. Detection of respiratory syncytial virus RNA in blood of neonates by polymerase chain reaction. J Med Virol. 1998;54:320–327. doi: 10.1002/(sici)1096-9071(199804)54:4<320::aid-jmv13>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 30.O'Donnell DR, McGarvey MJ, Tully JM, Balfour-Lynn IM, Openshaw PJ. Respiratory syncytial virus RNA in cells from the peripheral blood during acute infection. J Pediatr. 1998;133:272–274. doi: 10.1016/s0022-3476(98)70234-3. [DOI] [PubMed] [Google Scholar]
- 31.Iankevich OD, Dreizin RS, Makhlinovskaia NL, Gorodnitskaia NA. Viremia in respiratory syncytial virus infection [in Russian]. Vopr Virusol. 1975 Jul-Aug;:455–458. [PubMed] [Google Scholar]
- 32.Xatzipsalti M, Kyrana S, Tsolia M, et al. Rhinovirus viremia in children with respiratory infections. Am J Respir Crit Care Med. 2005;172:1037–1040. doi: 10.1164/rccm.200502-315OC. [DOI] [PubMed] [Google Scholar]
- 33.Anderson EJ, Guzman-Cottrill JA, Kletzel M, et al. High-risk adenovirus-infected pediatric allogeneic hematopoietic progenitor cell transplant recipients and preemptive cidofovir therapy. Pediatr Transplant. 2008;12:219–227. doi: 10.1111/j.1399-3046.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- 34.Echavarria M, Forman M, van Tol MJ, Vossen JM, Charache P, Kroes AC. Prediction of severe disseminated adenovirus infection by serum PCR. Lancet. 2001;358:384–385. doi: 10.1016/S0140-6736(01)05580-5. [DOI] [PubMed] [Google Scholar]
- 35.Erard V, Huang ML, Ferrenberg J, et al. Quantitative real-time polymerase chain reaction for detection of adenovirus after T cell-replete hematopoietic cell transplantation: viral load as a marker for invasive disease. Clin Infect Dis. 2007;45:958–965. doi: 10.1086/521851. [DOI] [PubMed] [Google Scholar]
- 36.Gimeno C, Solano C, Latorre JC, et al. Quantification ofDNAin plasma by an automated real-time PCR assay (cytomegalovirus PCR kit) for surveillance of active cytomegalovirus infection and guidance of preemptive therapy for allogeneic hematopoietic stem cell transplant recipients. J Clin Microbiol. 2008;46:3311–3318. doi: 10.1128/JCM.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustafson I, Lindblom A, Yun Z, et al. Quantification of adenovirus DNA in unrelated donor hematopoietic stem cell transplant recipients. J Clin Virol. 2008;43:79–85. doi: 10.1016/j.jcv.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Ljungman P, Perez-Bercoff L, onsson J, et al. Risk factors for the development of cytomegalovirus disease after allogeneic stem cell transplantation. Haematologica. 2006;91:78–83. [PubMed] [Google Scholar]
- 39.Tanaka Y, Kanda Y, Kami M, et al. Monitoring cytomegalovirus infection by antigenemia assay and two distinct plasma real-time PCR methods after hematopoietic stem cell transplantation. Bone Marrow Transplant. 2002;30:315–319. doi: 10.1038/sj.bmt.1703661. [DOI] [PubMed] [Google Scholar]
- 40.Kuypers J, Campbell AP, Cent A, Corey L, Boeckh M. Comparison of conventional and molecular detection of respiratory viruses in hematopoietic cell transplant recipients. Transpl Infect Dis. 2009;11:298–303. doi: 10.1111/j.1399-3062.2009.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics. 2007;119:e70–e76. doi: 10.1542/peds.2006-1406. [DOI] [PubMed] [Google Scholar]
- 42.Kuypers J, Wright N, Corey L, Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RTPCR. J Clin Virol. 2005;33:299–305. doi: 10.1016/j.jcv.2004.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuypers J, Wright N, Ferrenberg J, et al. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol. 2006;44:2382–2388. doi: 10.1128/JCM.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lumley T, Kronmal R, Ma S. Relative risk regression in medical research: models, contrasts, estimators, and algorithms. [Accessed 18 March 2010];University of Washington Biostatistics Working Paper Series. 2006 http://www.bepress.com/uwbiostat/paper293. [Google Scholar]
- 45.Ray CG, Icenogle TB, Minnich LL, Copeland JG, Grogan TM. The use of intravenous ribavirin to treat influenza virus-associated acute myocarditis. J Infect Dis. 1989;159:829–836. doi: 10.1093/infdis/159.5.829. [DOI] [PubMed] [Google Scholar]
- 46.Kuiken T, Taubenberger JK. Pathology of human influenza revisited. Vaccine. 2008;26(suppl 4):D59–D66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guarner J, Paddock CD, Shieh WJ, et al. Histopathologic and immunohistochemical features of fatal influenza virus infection in children during the 2003–2004 season. Clin Infect Dis. 2006;43:132–140. doi: 10.1086/505122. [DOI] [PubMed] [Google Scholar]
- 48.Fujimoto S, Kobayashi M, Uemura O, et al. PCR on cerebrospinal fluid to show influenza-associated acute encephalopathy or encephalitis. Lancet. 1998;352:873–875. doi: 10.1016/S0140-6736(98)12449-2. [DOI] [PubMed] [Google Scholar]
- 49.Modjarrad K, Chamot E, Vermund SH. Impact of small reductions in plasma HIV RNA levels on the risk of heterosexual transmission and disease progression. AIDS. 2008;22:2179–2185. doi: 10.1097/QAD.0b013e328312c756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spector SA, Hsia K, Crager M, Pilcher M, Cabral S, Stempien MJ. Cytomegalovirus (CMV) DNA load is an independent predictor of CMV disease and survival in advanced AIDS. J Virol. 1999;73:7027–7030. doi: 10.1128/jvi.73.8.7027-7030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]