Architecture of a nascent viral fusion pore (original) (raw)
Abstract
Enveloped viruses use specialized protein machinery to fuse the viral membrane with that of the host cell during cell invasion. In influenza virus, hundreds of copies of the haemagglutinin (HA) fusion glycoprotein project from the virus surface. Despite intensive study of HA and its fusion activity, the protein's modus operandi in manipulating viral and target membranes to catalyse their fusion is poorly understood. Here, the three-dimensional architecture of influenza virus–liposome complexes at pH 5.5 was investigated by electron cryo-tomography. Tomographic reconstructions show that early stages of membrane remodeling take place in a target membrane-centric manner, progressing from punctate dimples, to the formation of a pinched liposomal funnel that may impinge on the apparently unperturbed viral envelope. The results suggest that the M1 matrix layer serves as an endoskeleton for the virus and a foundation for HA during membrane fusion. Fluorescence spectroscopy monitoring fusion between liposomes and virions shows that leakage of liposome contents takes place more rapidly than lipid mixing at pH 5.5. The relation of ‘leaky' fusion to the observed prefusion structures is discussed.
Keywords: electron cryo-tomography, haemagglutinin, influenza virus, matrix, membrane fusion
Introduction
Membrane fusion is a fundamental biological process that lies at the heart of enveloped virus infection, synaptic signaling, intracellular vesicle trafficking, gamete fertilization, and cell–cell fusion. Despite intensive study, we have a limited mechanistic understanding of how fusion protein machinery manipulates lipid membranes to induce their fusion. The haemagglutinin (HA) envelope glycoprotein mediates influenza virus membrane fusion and hence has a key function in host cell invasion by this major human pathogen. It is the archetypal class-I fusion protein and shares core architectural elements with fusogens in retroviruses (e.g. HIV), filoviruses (e.g. ebola virus), coronaviruses (e.g. SARS virus), and paramyxoviruses (e.g. respiratory syncytial virus, measles virus) (Hughson, 1997; Lamb and Jardetzky, 2007).
HA ‘spikes' project outward from the viral envelope and become fusion active when exposed to the acidic pH found in endosomes. Activated proteins grab hold of the host membrane, and in concert with an energy-releasing conformational change, are able to draw the host (target) and viral membranes into apposition and induce them to fuse. To date, the organization of lipid and protein in the virus–target membrane complexes during fusion has eluded structural characterization. Numerous models of membrane deformation leading to fusion have been proposed (reviewed, for example, in Chernomordik and Kozlov, 2003, 2008; Tamm et al, 2003; Martens and McMahon, 2008). The majority of models describing HA-mediated membrane fusion focus on the properties of a hemifusion stalk in which the outer leaflets of the two previously distinct lipid bilayers have joined, the inner leaflets remain separate, and aqueous contents have not mixed. The existence of a hemifused state is supported by fluorescence and electrophysiology assays (reviewed in Chernomordik and Kozlov, 2003, 2005). The events leading up to and following the formation of the hemifusion stalk are less understood. It has been suggested that HA induces the formation of highly curved dimples in the target membrane to initiate the fusion process (Kuzmin et al, 2001; Efrat et al, 2007). Alternatively, some have proposed that the fusion peptides act primarily on the viral membrane, generating local bending and protrusions directed towards the target membrane (Kozlov and Chernomordik, 1998). Computational simulations have yet to resolve pre-stalk fusion intermediates including dimples. In the absence of structural characterization, our understanding of the mechanisms of membrane fusion is likely to remain nebulous.
The stoichiometry and organization of HA spikes at fusion loci is likewise unresolved. Several studies of inter-HA spike cooperativity have indirectly estimated the number of fusion proteins required to induce fusion, with figures ranging from one spike to several (Ellens et al, 1990; Clague et al, 1991; Blumenthal et al, 1996; Danieli et al, 1996; Gunther-Ausborn et al, 2000; Mittal et al, 2002; Imai et al, 2006); however, fusion proteins involved in virus–target membrane contact sites have not been directly imaged.
Despite significant gaps in our understanding of the mechanics of protein-mediated membrane fusion, much is known about the fusion proteins themselves (Skehel and Wiley, 2000; Eckert and Kim, 2001). Influenza HA is expressed as a single precursor polypeptide called HA0. Each 225 kDa HA spike is a homotrimer of HA0 subunits. Host proteases cleave the HA0 into two disulfide-bonded subunits, HA1 and HA2 (reviewed in Steinhauer, 1999). Cleavage is necessary for rendering HA fusion capable and thus for infectivity (Klenk et al, 1975; Lazarowitz and Choppin, 1975). HA1 contains the sialic acid receptor-binding site as well as the majority of antigenic features for influenza, whereas HA2 carries out membrane fusion. HA is bound to the viral envelope by a transmembrane anchor on the C-terminal tail of HA2; the N-terminus of HA2 generated by proteolysis of HA0 is the fusion peptide itself. Between 300 and 500 trimeric, HA spikes are arrayed on the surface of each virus (Inglis et al, 1976; Ruigrok et al, 1984; Harris et al, 2006). After attachment to the cell surface, influenza virus enters the cell by endocytosis (Figure 1). Endosomal maturation and the resulting decrease in endocytic pH trigger a multi-step, ‘spring-loaded' conformational change of HA to an extended coiled-coil conformation that grapples the viral and target membranes together and induces them to fuse (Skehel et al, 1982; Carr and Kim, 1993; Gruenke et al, 2002). X-ray crystal structures of influenza HA have elucidated the protein machinery's detailed structure in the neutral pH and post-fusion conformations, and in the precursor HA0 form (Wilson et al, 1981; Bullough et al, 1994; Chen et al, 1998). Fusion between the viral and endosomal membranes takes place optimally at pH 5 in late endosomes (Maeda and Ohnishi, 1980; Matlin et al, 1981; White et al, 1982; Puri et al, 1990; Krumbiegel et al, 1994; Korte et al, 1999; Lakadamyali et al, 2003); however, prefusion intermediates are populated at the pH values between 5.5 and 6.0 found in early and maturing endosomes (Doms et al, 1985; White and Wilson, 1987; Stegmann et al, 1990; Korte et al, 1999).
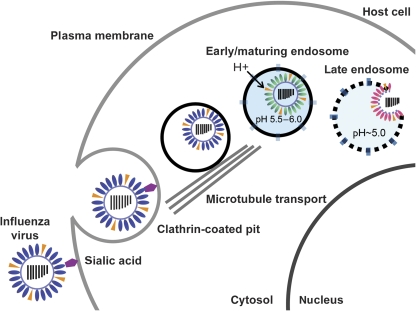
Figure 1.
Influenza A virus, an enveloped, negative-sense RNA virus, enters cells by endocytosis. Endocytic maturation exposes the virus to stages of acidification. Fusion of viral and endosomal membranes occurs most rapidly at pH 5; however, studies have shown that the HA fusion protein becomes active at pH 5.5–6 to begin the process of membrane remodeling that ultimately leads to fusion.
The earliest stage of conformational change at intermediate endosomal pH values involves release of the amphipathic, membrane-active fusion peptide from its sequestered position in the interior of the HA spike at neutral pH (White and Wilson, 1987; Stegmann et al, 1990). HA1 globular head domain dissociation is not required for fusion-peptide deployment, however, it is required for the refolding of HA2 into the TBHA2 extended coiled-coil conformation (Godley et al, 1992). The early conformational intermediates may dock the target membrane and be responsible for priming the membranes for fusion by inducing critical initial deformations.
Influenza virus entry and ultrastructure have been investigated by electron microscopy (EM) in a number of seminal studies (Matlin et al, 1981; Ruigrok et al, 1986; Fujiyoshi et al, 1994; Wharton et al, 1995; Kanaseki et al, 1997; Shangguan et al, 1998; Harris et al, 2006; Noda et al, 2006). These have provided a glimpse of the virus's detailed construction, the general nature of changes exhibited by the virus in response to fusion activation, and its trafficking into cells. This study is the first, however, to provide a detailed three-dimensional characterization of fusion intermediates in this intensively studied system. Here, electron cryo-tomography (ECT) was used to image the three-dimensional architecture of loci formed between authentic X31 influenza virions and liposomal membranes under fusogenic conditions. Flash freezing of unstained biological specimens enables transient intermediates to be trapped, and the organization of protein and membranes in fusion intermediates to be determined. ECT builds a three-dimensional image of unfixed biological structures preserved in vitreous ice by imaging the specimen over a range of axial angular tilts (Lucic et al, 2005). Conditions were chosen to recreate the fusogenic interactions between virus and target membrane that are present at the early and maturing endosomal stage of cell invasion (pH∼5.5–6.0), with the goal of imaging nascent fusion loci at the earliest stages of membrane remodeling. In parallel with the ECT studies, time-resolved fluorescence measurements at pH 5.5 and 5.0 were performed to validate that the liposomes are a reasonable facsimile of a target membrane and verify that the virions were capable of fusing with the liposomes.
Results
Fluorescence spectroscopy of fusion between DOPC liposomes and virions at acidic pH
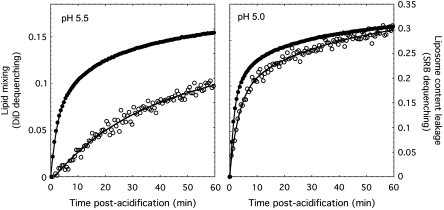
A series of fluorescence measurements at pH 5.0 and 5.5 were performed using 1,2-dioleoyl-_sn_-glycero-3-phosphatidylcholine (DOPC) liposomes that contained the water-soluble fluorophore sulforhodamine-B (SRB) at self-quenching concentrations and the lipophilic fluorophore 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD) incorporated into the bilayer, also at self-quenching concentrations. SRB and DiD have been used previously to monitor liposome permeabilization under acidic conditions (e.g. Lee et al, 1998; Odegard et al, 2009) and influenza fusion respectively (e.g. Lakadamyali et al, 2003). When the doubly labeled liposomes were mixed with unlabeled virus particles at acidic pH, dequenching of both SRB and DiD fluorescence was observed (Figure 2). At pH 5.5 DiD dequenching was a slow process, reaching only 9% after 60 min. Interestingly, SRB fluorescence dequenching took place significantly more rapidly compared with DiD fluorescence changes and reached 30% by 60 min. When the pH of the reaction was 5.0, changes in the two signals were nearly concomitant, and DiD dequenching was also more complete than at pH 5.5 reaching 17% by 60 min post-acidification.
Figure 2.
Fluorescence spectroscopy monitoring lipid exchange and liposomal content transfer and leakage. SRB/DiD-labeled DOPC liposomes were incubated with X31 influenza virions at pH 5.5 and 5.0. Fluorescence dequenching of the water-soluble (SRB, solid circles) and lipophilic (DiD, open circles) dyes was monitored as a function of time. The vertical scales for the two pH conditions are the same. Extent of dequenching was calculated as [F(t)−F(0)]/[F_TX-100−_F(0)], where _F_TX-100 was the fluorescence measured in the presence of 0.1% w/v TritonX-100 detergent. Fitted curves are only meant to guide the eye and are not intended to reflect a particular kinetic model.
The dequenching observed for SRB fluorescence was significantly larger in magnitude than dequenching of the lipidic dye. A reasonable explanation for this is that though some of the dequenching signal may be due to transfer of some SRB to the viral lumen, additional dye leaks out of the liposomes and diffuses into the entire volume of the cuvette. This appears to precede lipid mixing. By contrast, as lipid mixing takes place between virus and liposome, the two-dimensional surface area coverage of DiD only increases modestly (∼50%) as ∼150 nm virus envelopes fuse with membrane of the 100 nm liposomes, leading to a relatively small dequenching signal. Fluorescence experiments were carried out with an excess of liposomes to disfavour disruption of liposomes by multiple virus particles. As a result, the total extent of dequenching of the dyes did not reach 100% (as determined by complete dequenching in the presence of 0.1% TritonX-100) over the time frame examined. No qualitatively different fluorescence kinetics was observed when the amount of DiD incorporated in the DOPC membrane was varied by a factor of 4. A complementary set of experiments were performed in which the DiD fluorophore was incorporated into the virus membrane, and liposomes contained only encapsulated SRB (Supplementary Figure S1). The changes and pH effects closely paralleled the changes observed when the lipophilic DiD dye was in the liposomal bilayer. These data confirm that DOPC liposomes and viruses are capable of merging their membranes, which occurs more efficiently at pH 5.0, and the HA-mediated process is ‘leaky'.
Influenza A virus exhibits a complex envelope and glycoprotein coat
In the electron cryo-tomograms of X31 influenza A virions at pH 5.5, HA spikes are generally well ordered on the surface of the particles and resemble the neutral pH HA structure (Figure 3), although it is not possible to distinguish between the neutral pH structure and a similar fusogenic conformation that has been reported (Wilson et al, 1981; Bottcher et al, 1999). As in a previous ECT study of influenza virus ultrastructure, the HA and neuraminidase surface glycoproteins can often be distinguished (Harris et al, 2006). In most virions a thick (∼8–12 nm) lipoprotein envelope lies beneath the glycoprotein coat and encapsulates the virus's ribonucleoprotein material (the individual segments are unresolved). The envelope's braided appearance results from the compositional mixture of phospholipids, cholesterol, transmembrane protein anchors, M2 ion channels, and associated M1 matrix proteins. Three distinct layers of the envelope can be discerned in volume slices through many of the viruses (Supplementary Figure S2). The two outermost layers exhibit ∼4–6 nm separation, whereas the inner layer, presumably matrix protein, is ∼4–6 nm further below the central layer. Fujiyoshi et al (1994) have proposed that the inner portion of the envelope reflects a matrix protein–lipid composite, rather than a classical phospholipid bilayer leaflet with associated protein. Further study is required to elucidate the detailed construction and stratification of the envelope; however, for the purpose of this study, virus particles that exhibit thick envelopes are inferred to possess an intact matrix layer. Density that extends beneath some glycoprotein spikes and bores completely through the virus envelope to the matrix layer may correspond to the glycoproteins' transmembrane anchors (Figure 3A). A direct association between glycoprotein anchors and matrix protein is consistent with previous reports that have described the importance of this interaction for virus morphology (Enami and Enami, 1996; Jin et al, 1997; Ali et al, 2000). The leaflets of the DOPC liposomes tend to be difficult to resolve under the imaging conditions, although in some tomograms, two leaflets with ∼4 nm spacing are evident (Supplementary Figure S2).
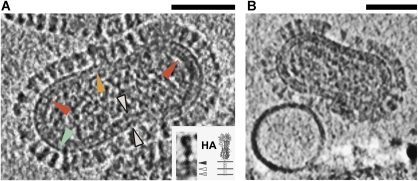
Figure 3.
Electron cryo-tomograms of influenza virions at pH 5.5. (A) 5.3 nm-thick section through the reconstructed tomographic density for an X31 influenza A virion at pH 5.5 after 5 min acidification. The glycoprotein fringe is evident on the exterior. Most particles bear an ∼8–12 nm-thick viral envelope. Density beneath the exterior spikes (red arrowheads), which bores through the viral envelope (white arrowheads), may be HA and NA transmembrane anchors. HA (green arrowhead) and NA (orange arrowhead) ectodomains are distinguishable. Inset: HA spike density, cropped from the tomogram is in good agreement with the HA ectodomain crystal structure; anchors are modeled into the structure (Wilson et al, 1981). The spike is tethered to three layers of the virus envelope (postulated strata: black arrowhead outer leaflet, white, inner leaflet, grey, M1 matrix protein layer). Microscope defocus 3 μm. Scalebar 50 nm. (B) At early timepoints, DOPC liposomes do not appear to be deformed by virions even when they are in close proximity. Sample acidified at pH 5.5 for 2.5 min. Microscope defocus 3 μm. Scalebar 50 nm.
Summary of liposome–virion complexes
To produce fusion-active complexes of influenza virus and target membrane for ECT, whole X31 strain virions were combined with DOPC liposomes at pH 5.5 and flash frozen. For samples that were acidified for 2.5 min, few changes in virus or liposome structure were observed (Figure 3B). In this case, virus particles and liposomes were sometimes found close to each other but with no apparent deformation of the liposome, suggesting that the majority of HA on the virus surface was still inactive.
Between 5 and 15 min post-acidification, more significant numbers of virions and liposomes were observed to form a range of complexes, with the liposomal bilayer typically showing clear signs of perturbation (Figures 4, 5 and 6; Supplementary Movies 1–3). Identification of relative concentrations of virus and liposome that produce significant numbers of discrete pairings of the particle types and also vitreous ice suitable for low-dose ECT has been a challenge. The majority (_n_>100) of virions in the ECT specimens for this study were not participating in complexes. Of the 53 loci observed between virions and liposomes at pH 5.5, nine exhibited dimple-like features on the target membranes without apparent disruption of the liposomal bilayer density (Figure 4). 23 exhibited a funnel-shaped deformation of the target liposomal membrane with what appears to be a break in the target membrane (Figures 5 and 6). Two instances of intermediates further along the pathway were observed (Figure 7), the low count number being consistent with the relatively low efficiency of lipid mixing and fusion at pH 5.5 (Figure 2). The features are described further below. The remainder of contacts involved various types of flat contacts in which the target membrane runs parallel to the outer viral envelope and otherwise does not exhibit any detectable deformation. In some cases, these appear to be mediated by HA (Figure 4D), in others, the virus and liposome may simply have been brought into contact because of surface tension effects in the thin water film that spans the 2 μm grid hole. A limited number of putative post-fusion complexes were observed (Supplementary Figure S3), again consistent with the relatively low efficiency of lipid mixing and fusion at pH 5.5. These complexes tend to be large, on the order of 200–250 nm in diameter; they exhibit granular contents that are localized on primarily one side of the vesicle near concentrations of glycoprotein spikes; other regions on the vesicle surface are bare, with low density of associated spikes.
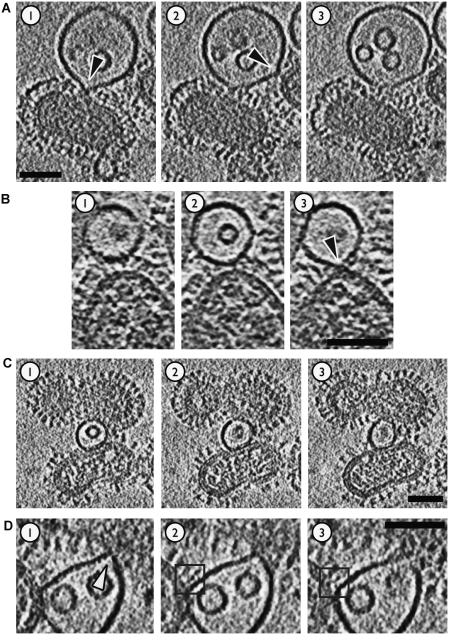
Figure 4.
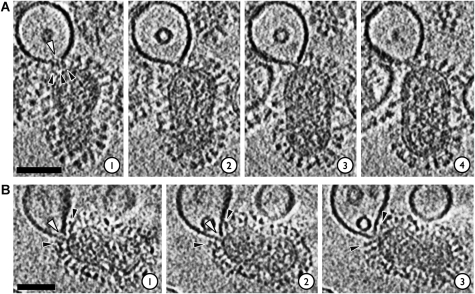
Target membrane deformations in virion–liposome complexes observed by ECT. Samples had been acidified at pH 5.5 for 5–8 min. Microscope defocus was set at 3–5 μm; 5.3 nm-thick slices through reconstructed cryo-tomographic density are shown. Scalebars 50 nm. (A) At early stages of docking, the liposomal bilayer is pulled into ∼5 nm-wide dimples (black arrowheads). (B) Some liposomes make glancing contacts (arrowhead in slice 3) with the viral envelope. (C) Liposomes can form extended planar contacts with virus envelope. (D) The liposome in this complex is engaged by two virions, one above and one to the left hand side. The liposome in this case forms a nearly planar contact with the virus on the left and appears to be coordinated by HA. The boxed HA spike in slices 2 and 3 exhibits density in a ‘Y' shape bridging the virus surface and liposome. This resembles features seen at virus membrane junctions in HSV-1 entry studies (Maurer et al, 2008). The interaction with the upper virus is more localized, with the liposomal membrane being pinched into an acute angle (arrowhead, slice 1).
Figure 5.
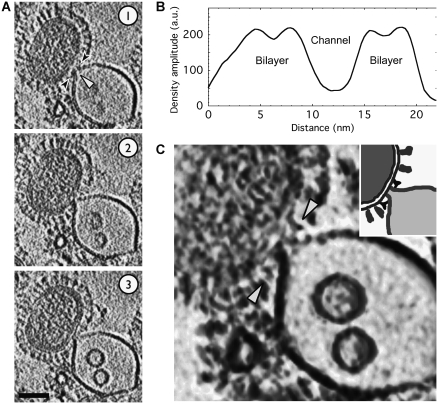
Electron cryo-tomograms of deformations in the liposomal membrane induced by interaction with virions at pH 5.5. Also see Supplementary Movies 1 and 2. (A) In this complex, the liposomal membrane appears to have been breached (white arrowhead, slice 1) at the contact zone where it is coordinated by HA spikes (black arrowheads, slice 1). Sample acidified for 5 min. Microscope defocus 3 μm. (B) In this tomogram, the liposome has been remodeled into a funnel that is apposed to the viral envelope; a putative opening at the funnel's mouth is indicated by the white arrowheads in slices 1 and 2. The virus envelope by contrast is intact. HA spikes (black arrowheads) ring the prefusion locus. Sample acidified for 8 min. Microscope defocus 5 μm; 5.3 nm virtual serial sections through the reconstructed tomograms are shown. Scalebars 50 nm.
Figure 6.
Electron cryo-tomograms of a funnel-shaped deformation in the liposomal membrane induced by interaction with virions at pH 5.5. Also see Supplementary Movie 3. (A) The virus has grabbed hold of a liposome and drawn its membrane towards the viral envelope. A putative opening at the funnel's mouth is indicated by the white arrowheads in slice 1. The viral envelope appears to be intact; 5.3 nm-thick serial sections are shown. Samples have been acidified to pH 5.5 for 5 min; 50 nm scalebar. (B) A cross-sectional density plot (path indicated by black arrowheads in (A), slice 1) maps the ∼5 nm-wide channel through the lipid funnel. The channel is lined by the liposomal bilayer with 3–4 nm leaflet spacings. (C) A 10 nm-thick volume section shows density that is interpreted to be HA spikes in a ‘Y' shaped conformation (white arrowheads) coordinating the liposomal funnel. Density rendered using Chimera. Inset shows a schematic of the key density features.
Figure 7.
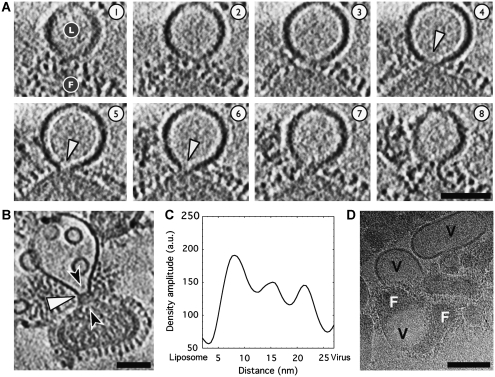
Constrained necks can join virions and liposomes at pH 5.5. (A) Serial 5.3 nm-thick sections through a liposome–virion complex; liposome marked with ‘L' and virus with ‘F' in slice 1. A neck with 10–15 nm-wide mouth is seen in slices 4–6. Liposomal membrane and viral envelope density appear continuous, although the strong liposomal density exhibits a pronounced boundary at the base of the neck. Sample acidified for 8 min. Microscope defocus 5 μm. Scalebar 50 nm. (B) In the 10.6 nm-thick section through another complex, an ∼15 nm-wide neck connects a virion and liposome. Sample acidified for 5 min. Microscope defocus 3 μm. Scalebar 50 nm. (C) A density plot along the channel axis (path indicated by black arrowheads in (B)) indicates that three ridges of density traverse the central section of the neck. The density may be due to an intact matrix layer or liposome and virus membrane leaflets. (D) Low-dose cryo-EM image of influenza X31 virions at pH 5.0 in the absence of liposomes suggests that the viral envelopes are more labile at pH 5.0 and give rise to envelope-derived lipidic vesicles (marked ‘V'); no comparable vesicles were observed in identical samples that were kept at pH 7.5. Two virions (marked ‘F') seen fusing with a vesicle in the lower left quadrant appear to be undergoing expansive pore dilation; an alternative explanation for this feature is that the virions may be caught in the process of shedding or blebbing their membranes. Sample acidified at pH 5.0 for 10 min. Microscope defocus 3 μm. Scalebar 100 nm.
A number of ∼20 nm diameter unilamellar vesicles were observed on the interior of nearly every liposomes that was in complex with a virus particle. These were comparatively less abundant in liposomes that had not been exposed to influenza virus, with ∼15% of virus-naive liposomes exhibiting similar features (Supplementary Figure S4). It is possible the small vesicles seen in virion–liposome complexes ‘blebbed' off the parent liposome in response to the bending stresses on the liposomal membrane induced by interaction with HA. Similar vesicles have been observed by freeze-fracture EM, leading Kanaseki et al (1997) to postulate that the vesicles originate from a virion making multiple points of contact with its target membrane. SNARE protein-mediated vacuole fusion has also been shown to yield pinched off membrane fragments that end up inside the fused organelle (Wang et al, 2002).
Target membrane remodeling by influenza virus
Liposome membrane deformation is localized around points of contact between virus and liposome (Figure 4). For complexes at early stages of docking, individual, localized dimples on the liposomal bilayers were observed (Figure 4A and B). These resemble ‘dimples' that have been proposed in physical modeling schemes (Efrat et al, 2007) and may correspond to similar punctate features observed by Kanaseki et al (1997) using freeze-fracture EM. Other liposomes are pinched by a small set of HA spikes as shown in Figure 5A (see also Supplementary Movie 1) and appear to be breached (white arrowhead, slice 1) at the point of contact with the HA spikes (black arrowheads, slice 1). A scenario in which target membrane scission is induced by fusogenic HA is consistent with the fluorescence observations that significant liposome content leakage precedes lipid mixing (Figure 2). Others have documented HA-induced content leakage through a range of target membranes and have found that the fusion peptide of HA can lead to significant changes in lipid order and membrane curvature (Epand and Epand, 1994, 2000; Shangguan et al, 1996; Jiricek et al, 1997; Longo et al, 1997; Bonnafous and Stegmann, 2000; Frolov et al, 2003; Lau et al, 2004; Ge and Freed, 2009).
One of the most striking and highly populated (23 out of 53 loci) features observed in liposome–virion complexes is a state in which the target membrane has been drawn and pinched towards the viral envelope such that it is in close apposition to the envelope's face (Figures 5 and 6; Supplementary Movies 2 and 3). Nearly all such funnel-shaped target membrane structures show an apparent lack of density, ∼5 nm wide, at the point of closest approach to the virus particle, suggesting that the liposome may present an open mouth towards the virus envelope. The proximal virus membrane by comparison does not appear to exhibit similar breaks or deformations. Given the limitations in ECT resolution, partial information because of the missing wedge effect, as well as the potential for weak density to be obscured by contrast transfer function (CTF) artefacts (seen as white Fresnel fringes) around liposomal bilayers, one cannot definitively rule out the possibility that tenuous connectivity with the viral envelope has been established or that membrane in some non-bilayer form may be present in the funnel mouth. The putative membrane breaches are observed not only at higher microscope defocus settings (e.g. 5 μm in Figure 5B), but also closer to true focus as well (3 μm in Figure 6) where fringing is less pronounced and where individual liposomal leaflets (still associated as a bilayer) can be observed lining the channel through the liposome's pinched neck (see Figure 6A slice 1, and Figure 6B). An open-mouthed funnel would be consistent with observations of significant leakage of liposomal contents before lipid mixing. Once a hemifused state is reached, the joining of target membrane and viral lipids into a continuous barrier would inhibit further leakage of contents from the liposomal lumen.
In Figure 7A, a liposome and virus are seen to be bridged by a neck that exhibits a 10–15 nm-wide central channel (indicated by the white arrowheads in slices 4–6). In this case, within resolution limitations, the liposomal membrane appears to exhibit density continuity with the viral envelope (particularly evident in slices 4–6), however the lipids may not have mixed extensively. Specifically, the high contrast DOPC liposomal membrane exhibits an abrupt boundary at the base of the neck. Previous studies of HA-mediated fusion have indicated that at early stages of membrane fusion, lipid transfer may be restrained (Zimmerberg et al, 1994; Chernomordik et al, 1998). The tomogram is not sharp enough to distinguish individual membrane leaflets, thus it is not possible to discern whether the complex exists as hemifused stalk-like structure or possibly as a constrained pore with joined lumens. Future efforts will seek to increase the acuity of the reconstructed density to clarify membrane leaflet continuity by working closer to focus, at higher magnifications, and by applying recently developed CTF-correction algorithms for tomography (Fernandez et al, 2006; Xiong et al, 2009; Zanetti et al, 2009).
Matrix layer stabilizes the viral envelope during fusion
Although the liposomal bilayer is often observed to be pinched and drawn out like a funnel, the thick viral envelope generally remains unperturbed (Figures 4, 5 and 6). This may result from an essential stabilization conferred by the intact matrix layer to the viral membrane. An intact matrix protein layer may also constrain the transition to an open fusion pore and its expansion. The complex shown in Figure 7B shows a liposome–virus complex in which, based on the continuous curvature of the perimeter of the two particles, the membranes appear to have joined to some degree. A neck with a ∼15 nm-wide inner diameter links the virion and liposome. As in the complex shown in Figure 7A, the high-density liposomal bilayer exhibits an abrupt boundary midway along the neck suggesting that lipid exchange was not complete (white arrowhead). Three peaks of density arranged in roughly parallel striations across the neck are observed (Figure 7B and C). The density within the neck may be membrane leaflets or viral contents such as the matrix layer, which is intact as evidenced by the thick envelope around the virus's periphery. This type of later-stage fusion complex will be investigated more fully when conditions have been identified that enrich the population of this intermediate.
Efforts to perform ECT studies at pH 5.0 and identify whether low pH fusion loci differ substantially from structures observed at pH 5.5 have been stymied by the tendency of virions and liposomes to form aggregates that are refractory to characterization by ECT. Preliminary studies of X31 influenza virions in the absence of liposomes at pH 5.0 have yielded three notable features (Figure 7D): (1) the thick viral envelopes are less prevalent, having apparently reverted in large part to a standard bilayer; (2) unilamellar vesicles that are not observed in neutral virus samples from the identical preparation are abundant, possibly reflecting that significant amounts of lipid can be stripped from virions at pH 5.0; (3) a few instances of virions fusing with the virus-derived vesicles are observed and fusion pores are dilated and expansive in contrast to the tight connections observed at pH 5.5 (alternatively, this may reflect a virion in the process of shedding its membrane).
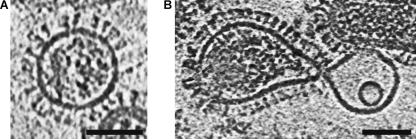
An additional observation that supports the hypothesis that matrix has a skeletal role during early stages of fusion involves a subset of the total population of particles that lack the matrix layer (Class-III particles by the nomenclature of Harris et al (2006)). Matrixless particles present a similar surface density of glycoprotein spikes to matrix-bearing particles, however they tend to be spherical rather than capsular. This suggests that in the absence of matrix, particles adopt the lower energy, isotropic shape. The envelope layer in reconstructed tomograms of these particles at pH 5.5 is similar in simplicity to the DOPC bilayers. In contrast to the robust, matrix-bearing virions, the matrixless virus and liposomes deform symmetrically around the point of contact (Figure 8).
Figure 8.
Matrixless particles are highly deformable. Matrixless virus particles are ordinarly spherical (A), but they deformed to the same extent as the liposome during fusion (B); 5.3 nm-thick section shown. Sample acidified to pH 5.5 for 6.25 min. Microscope defocus 3 μm. Scalebar 50 nm.
HA is localized around viral and target membrane contact sites
Density that may be attributable to HA spikes surrounds prefusion liposome:virion loci. In general, is not possible to definitively assign the fringe of density to a specific number of glycoprotein spikes, given the low signal-to-noise levels, lack of complete density information because of the missing wedge effect, and the likely non-canonical HA spike morphology. A number of studies have inferred that HA spikes form functional aggregates at the onset of fusion (Ellens et al, 1990; Clague et al, 1991; Blumenthal et al, 1996; Danieli et al, 1996; Mittal et al, 2002). This evidence largely comes from HA-expressing cell fusion assays rather than from direct structural characterization. The tomographic density at virion–liposome loci is generally consistent with previous estimations of two to eight spikes per fusion site.
Some of the density features that appear to coordinate the funnel neck have a ‘V' or ‘Y' shape (Figures 4D and 6C). These features resemble structures observed by ECT that were presumed to be herpes simplex virus-1 (HSV-1) fusion glycoproteins (gH/gL/gB) bridging the HSV-1 envelope and target membranes during cell entry (Maurer et al, 2008). The observed density also resembles a bent HA organization that is hypothesized to precede the formation of the extended coiled-coil state, as described by Gruenke et al (2002) in their leash-in-groove model of HA refolding.
Discussion
During entry, influenza virions are exposed to stages of endosomal acidification (Lakadamyali et al, 2003). Although many studies have focused on fusion at pH 5.0, the ‘optimal' pH for fusion found in late endosomes, changes in HA at more elevated pH∼5.5–6.0 values render the virion fusion active and appear to initiate membrane remodeling that subsequently leads to efficient fusion as the endosomes continue to mature. The results show that early stages of membrane deformation are highly localized and focused on the target membrane, while the viral envelope appears to remain essentially unperturbed.
The target membrane was chosen to be simple, without sialic acid present, to ensure that any liposome–virus interaction was mediated by the membrane-active fusion peptides of HA (Stegmann et al, 1987, 1991). Previous reports have shown that fusion can take place in the absence of receptor (White et al, 1982; Stegmann et al, 1985, 1989). Although high avidity sialic acid binding by multiple HA on the virus surface is likely to contribute to determining the contact zone between virus and target membrane, studies have argued that the HA spikes that participate in sialic acid coordination are not themselves involved in productive fusion loci (Ellens et al, 1990; Alford et al, 1994). Others studies have found evidence for a significant effect on fusion kinetics resulting from the presence of receptor, but with still undetermined mechanistic origins (Stegmann et al, 1989, 1995; Ohuchi et al, 2002).
It is also important to note that though many membrane fusion studies including this one have used synthetic liposomes, a virus in an endosome may encounter a somewhat different situation including facing a target membrane with concave rather than convex curvature as well as a more complex mixture of lipids and associated proteins that are distributed asymmetrically between the two endosomal leaflets. Phosphatidylcholine (PC) is the dominant component of the proximal endosomal membrane leaflet. The distal endosomal leaflet is likely to be enriched in phospholipids such as phosphatidylethanolamine and phosphatidylserine that have different curvature and fusion propensities. Understanding the role that intrinsic curvature of these lipids has in fusion is critical. It is likely, for example, that some of the membrane remodeling observed in this study resulted from the curvature-neutral PC bilayer reorganizing under HA's influence to relieve strain present in the curved, ∼100 nm liposome. Future studies will examine HA-mediated fusion with target membranes that more closely mimic the complex conditions found in endosomes.
Implications for cell invasion and fusion mechanisms
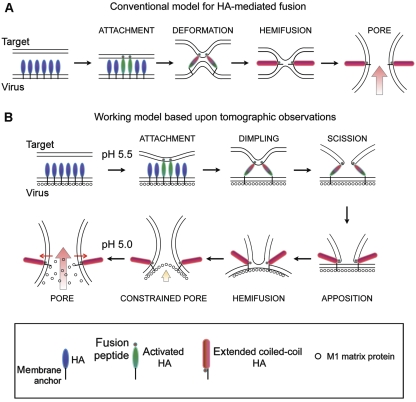
The target membrane-centric process described in this report is summarized in Figure 9. The working model proposes the following sequence of events during fusion: (1) in the mildly acidic early endosomes, at pH∼5.5–6, HA spikes deploy their fusion peptides; (2) a localized cluster of fusogenic HA grapples to the target membrane, pulling it into a dimple; (3) possibly as a result of the stressed configuration or destabilizing effect of fusion-peptide insertion, a hole is opened through the target membrane (scission). The virus envelope remains unperturbed; (4) the open-mouthed lipid funnel is brought into apposition with the envelope because of HA refolding into its low pH, ‘hairpin' conformation; (5) the funnel may insert into the virus envelope to initiate lipid mixing, leading to a hemifused intermediate; (6) at pH∼5.5, the matrix layer remains intact constraining pore expansion, but it is hypothesized that as pH is lowered to 5, such as in the mature endosomes or lysosomes, the matrix layer may weaken or disintegrate, allowing the fusion pore to dilate. The primary evidence in this report supports the dimpling stage as well as the existence of a funnel-shaped intermediate that may reflect a combination of the subsequent three steps (scission, apposition, and possibly hemifusion).
Figure 9.
Working model of fusion between influenza virus and a target membrane. Most canonical models of protein-mediated fusion resemble the sequence shown in (A). The models have lacked the framework of concrete structural detail. On the basis of the combined electron tomography and fluorescence spectroscopy observations, the sequence of events shown in (B) is proposed to take place; see main text for detailed description.
A process of membrane deformation that is focused on the target membrane resolves a fundamental paradox that had been implicit in previously proposed models for fusion. Many models invoke joint target and viral membrane deformation even at early stages of the fusion reaction (Chernomordik and Kozlov, 2003; Tamm et al, 2003); in one case it was proposed that the fusion peptides act primarily on the viral envelope to pull it towards the target membrane (Kozlov and Chernomordik, 1998). Such schemes would likely lead to disruption of the protective viral envelope before the formation of a productive fusion pore because of the destabilizing effect of the fusion peptides. By concentrating membrane remodeling on the target membrane as proposed here, the virus does not risk premature release and entrapment of its RNP cargo in the degradative endosomal and lysosomal lumens.
On the basis of the complementary tomography and fluorescence data, the most likely organization of lipid and protein at the prefusion loci appears to be an open-mouthed lipidic funnel coordinated by the fusion peptides from a number of HA spikes, with the mouth held in close apposition to the viral envelope (Figures 5, 6 and 7). Bonnafous and Stegmann (2000) proposed a model in which the target membrane is punctured before mixing of viral and target membrane lipids. The observation of prefusion loci with apparent membrane breaks at the HA-contact zone is consistent with their proposed model. A 5 nm-wide conduit is also consistent in dimension with hypothesized pore diameters (4 nm) from patch clamp measurements of individual fusion events between HA-expressing fibroblasts and erythrocytes (Spruce et al, 1989), although in the present case continuity between the viral and liposomal lumens has yet to be established. The narrow phospholipid funnel induced in the target membrane would minimize the area of contact between membranes that ordinarily resist apposition because of electrostatic repulsion and hydration effects (Cowley et al, 1978). The tight curvature of the funnel neck may also facilitate the next stages of fusion after insertion of this tube into the viral envelope. Several studies have highlighted the importance of highly curved membranes during fusion (Lentz et al, 2000; Chernomordik and Kozlov, 2003). The fine structure of the funnel at its virus-proximal site cannot be resolved in the tomograms given the ECT resolution, however, one possibility envisages a mouth opening ringed by fusion peptides from multiple spikes that are embedded in a micellar cap or similar non-bilayer structure. Interestingly, such a configuration could potentially lead to lipid mixing between the inner and outer leaflets of the target membrane before mixing with lipids in the virus membrane.
Role of the matrix layer during fusion
The robustness of the viral envelope during the early stages of fusion appears to derive from the presence of an intact matrix protein layer that undergirds the membrane. The matrix layer maintains the integrity and shape of the influenza virus, acting in essence like an endoskeleton, while also providing a foundation and anchor point for the HA fusion machinery. As a result, the fusion protein's membrane bending and remodeling actions appear to be concentrated on the target membrane. As a corollary to the buttressing role played by matrix at pH 5.5, it is plausible that dissolution of the matrix layer follows at late endosomal or lysosomal pH<5, loosening the restraints on the viral envelope and enabling it to complete the fusion process. As pH drops, the protonation state of the phosphate head groups and/or matrix protein may change, altering the protein's electrostatic interaction with the inner leaflet and stabilization of the envelope as a whole (Ruigrok et al, 2000). Indeed, the fluorescence results and preliminary cryo-EM images of X31 virions at pH 5.0 suggest that the viral envelope may be more labile and predisposed to fuse at pH 5.0 than at pH 5.5, possibly because of dissolution or weakening of the M1 matrix layer (Figure 7D).
A significant number of studies of HA-mediated fusion to date have used cell lines that express the protein on their surface, rather than whole virions. These HA expression systems lack the M1 matrix layer, and hence may exhibit different mechanics of fusion relative to the virus. One might anticipate that fusion of cell-expressed HA would more closely resemble the changes observed with the matrixless particles (Figure 8), and not show the same asymmetry of membrane deformation as is observed with matrix-bearing virions. Interestingly, in the only other case of a viral fusion intermediate imaged by ECT, the envelope of HSV-1 and target cellular membrane appears to exhibit symmetrical deformation during fusion, similar to what is observed for matrixless influenza virions (Maurer et al, 2008). This may relate to the fact that HSV-1 has an amorphous tegument lying between the envelope and icosahedral capsid, rather an ordered matrix layer.
Many enveloped RNA viruses including the orthomyxoviruses such as influenza, retroviruses, arenaviruses, filoviruses, and paramyxoviruses possess class-I fusion proteins that share common architecture and undergo similar refolding events as they mediate the fusion of host and viral membranes (Hughson, 1997; Lamb and Jardetzky, 2007). These viruses also possess matrix proteins that have a critical role in virion morphogenesis during budding from host cells. Some of the matrix proteins, such as that from HIV, may be homologous to the influenza M1 protein (Harris et al, 1999). Immature HIV particles exhibit a well-ordered albeit partial matrix layer (Wright et al, 2007; Briggs et al, 2009); a distinct matrix layer is not readily apparent in electron cryo-tomograms of mature virions, although it is inferred that matrix may connect envelope and capsid (Briggs et al, 2003, 2006). Interaction of the Env protein's cytoplasmic domain and matrix protein in HIV-1 has been shown (Yu et al, 1992; Cosson, 1996), thus it is conceivable that matrix may serve as an anchor for the Env protein and support the viral envelope during membrane fusion. Initiation of membrane fusion through asymmetrical membrane deformation may be a means of ordering a complex physical process that is crucial to the life cycle of many enveloped RNA viruses.
Materials and methods
Virus and liposome preparation
Gradient-purified X31 (H3N2) influenza A virus purchased from Charles River Laboratories was concentrated by ultracentrifugation to 6–10 mg/ml and stored in 250 mM NaCl, 10 mM HEPES pH 7.5 buffer; 3–5 mg/ml DOPC (Avanti Polar Lipids) liposomes containing 25 mM SRB fluorophore (Invitrogen Corp.) were produced by drying a 25 mg/ml DOPC chloroform solution under nitrogen gas. The dried lipid was resuspended in 225 mM NaCl, 10 mM HEPES pH 7.5 buffer containing 25 mM SRB dye; the lower salt concentration was used to equalize the osmotic balance on the two sides of the membrane with 250 mM NaCl, 10 mM HEPES pH 7.5 storage buffer. The resuspended lipid was subjected to 5 sequential freeze-thaw cycles in liquid nitrogen, followed by 21 extrusions through polycarbonate filters with 100 nm pore size (Avanti Polar Lipids); 100 nm liposomes were selected for the study because larger sizes (e.g. 200 or 400 nm diameter) would likely be as thick as or thicker than typical ice thicknesses in the cryo-EM samples. One concern in using the larger liposomes is that they would be squashed in the thin ice layer and subject to stresses because of the compression. The dye-encapsulating liposomes eluted as a single band from PD-10 gel filtration columns (GE Healthcare) and were stored in the same pH 7.5 HEPES buffer used for virus storage.
Dynamic light scattering (DLS) with a Brookhaven 90+ Nanoparticle Size Analyzer was used to characterize the polydispersity and average dimension of the liposomes at room temperature. Liposome diameters from individual preparations ranged from 125 to 145 nm by DLS, and polydispersity was typically very low, 0.002–0.020. DLS also confirmed that no significant change in liposome dimensions resulted from incubating the liposomes at pH 5.0–5.5 for up to an hour (longest duration tested). Likewise, when liposomes were acidified to pH 5.0 and 5.5, leakage of SRB was not detected by fluorescence monitoring. It was concluded that the liposomes are stable under the acidic pH conditions used in the fluorescence and ECT studies. Liposomes were used within 24 h of their production.
Doubly labeled DOPC liposomes containing 25 mM encapsulated SRB and the lipophilic carbocyanine dye DiD (Invitrogen Corp.) incorporated into the membrane were produced by a nearly identical procedure as described above. The liposomes were made by adding a 5–20 μl aliquots of ethanol-based DiD Vybrant solution (Invitrogen Corp.) directly to the DOPC chloroform stock solution before drying of the lipids; then subjected to freeze-thaw and extrusion steps as described above.
X31 virions for fluorescence measurements were labeled with DiD by adding 5–20 μl of DiD Vybrant solution (Invitrogen Corp.) directly to 500 μl of 2 mg/ml virus stock solutions in HEPES-buffered saline. The range of DiD added in the reaction gave rise to a range of DiD incorporation into the viral envelopes. Labeling reactions were incubated for 2 h at 30°C with gentle rocking. The reaction solution was passed over a PD-10 column equilibrated with 250 mM NaCl, 10 mM HEPES, pH 7.5. A single blue band containing the DiD-labeled virions was collected and stored at 4°C.
Generation of the fusogenic complexes for ECT
To produce the fusion-active complex of virus and liposome for ECT, equal volumes of influenza A virus, DOPC liposomes containing 25 mM internal SRB and BSA-blocked 10 nm gold beads in 250 mM NaCl, 10 mM HEPES, pH 7.5 were first mixed together. The combined solution was then mixed with a pH 5.0 citric acid solution to produce a final pH of 5.5. Samples were incubated at 25°C. At 2.5, 5, 6.25, 8, and 15 min after acidification, 3 μl of the acidified mixture was transferred onto plasma-cleaned, C-flat holey carbon grids (Protochips Inc.), manually blotted, and plunge frozen in liquid ethane. The samples that had been acidified for longer periods tended to produce less optimal specimens for low-dose imaging apparently because of aggregation of liposomes and virions.
Cryo-EM and tomography processing
EM was conducted at the National Resource for Automated Molecular Microscopy (NRAMM). Tilt series were gathered using an FEI Company T12 cryo-EM operating at 120 kV. A 2K × 2K TVIPS F224 CCD camera with 24 μm pixel size was used for imaging. Image magnification was set to × 26 000, which gave rise to 0.525 nm/pixel sampling. Nominal microscope defocus values of 3 and 5 μm were used; the first node of the microscope's CTF is at 3.1 and 3.9 nm for the two defocus values, respectively. Total electron dose for the entire tilt series was estimated ∼100 electrons/Å2; under these imaging conditions no bubbling was observed. Tilt series data collection was controlled by the Leginon software package with images gathered at 2° increments from −58 to +58° (Suloway et al, 2009). Tomograms were reconstructed using the weighted back-projection method implemented in IMOD (Kremer et al, 1996). The tomograms were then filtered using one iteration of a three-dimensional median filter followed by a real-space Gaussian filter, width 1.5σ. This process appears to preserve fine features reasonably well while reducing the amplitude of noise relative to the specimen density. Representative unfiltered and filtered images are shown in Supplementary Figure S5. Images were visualized either in ImageJ (Abramoff et al, 2004) or Chimera (Pettersen et al, 2004). Some particle tomograms appear sharper than others largely because of the different defocus settings, but also because of a combination of proximity in the field of view relative to the tilt axis, more stable cryo-stage performance during tilt series collection, more accurate true focus and eucentric height determination, thinner ice, and better alignment during image reconstruction.
Fluorescence spectroscopy
In one series of fluorescence experiments, SRB/DiD doubly labeled liposomes were combined with unlabeled virions. In a second series of measurements, SRB-labeled liposomes were combined with DiD-labeled virions. Over the range of DiD incorporation examined, dye-dependent effects were not observed. Virus and liposomes in 250 mM NaCl, 10 mM HEPES pH 7.5 were diluted to ∼50 and ∼20 μg/ml, respectively, in 250 mM NaCl, 50 mM sodium citrate buffer pH 5.5 and 5.0. Note the relative concentrations present in the fluorescence measurements were ∼2 orders of magnitude lower than the concentrations used in the ECT sample preparations. This was necessitated by the very different sample requirements of each method. Fluorescence spectroscopy was carried out in a Varian Cary Eclipse spectrophotometer using the following excitation/emission spectral pairs λEx=565 nm/λEM=585 nm (SRB) and λEx=644 nm/λEx=665 nm (DiD) with 2.5 nm slit widths. The fluorescence cuvette was thermostated at 25°C.
Supplementary Material
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Review Process File
Acknowledgments
I thank Dr Anchi Cheng of the National Resource for Automated Molecular Microscopy (NRAMM) for providing Leginon training. I thank Drs Kelly Dryden (The Scripps Research Institute) and Kristin Parent (University of California, San Diego) for assistance in plunge-freezing samples. Ms Connie Lu (University of Washington) performed preliminary fluorescence measurements. Dr Lu Gan provided valuable discussions and critical evaluation of the paper. I thank Dr Jack Johnson for providing lab space during the early stages of this work. The work is supported by the NIH Grant R00GM080352 to KKL. Some of the work presented here was conducted at the NRAMM, which is supported by the National Institutes of Health though the National Center for Research Resources' P41 program (RR17573). Some of the molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR01081).
Footnotes
The author declares that he has no conflict of interest.
References
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton International 11: 36–42 [Google Scholar]
- Alford D, Ellens H, Bentz J (1994) Fusion of influenza virus with sialic acid-bearing target membranes. Biochemistry 33: 1977–1987 [DOI] [PubMed] [Google Scholar]
- Ali A, Avalos RT, Ponimaskin E, Nayak DP (2000) Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol 74: 8709–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R, Sarkar DP, Durell S, Howard DE, Morris SJ (1996) Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J Cell Biol 135: 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnafous P, Stegmann T (2000) Membrane perturbation and fusion pore formation in influenza hemagglutinin-mediated membrane fusion. A new model for fusion. J Biol Chem 275: 6160–6166 [DOI] [PubMed] [Google Scholar]
- Bottcher C, Ludwig K, Herrmann A, van Heel M, Stark H (1999) Structure of influenza haemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett 463: 255–259 [DOI] [PubMed] [Google Scholar]
- Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD (2006) The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14: 15–20 [DOI] [PubMed] [Google Scholar]
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG (2009) Structure and assembly of immature HIV. Proc Natl Acad Sci USA 106: 11090–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD (2003) Structural organization of authentic, mature HIV-1 virions and cores. EMBO J 22: 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371: 37–43 [DOI] [PubMed] [Google Scholar]
- Carr CM, Kim PS (1993) A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73: 823–832 [DOI] [PubMed] [Google Scholar]
- Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC (1998) Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95: 409–417 [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Frolov VA, Leikina E, Bronk P, Zimmerberg J (1998) The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol 140: 1369–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM (2003) Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem 72: 175–207 [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM (2005) Membrane hemifusion: crossing a chasm in two leaps. Cell 123: 375–382 [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM (2008) Mechanics of membrane fusion. Nat Struct Mol Biol 15: 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Schoch C, Blumenthal R (1991) Delay time for influenza virus hemagglutinin-induced membrane fusion depends on hemagglutinin surface density. J Virol 65: 2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P (1996) Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J 15: 5783–5788 [PMC free article] [PubMed] [Google Scholar]
- Cowley AC, Fuller NL, Rand RP, Parsegian VA (1978) Measurement of repulsive forces between charged phospholipid bilayers. Biochemistry 17: 3163–3168 [DOI] [PubMed] [Google Scholar]
- Danieli T, Pelletier SL, Henis YI, White JM (1996) Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol 133: 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms RW, Helenius A, White J (1985) Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J Biol Chem 260: 2973–2981 [PubMed] [Google Scholar]
- Eckert DM, Kim PS (2001) Mechanisms of viral membrane fusion and its inhibition. Ann Rev Biochem 70: 777–810 [DOI] [PubMed] [Google Scholar]
- Efrat A, Chernomordik LV, Kozlov MM (2007) Point-like protrusion as a prestalk intermediate in membrane fusion pathway. Biophys J 92: L61–L63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellens H, Bentz J, Mason D, Zhang F, White JM (1990) Fusion of influenza hemagglutinin-expressing fibroblasts with glycophorin-bearing liposomes: role of hemagglutinin surface density. Biochemistry 29: 9697–9707 [DOI] [PubMed] [Google Scholar]
- Enami M, Enami K (1996) Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol 70: 6653–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM, Epand RF (1994) Relationship between the infectivity of influenza virus and the ability of its fusion peptide to perturb bilayers. Biochem Biophys Res Commun 202: 1420–1425 [DOI] [PubMed] [Google Scholar]
- Epand RM, Epand RF (2000) Modulation of membrane curvature by peptides. Biopolymers 55: 358–363 [DOI] [PubMed] [Google Scholar]
- Fernandez JJ, Li S, Crowther RA (2006) CTF determination and correction in electron cryotomography. Ultramicroscopy 106: 587–596 [DOI] [PubMed] [Google Scholar]
- Frolov VA, Dunina-Barkovskaya AY, Samsonov AV, Zimmerberg J (2003) Membrane permeability changes at early stages of influenza hemagglutinin-mediated fusion. Biophys J 85: 1725–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyoshi Y, Kume NP, Sakata K, Sato SB (1994) Fine structure of influenza A virus observed by electron cryo-microscopy. EMBO J 13: 318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge M, Freed JH (2009) Fusion peptide from influenza hemagglutinin increases membrane surface order: an electron-spin resonance study. Biophys J 96: 4925–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godley L, Pfeifer J, Steinhauer D, Ely B, Shaw G, Kaufmann R, Suchanek E, Pabo C, Skehel JJ, Wiley DC, Wharton S (1992) Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68: 635–645 [DOI] [PubMed] [Google Scholar]
- Gruenke JA, Armstrong RT, Newcomb WW, Brown JC, White JM (2002) New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J Virol 76: 4456–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther-Ausborn S, Schoen P, Bartoldus I, Wilschut J, Stegmann T (2000) Role of hemagglutinin surface density in the initial stages of influenza virus fusion: lack of evidence for cooperativity. J Virol 74: 2714–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC (2006) Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci USA 103: 19123–19127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Sha B, Luo M (1999) Structural similarities between influenza virus matrix protein M1 and human immunodeficiency virus matrix and capsid proteins: an evolutionary link between negative-stranded RNA viruses and retroviruses. J Gen Virol 80: 863–869 [DOI] [PubMed] [Google Scholar]
- Hughson FM (1997) Enveloped viruses: a common mode of membrane fusion? Curr Biol 7: R565–R569 [DOI] [PubMed] [Google Scholar]
- Imai M, Mizuno T, Kawasaki K (2006) Membrane fusion by single influenza hemagglutinin trimers: Kinetic evidence from image analysis of hemagglutinin-reconstituted vesicles. J Biol Chem 281: 12729–12735 [DOI] [PubMed] [Google Scholar]
- Inglis SC, Carroll AR, Lamb RA, Mahy BW (1976) Polypeptides specified by the influenza virus genome I. Evidence for eight distinct gene products specified by fowl plague virus. Virology 74: 489–503 [DOI] [PubMed] [Google Scholar]
- Jin H, Leser GP, Zhang J, Lamb RA (1997) Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J 16: 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricek R, Schwarz G, Stegmann T (1997) Pores formed by influenza hemagglutinin. Biochim Biophys Acta 1330: 17–28 [DOI] [PubMed] [Google Scholar]
- Kanaseki T, Kawasaki K, Murata M, Ikeuchi Y, Ohnishi S (1997) Structural features of membrane fusion between influenza virus and liposome as revealed by quick-freezing electron microscopy. J Cell Biol 137: 1041–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk HD, Rott R, Orlich M, Blodorn J (1975) Activation of influenza A viruses by trypsin treatment. Virology 68: 426–439 [DOI] [PubMed] [Google Scholar]
- Korte T, Ludwig K, Booy FP, Blumenthal R, Herrmann A (1999) Conformational intermediates and fusion activity of influenza virus hemagglutinin. J Virol 73: 4567–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, Chernomordik LV (1998) A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys J 75: 1384–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Krumbiegel M, Herrmann A, Blumenthal R (1994) Kinetics of the low pH-induced conformational changes and fusogenic activity of influenza hemagglutinin. Biophys J 67: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin PI, Zimmerberg J, Chizmadzhev YA, Cohen FS (2001) A quantitative model for membrane fusion based on low-energy intermediates. Proc Natl Acad Sci USA 98: 7235–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, Zhuang X (2003) Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA 100: 9280–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Jardetzky TS (2007) Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol 17: 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WL, Ege DS, Lear JD, Hammer DA, DeGrado WF (2004) Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys J 86: 272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Choppin PW (1975) Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68: 440–454 [DOI] [PubMed] [Google Scholar]
- Lee RJ, Wang S, Turk MJ, Low PS (1998) The effects of pH and intraliposomal buffer strength on the rate of liposome content release and intracellular drug delivery. Biosci Rep 18: 69–78 [DOI] [PubMed] [Google Scholar]
- Lentz BR, Malinin V, Haque ME, Evans K (2000) Protein machines and lipid assemblies: current views of cell membrane fusion. Curr Opin Struct Biol 10: 607–615 [DOI] [PubMed] [Google Scholar]
- Longo ML, Waring AJ, Hammer DA (1997) Interaction of the influenza hemagglutinin fusion peptide with lipid bilayers: area expansion and permeation. Biophys J 73: 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W (2005) Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem 74: 833–865 [DOI] [PubMed] [Google Scholar]
- Maeda T, Ohnishi S (1980) Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett 122: 283–287 [DOI] [PubMed] [Google Scholar]
- Martens S, McMahon HT (2008) Mechanisms of membrane fusion: disparate players and common principles. Nat Rev 9: 543–556 [DOI] [PubMed] [Google Scholar]
- Matlin KS, Reggio H, Helenius A, Simons K (1981) Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol 91: 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer UE, Sodeik B, Grunewald K (2008) Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci USA 105: 10559–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A, Shangguan T, Bentz J (2002) Measuring pKa of activation and pKi of inactivation for influenza hemagglutinin from kinetics of membrane fusion of virions and of HA expressing cells. Biophys J 83: 2652–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y (2006) Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439: 490–492 [DOI] [PubMed] [Google Scholar]
- Odegard AL, Kwan MH, Walukiewicz HE, Banerjee M, Schneemann A, Johnson JE (2009) Low endocytic pH and capsid protein autocleavage are critical components of Flock House virus cell entry. J Virol 83: 8628–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi M, Ohuchi R, Sakai T, Matsumoto A (2002) Tight binding of influenza virus hemagglutinin to its receptor interferes with fusion pore dilation. J Virol 76: 12405–12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Puri A, Booy FP, Doms RW, White JM, Blumenthal R (1990) Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J Virol 64: 3824–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RW, Andree PJ, Hooft van Huysduynen RA, Mellema JE (1984) Characterization of three highly purified influenza virus strains by electron microscopy. J Gen Virol 65: 799–802 [DOI] [PubMed] [Google Scholar]
- Ruigrok RW, Barge A, Durrer P, Brunner J, Ma K, Whittaker GR (2000) Membrane interaction of influenza virus M1 protein. Virology 267: 289–298 [DOI] [PubMed] [Google Scholar]
- Ruigrok RW, Wrigley NG, Calder LJ, Cusack S, Wharton SA, Brown EB, Skehel JJ (1986) Electron microscopy of the low pH structure of influenza virus haemagglutinin. EMBO J 5: 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan T, Alford D, Bentz J (1996) Influenza-virus-liposome lipid mixing is leaky and largely insensitive to the material properties of the target membrane. Biochemistry 35: 4956–4965 [DOI] [PubMed] [Google Scholar]
- Shangguan T, Siegel DP, Lear JD, Axelsen PH, Alford D, Bentz J (1998) Morphological changes and fusogenic activity of influenza virus hemagglutinin. Biophys J 74: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Bayley PM, Brown EB, Martin SR, Waterfield MD, White JM, Wilson IA, Wiley DC (1982) Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA 79: 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69: 531–569 [DOI] [PubMed] [Google Scholar]
- Spruce AE, Iwata A, White JM, Almers W (1989) Patch clamp studies of single cell-fusion events mediated by a viral fusion protein. Nature 342: 555–558 [DOI] [PubMed] [Google Scholar]
- Stegmann T, Bartoldus I, Zumbrunn J (1995) Influenza hemagglutinin-mediated membrane fusion: influence of receptor binding on the lag phase preceding fusion. Biochemistry 34: 1825–1832 [DOI] [PubMed] [Google Scholar]
- Stegmann T, Booy FP, Wilschut J (1987) Effects of low pH on influenza virus. Activation and inactivation of the membrane fusion capacity of the hemagglutinin. J Biol Chem 262: 17744–17749 [PubMed] [Google Scholar]
- Stegmann T, Delfino JM, Richards FM, Helenius A (1991) The HA2 subunit of influenza hemagglutinin inserts into the target membrane prior to fusion. J Biol Chem 266: 18404–18410 [PubMed] [Google Scholar]
- Stegmann T, Hoekstra D, Scherphof G, Wilschut J (1985) Kinetics of pH-dependent fusion between influenza virus and liposomes. Biochemistry 24: 3107–3113 [DOI] [PubMed] [Google Scholar]
- Stegmann T, Nir S, Wilschut J (1989) Membrane fusion activity of influenza virus. Effects of gangliosides and negatively charged phospholipids in target liposomes. Biochemistry 28: 1698–1704 [DOI] [PubMed] [Google Scholar]
- Stegmann T, White JM, Helenius A (1990) Intermediates in influenza induced membrane fusion. EMBO J 9: 4231–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA (1999) Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258: 1–20 [DOI] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ (2009) Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol 167: 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm LK, Crane J, Kiessling V (2003) Membrane fusion: a structural perspective on the interplay of lipids and proteins. Curr Opin Struct Biol 13: 453–466 [DOI] [PubMed] [Google Scholar]
- Wang L, Seeley ES, Wickner W, Merz AJ (2002) Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108: 357–369 [DOI] [PubMed] [Google Scholar]
- Wharton SA, Calder LJ, Ruigrok RW, Skehel JJ, Steinhauer DA, Wiley DC (1995) Electron microscopy of antibody complexes of influenza virus haemagglutinin in the fusion pH conformation. EMBO J 14: 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Kartenbeck J, Helenius A (1982) Membrane fusion activity of influenza virus. EMBO J 1: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Wilson IA (1987) Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J Cell Biol 105: 2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC (1981) Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289: 366–373 [DOI] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ (2007) Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26: 2218–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN (2009) CTF determination and correction for low dose tomographic tilt series. J Struct Biol 168: 378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yuan X, Matsuda Z, Lee TH, Essex M (1992) The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol 66: 4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Riches JD, Fuller SD, Briggs JA (2009) Contrast transfer function correction applied to cryo-electron tomography and sub-tomogram averaging. J Struct Biol 168: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Blumenthal R, Sarkar DP, Curran M, Morris SJ (1994) Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J Cell Biol 127: 1885–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Review Process File