CARD9 Mediates Dectin-2-induced IκBα Kinase Ubiquitination Leading to Activation of NF-κB in Response to Stimulation by the Hyphal Form of Candida albicans (original) (raw)
Abstract
The scaffold protein CARD9 plays an essential role in anti-fungus immunity and is implicated in mediating Dectin-1/Syk-induced NF-κB activation in response to Candida albicans infection. However, the molecular mechanism by which CARD9 mediates _C. albicans_-induced NF-κB activation is not fully characterized. Here we demonstrate that CARD9 is involved in mediating NF-κB activation induced by the hyphal form of C. albicans hyphae (Hyphae) but not by its heat-inactivated unicellular form. Our data show that inhibiting Dectin-2 expression selectively blocked Hyphae-induced NF-κB, whereas inhibiting Dectin-1 mainly suppressed zymosan-induced NF-κB, indicating that Hyphae-induced NF-κB activation is mainly through Dectin-2 and not Dectin-1. Consistently, we find that the hyphae stimulation induces CARD9 association with Bcl10, an adaptor protein that functions downstream of CARD9 and is also involved in _C. albicans_-induced NF-κB activation. This association is dependent on Dectin-2 but not Dectin-1 following the hyphae stimulation. Finally, we find that although both CARD9 and Syk are required for Hyphae-induced NF-κB activation, they regulate different signaling events in which CARD9 mediates IκBα kinase ubiquitination, whereas Syk regulates IκBα kinase phosphorylation. Together, our data demonstrated that CARD9 is selectively involved in Dectin-2-induced NF-κB activation in response to C. albicans hyphae challenging.
Keywords: Adaptor Proteins, Fungi, Innate Immunity, NF-κB, Signal Transduction
Introduction
Candida albicans is a major opportunistic fungal pathogen that predominantly causes infection to cancer patients and immunocompromised individuals. During C. albicans infection, macrophages and dendritic cells recognize components from the fungal cell wall through their pattern recognition receptors (1, 2), which triggers a series of signaling cascades leading to activation of various transcription factors including NF-κB (1). The activation of NF-κB and other transcription factors further induce the expression of various cytokines and chemokines and inflammatory responses. However, the pattern recognition receptors that recognize fungal cell wall components are not fully defined (3).
NF-κB is a family of transcription factors that control the expression of pro-inflammatory genes in immune cells (4). In resting cells, the activity of NF-κB is tightly controlled by the IκB family of proteins, which bind to NF-κB dimers and keep these dimers in the cytoplasm. The canonical NF-κB activation pathway by most of NF-κB-inducing stimuli activates the IκBα kinase (IKK)2 complex. The IKK complex is controlled by signal-induced phosphorylation of IKKα and IKKβ subunits (5) and signal-induced K63-linked ubiquitination of the regulatory subunit NEMO (6). The activated IKK complex in turn phosphorylates IκBα proteins on N-terminal conserved serine residues to target them for ubiquitination-dependent degradation (5). This process releases NF-κB and allows its translocation into the nucleus for the activation of its target genes (4). Although it has been shown that bacterial and viral infections induce IKK activation by Toll-like receptors (TLRs), the molecular mechanism by which fungal infection induces NF-κB activation is not fully defined.
Dectin-1 is a glycosylated type II transmembrane receptor and is mainly expressed in myeloid cells (7). It contains a single extracellular C-type lectin-like domain and a cytoplasmic domain containing an immunoreceptor tyrosine-based activation-like motif (7, 8). The ligand for Dectin-1 is β-glucan (9,10), a carbohydrate found in the cell wall of plant and fungi. Upon binding to β-glucan, Dectin-1 recruits and activates Syk (11, 12), an intracellular tyrosine kinase, through its immunoreceptor tyrosine-based activation-like motif, which triggers several intracellular signaling cascades leading to induction of various cytokines (10, 13, 14). In addition, it has been shown that Dectin-1 collaborates with TLRs to activate inflammatory responses following fungal infection (14). Therefore, it has been proposed that Dectin-1 functions as a pattern recognition receptor for fungal infection and mediates anti-fungus immune responses (8, 14, 15). Although the definite role of Dectin-1 in anti-fungus immunity remains to be fully determined (16, 17), the deficiency in human Dectin-1 expression results in a defect of mucosal anti-fungal defense (18). In addition, it has been shown that β-glucan on the surface of C. albicans is predominantly buried beneath a monoprotein coat upon transforming C. albicans into its hyphal form under the infection condition (19). Therefore, the β-glucan moiety on the cell wall of C. albicans is invisible for the host, suggesting that the host innate immunity is also induced by other components rather than β-glucan on the surface of C. albicans.
Dectin-2 is another C-type lectin-like receptor and is expressed in myeloid cells (20). In contrast to Dectin-1, Dectin-2 does not contain an immunoreceptor tyrosine-based activation-like motif in its cytoplasmic tail (20, 21). The extracellular carbohydrate-recognition domain of Dectin-2 appears to have specificity for high mannose such as Man9GlcNAc2 and Man8GlcNAc2 (22), suggesting that these molecules may be the ligand for Dectin-2. It has been suggested that Dectin-2 recruits Fcγ triggering intracellular signaling cascades, leading to activation of NF-κB upon encountering the hyphal form of C. albicans (23, 24). However, the molecular mechanism by which Dectin-2 mediates anti-fungus immunity is not fully characterized. In addition, the nature ligand for Dectin-2 and the signaling pathway induced by Dectin-2 remain to be determined.
CARD9 is an adaptor protein that contains an N-terminal caspase recruitment domain and a C-terminal coiled-coil domain and is mainly expressed in myeloid cells (25, 26). Recent studies demonstrate that CARD9 plays important roles against bacterial and fungal infection, and Card9-deficient mice are more susceptible to Listeria monocytogenes and C. albicans infection (26–28). More recently, it has been shown that human mutation in Card9 gene results in a defect in anti-fungal defense (29). Although the molecular mechanism by which CARD9 is involved in anti-fungal responses is not fully characterized, it has been shown that CARD9-deficient cells are defective in zymosan-induced NF-κB activation (27). Zymosan is a β-glucan, a component of yeast cell wall, and a ligand for Dectin-1 (9). Therefore, the current model for fungal infection-induced NF-κB activation is that fungal cell wall components, such as β-glucan, activate Dectin-1, and then Dectin-1 activates NF-κB through a Syk- and CARD9-dependent pathway (2, 27, 30).
In this study, we demonstrated that CARD9-deficient macrophages were defective for _C. albicans_-induced NF-κB activation. Our data indicate that NF-κB activation induced by live _C. albican_s is mainly through its hyphal form, which activates Dectin-2 but not Dectin-1. In contrast, NF-κB activation induced by zymosan, a β-glucan from yeast, is mainly dependent on Dectin-1 but not Dectin-2. Furthermore, our results indicate that CARD9 mediates Dectin-2-induced IKK ubiquitination, but not IKK phosphorylation, following Hyphae stimulation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Curdlan, zymosan, and LPS were purchased from Sigma. Piceatanol was purchased from Calbiochem. Antibodies to phospho-Syk (antibody 2701), Syk (antibody 2712), phospho-p38 (antibody 9211), p38 (antibody 9212), phospho-ERK (antibody 4307), phospho-IKKα/β (Ser180/181) (antibody 2681), phospho-IKKα/β (Ser176/180) (antibody 2697), and phospho-IκBα (antibody 9246) were from Cell Signaling Technology; antibodies for ERK (antibody sc-154), IKKα (antibody sc7218), IKKβ (antibody sc8014), Bcl10 (antibody sc5213), IKKγ (NEMO) (antibody sc8032), Ub (antibody sc8017), IκBα (antibody sc371), and α-actin (antibody sc8432) were from Santa Cruz Biotechnology. CARD9 antibody was described previously (26).
Mice and Cell Cultures
CARD9 and Bcl10 null mice were described previously (26, 31), and MyD88 null mice were kindly provided by Dr. S. Akira (32). They were housed under specific pathogen-free conditions in the M. D. Anderson Cancer Center animal facility. All of the animal experiments were performed in compliance with the institutional guidelines and according to the protocol approved by the Institutional Animal Use and Care Committee of the University of Texas M. D. Anderson Cancer Center. RAW264.7 and THP-1 cells were cultured in RPMI 1640 medium with 100 unit/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum. Primary cultures of bone marrow-derived macrophages (BMDMs) were prepared as previously described (26).
Fungus Strains
C. albicans (SC5314) was kindly provided by Dr. Michael C. Lorenz (Department of Microbiology and Molecular Genetics, University of Texas Medical School, Houston, TX). A single colony of C. albicans was grown overnight at 30 °C in yeast peptone dextrose medium. For preparing the hyphal form, washed C. albicans was resuspended in RPMI 1640 complete medium and grown for 3 h or were heat-inactivated by boiling for 45 min. After washing in phosphate-buffered saline twice, the hyphae and heat-inactivated yeast were used for stimulation.
Knockdown of Dectin-1 and Dectin-2
Lentiviral vectors for Dectin-1 and Dectin-2 shRNA were purchased from Sigma. Infectious lentiviral particles were prepared by co-transfection of shRNA plasmids together with packaging plasmids into HEK293T cells according to the manufacturer's protocol. Mouse bone marrow cells were collected and infected with viral stocks at day 1 and again at day 2. The infected bone marrow cells were cultured to develop into BMDMs as previously described (26). Lentiviral knockdown efficiency by each shRNA was analyzed by quantitative PCR. The most efficient shRNA was used to knock down Dectin-1 and Dectin-2 in mouse bone marrow cells. THP-1 and RAW264.7 cells were infected with lentivirus encoding Dectin-2 or Dectin-1 together with lentivirus-encoding green fluorescent protein. The infection rate was analyzed by flow cytometry.
Cytokine Measurements
The enzyme-linked immunosorbent assay kit for mouse tumor necrosis factor α and IL-12p40 was purchased from eBioscience, and that for mouse IL-10 was from R & D Biosciences. All of the samples were measured in triplicate according to the manufacturer's protocol.
Western Blot and Immunoprecipitation
BMDMs were serum-starved for overnight, stimulated, and lysed in lysis buffer (150 mm NaCl, 50 mm HEPES, pH 7.4, 1 mm EDTA, 1% Nonidet P-40, protease inhibitors). The cell lysates were immunoprecipitated with indicated antibody-conjugated agarose. The resulting immunoprecipitates and lysates were subjected to SDS-PAGE and then blotted using the indicated antibodies.
Electrophoretic Mobility Shift Assay (EMSA)
BMDMs were stimulated with various inducers, and nuclear extracts were prepared after the stimulation. Nuclear extracts (5–10 μg) were incubated for 15 min at room temperature with 32P-labeled NF-κB or Oct-1 probe (Promega). The mixtures were then separated by 6% PAGE and viewed by autoradiography.
RESULTS
CARD9 and Bcl10 Are Required for NF-κB Activation Induced by C. albicans but Not by Zymosan
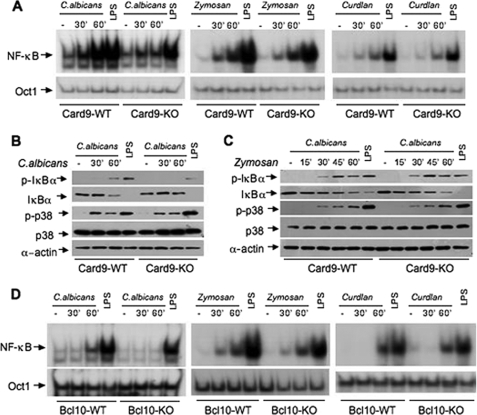
A recent report indicates that CARD9 is involved in the immune response against fungal infection and suggests that CARD9 plays an essential role in zymosan-induced NF-κB activation (27). To further investigate the molecular mechanism by which CARD9 is involved in anti-fungal responses, we directly examined the NF-κB activation induced by C. albicans. We found that C. albicans could effectively activate NF-κB in wild-type macrophages, but this activation was defective in CARD9-deficient macrophages (Fig. 1A). However, we surprisingly found that zymosan could activate NF-κB in both wild-type and CARD9-deficient macrophages (Fig. 1A), suggesting that zymosan can still activate NF-κB through a CARD9-independent mechanism. Although it has been suggested that CARD9 may be differentially required for zymosan-induced signaling (33), we found that zymosan could effectively induce NF-κB activation in both wild-type and CARD9-deficinet bone marrow-derived dendritic cells (supplemental Fig. S1). Consistent with these results, C. albicans failed to induce IκBα phosphorylation and degradation in CARD9-deficient macrophages (Fig. 1B), whereas zymosan could effectively induce IκBα phosphorylation and degradation in both wild-type and CARD9-deficient macrophages (Fig. 1C). To further examine the requirement of CARD9 on β-glucan-induced NF-κB activation, we stimulated wild-type and CARD9-deficient macrophages with curdlan, another β-glucan that was shown to activate cytokine production in a CARD9-dependent manner (30), and found that curdlan could also effectively activate NF-κB in wild-type and CARD9-deficient macrophages (Fig. 1A). Because zymosan is the ligand for Dectin-1, which can also collaborate with TLRs to activate inflammatory responses (13), we stimulated wild-type and MyD88 knock-out macrophages with zymosan and then examined NF-κB activation. We found that zymosan could effectively activate NF-κB in both of these cells (supplemental Fig. S2), whereas LPS failed to activate NF-κB in MyD88-deficient cells, suggesting that zymosan may be able to use a CARD9- and MyD88-independent pathway to activate NF-κB.
FIGURE 1.
CARD9 and Bcl10 are required for NF-κB activation induced by C. albicans but not activation induced by zymosan and curdlan. A, wild-type (WT) and Card9−/− (KO) BMDMs were stimulated with C. albicans (MOI = 1), zymosan (50 μg/ml), curdlan (50 μg/ml), or LPS (100 ng/ml) for the indicated times. The nuclear extracts were prepared from these cells and subjected to EMSA using 32P-labeled NF-κB or Oct-1 probe. B and C, WT and Card9−/− (KO) BMDMs were stimulated with live C. albicans (MOI = 1) and LPS (100 ng/ml) (B) or zymosan (50 μg/ml) and LPS (100 ng/ml) (C) for the indicated times. The cell lysates were subjected to immunoblotting analysis using the indicated antibodies. D, WT and Bcl10−/− (KO) BMDMs were stimulated with C. albicans (MOI = 1), zymosan (50 μg/ml), curdlan (50 μg/ml), or LPS (100 ng/ml) at the indicated times. The nuclear extracts were prepared from these cells and subjected to EMSA using 32P-labeled NF-κB or Oct-1 probe. A set of representative results from three independent experiments was presented.
Because CARD9 associates with Bcl10 (25), another CARD-containing protein that is also involved in anti-fungal immunity (27), we examined whether Bcl10 is required for _C. albicans_- and β-glucan-induced NF-κB activation. Similarly, we found that _C. albicans_-induced NF-κB activation (Fig. 1D) and IκBα phosphorylation and degradation (supplemental Fig. S3) were defective in Bcl10-deficient macrophages. However, both zymosan and curdlan could effectively induce NF-κB activation in Bcl10-deficient macrophages (Fig. 1D). Together, these results indicate that CARD9 and Bcl10 are required for _C. albicans_-induced NF-κB activation, but β-glucan-induced NF-κB activation is not fully dependent on CARD9 and Bcl10.
C. albicans Induces Two Independent Pathways Leading to NF-κB Activation
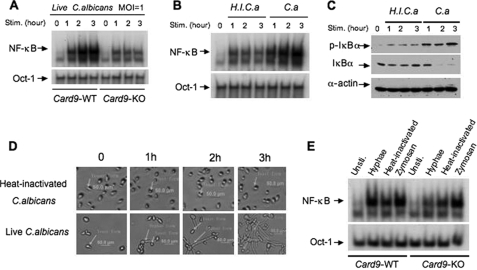
To determine how CARD9 is involved in _C. albicans_-induced NF-κB activation, we examined NF-κB activation following C. albicans stimulation at different time points. Interestingly, we found that although _C. albicans_-induced NF-κB activation was significantly defective in CARD9-deficient macrophages (Fig. 2A), a low level of activated NF-κB could still be induced in these cells (Fig. 2A). Because C. albicans switches from its usual unicellular yeast-like form (Yeast) into the invasive, multicellular filamentous forms, hyphae, under vegetative growth condition during infection, the host immune system mainly encountered the hyphal form of C. albicans (Hyphae) in vivo. To distinguish whether NF-κB is mainly induced by the multicellular hyphae or by the yeast-like, unicellular form, we blocked the hyphal transformation by heat-inactivating C. albicans. We then challenged bone marrow-derived macrophages with live or heat-inactivated C. albicans. Although the nuclear translocation of NF-κB (Fig. 2B) and IκBα phosphorylation (Fig. 2C) could be induced by the heat-inactivated form, which mainly consists of unicellular yeast form (Fig. 2D, top panels), the level of this induction was relatively low and did not further increase after 1 h of stimulation. In contrast, the level of NF-κB activation was continuously increased with the time following the stimulation by live C. albicans, which was correlated with the increased formation of hyphae during this period of time (Fig. 2D, bottom panels). Therefore, this result suggests that the time-dependent increase of NF-κB nuclear translocation may be caused by the newly formed hyphae.
FIGURE 2.
Both hyphal and yeast forms of C. albicans can induce NF-κB activation. A, wild-type (WT) and _Card9_−/− BMDMs were stimulated with C. albicans at MOI = 1 for the indicated time points. Nuclear extracts were prepared and subjected to EMSA using 32P-labeled NF-κB and Oct-1 probes. B, wild-type BMDMs were stimulated with heat-inactivated C. albicans (H.I.C.a) or live C. albicans (C.a) for the indicated time points. The nuclear extracts were prepared and subjected to EMSA as in A. C, cell lysates from the samples in B were subjected to immunoblotting analysis using the indicated antibodies. The results are representative of three independent experiments. D, the morphological changes of both heat-inactivated and live C. albicans in the condition for mammalian cell culture at the indicated time points were examined with microscopy. E, wild-type and _Card9_−/− BMDMs stimulated with hyphae or the heat-inactivated form of C. albicans at MOI = 1 or zymosan (50 μg/ml) for 1 h. The nuclear extracts were prepared from these cells and subjected to EMSA using 32P-labeled NF-κB and Oct-1 probes. Stim., unstimulated; Unsti., unstimulated; KO, knock-out.
To determine the requirement of CARD9 for the NF-κB activation induced by the heat-inactivated yeast or hyphal form of C. albicans, we challenged wild-type or CARD9-deficient macrophages with hyphal or heat-inactivated C. albicans, as well as zymosan. We found that CARD9 deficiency mainly affected the NF-κB activation induced by Hyphae but not by heat-inactivated yeast or zymosan (Fig. 2E). Consistent with this result, Hyphae failed to induce IκBα degradation in CARD9-deficient macrophages (supplemental Fig. S4). This defect is specific for NF-κB signaling, because CARD9 deficiency did not affect Hyphae-induced ERK activation (supplemental Fig. S4).
Dectin-2 Mediates Signaling Induced by C. albicans Hyphae, but Dectin-1 Responds to Heat-inactivated C. albicans and Zymosan
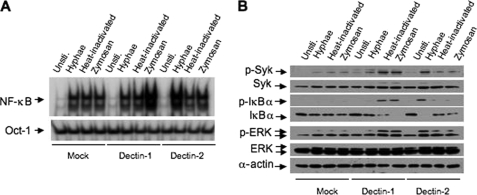
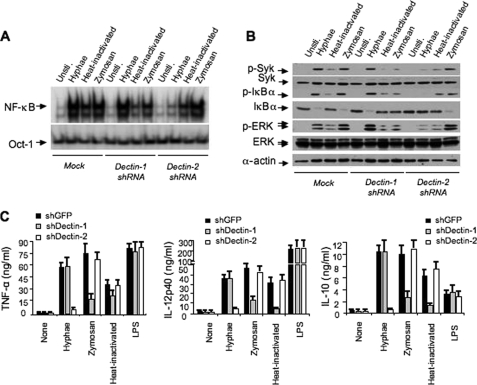
It has been suggested that Dectin-2 mediates C. albicans hyphae-induced NF-κB activation, whereas Dectin-1 is also shown to be involved in _C. albicans_-induced NF-κB activation. To confirm that hyphae mainly utilize Dectin-2 to activate NF-κB, we used shRNA specific for Dectin-1 or Dectin-2 (supplemental Fig. S5) to knock down these receptors in murine bone marrow-derived macrophages. Although inhibiting the expression of Dectin-1 partially suppressed the NF-κB activation induced by zymosan and heat-inactivated C. albicans, it had no inhibitory effects for Hyphae-induced NF-κB activation (see Fig. 4A). In contrast, inhibiting the expression of Dectin-2 significantly suppressed Hyphae-induced NF-κB activation but without any effect on zymosan- or heat-inactivated _C. albicans_-induced NF-κB activation (Fig. 3A). Consistent with these results, phosphorylation and degradation of IκBα by zymosan were sensitive to the Dectin-1 but not Dectin-2 knockdown, whereas Hyphae-induced IκBα phosphorylation and degradation were sensitive to the Dectin-2 but not Dectin-1 knockdown (Fig. 3B). Together, these results suggest that Dectin-2 is selectively involved in Hyphae-induced signaling, whereas Dectin-1 is involved in β-glucan and heat-inactivated yeast-induced signaling.
FIGURE 4.
Expression of Dectin-2 significantly enhanced C. albicans hyphae-induced NF-κB activation. A, THP-1 cells stably expressing Dectin-1, Dectin-2, or control vector were stimulated with C. albicans hyphae, C. albicans yeast, or zymosan for 1 h. The nuclear extracts were prepared and subjected to EMSA as described in A. B, lysates prepared from samples in the A were subjected to immunoblotting analysis using the indicated antibodies. Unsti., unstimulated.
FIGURE 3.
Dectin-2 but not Dectin-1 is required for NF-κB activation induced by C. albicans hyphae. A, BMDMs with stable knockdown of Dectin-1, Dectin-2, or green fluorescent protein (shGFP) control (Mock) were stimulated with hyphae or heat-inactivated (Yeast) C. albicans (MOI = 1), or zymosan (50 μg/ml) for 1 h. The nuclear extracts were prepared and subjected to EMSA using 32P-labeled NF-κB or Oct-1 probes. B, lysates prepared from the cells of A were subjected to immunoblotting analysis using the indicated antibodies. C, the levels of tumor necrosis factor tumor necrosis factor α (_TNF_-α), IL-12p40, and IL-10 in the supernatants from BMDMs with stable knockdown of Dectin-1, Dectin-2, or vector control were stimulated with C. albicans hyphae (MOI = 1), C. albicans yeast (MOI = 1), zymosan (50 μg/ml), or LPS (100 g/ml) overnight (error bars, standard deviation of triplicate each sample). Unsti., unstimulated.
To determine the physiological outcome of inhibiting Dectin-1 and Dectin-2 expression, we measured the cytokine production in macrophages with Dectin-1 or Dectin-2 knockdown (Fig. 3C). With Dectin-1 knockdown, it inhibited the expression of tumor necrosis factor α, IL-12, and IL-10 following zymosan or heat-inactivated C. albicans but not Hyphae stimulation (Fig. 3C). In contrast with Dectin-2 knockdown, it specifically inhibited the expression of these cytokines following Hyphae stimulation (Fig. 3C). These inhibitory effects were highly specific for fungal infection, because inhibiting the expression of Dectin-1 or Dectin-2 did not have any effect on LPS-induced cytokine production (Fig. 3C).
Because the expression levels of Dectin-1 and Dectin-2 in THP-1 and RAW264.7 cells are significantly lower than those in BMDMs (supplemental Fig. S6_A_) and the level of NF-κB activation in these cells was also lower than that in BMDMs in response to C. albicans challenging (supplemental Fig. S6_B_), we expressed Dectin-1 or Dectin-2 in THP-1 and RAW264.7 cells by lentiviral infection (supplemental Fig. S7). We found that expression of Dectin-1 significantly enhanced zymosan-induced NF-κB activation in both THP-1 cells (Fig. 4A) and RAW264.7 cells (supplemental Fig. S8). Although it was weaker than zymosan, this expression of Dectin-1 also slightly enhanced heat-inactivated _C. albicans_-induced NF-κB activation in these cells (Fig. 4A and supplemental Fig. S8). In contrast, expression of Dectin-2 selectively enhanced Hyphae-induced NF-κB activation (Fig. 4 and supplemental Fig. S8). Consistently, phosphorylation and degradation of IκBα induced by zymosan and heat-inactivated C. albicans were increased when Dectin-1 was expressed in THP-1 cells (Fig. 4B), whereas expression of Dectin-2 enhanced Hyphae-induced IκBα phosphorylation and degradation (Fig. 4B). Together, these results further confirm that Dectin-2 is selectively involved in Hyphae but that zymosan-induced NF-κB activation is at least partially dependent on Dectin-1. Altogether, these results demonstrate that Dectin-2 is the pattern recognition molecule for the hyphal form of C. albicans.
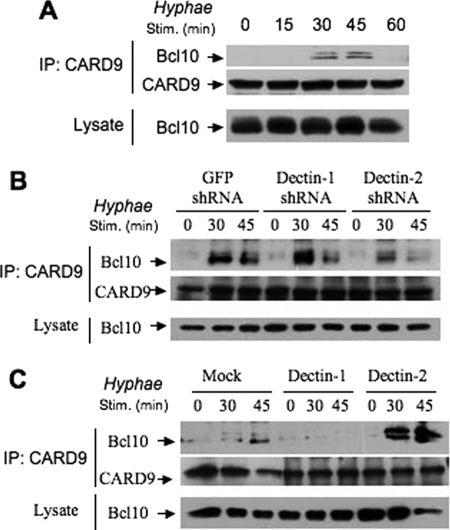
Dectin-2 Mediates a CARD9-Bcl10 Complex Formation Following Hyphae Stimulation
Because CARD9 is required for Hyphae-induced NF-κB activation, we next examined whether hyphae could activate the CARD9-dependent signaling pathway. CARD9 is an adaptor protein, and its main function is likely to recruit downstream signaling components. Therefore, we examined the complex formation of CARD9 with other proteins and found that CARD9 inducibly formed a complex with Bcl10 following Hyphae stimulation (Fig. 5A). To determine whether the inducible formation of the CARD9-Bcl10 complex is dependent on Dectin-2, we used the bone marrow-derived macrophages with Dectin-1 or Dectin-2 knockdown and found that suppressing the expression of Dectin-2 but not Dectin-1 inhibited the CARD9-Bcl0 complex formation following Hyphae stimulation (Fig. 5B). To further confirm that Dectin-2 mediates Hyphae-induced activation of CARD9, we examined the inducible formation of the CARD9-Bcl10 complex in THP-1 cells or THP-1 cells with overexpression of Dectin-1 or Dectin-2 (Fig. 5C). We found that overexpression of Dectin-2, but not Dectin-1, significantly enhanced the Hyphae-induced CARD9-Bcl10 association (Fig. 5C). Together, these results demonstrate that Dectin-2 links Hyphae-induced signaling to CARD9.
FIGURE 5.
Dectin-2 is required for the CARD9-Bcl10 interaction upon the stimulation by C. albicans hyphae. A, wild-type BMDMs were challenged with C. albicans hyphae at the indicated time points. The cell lysates were collected and subjected to the immunoprecipitation (IP) using CARD9 antibodies. The precipitates and lysates were subjected to immunoblotting analysis using the indicated antibodies. B, BMDMs with stable knockdown of Dectin-1, Dectin-2, or vector control were stimulated with C. albicans hyphae for the indicated time points. Cell lysates from these samples were subjected to immunoprecipitation analysis as in A. C, THP-1 cells stably expressing Dectin-1, Dectin-2, or its control vector were challenged with C. albicans hyphae at the indicated time points. Immunoprecipitation analysis was performed as in A and B.
CARD9 and Syk Mediate Hyphae-induced Signaling Cascade through Independent Pathways
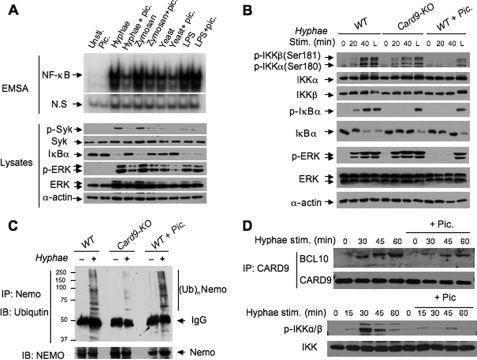
Syk is a key kinase for phagocytic cell activation, and recent studies suggested that Syk may also function downstream of Dectin-2 and upstream of CARD9 (24). To determine how Syk is involved in the _C. albicans_-induced NF-κB activation, we used the Syk inhibitor piceatanol to determine whether Syk is involved in Hyphae-induced NF-κB activation, and we treated the BMDMs with Syk inhibitors. We found that Syk inhibitor (Fig. 6A) specifically inhibited NF-κB activation and IκBα degradation induced by Hyphae and zymosan but not by LPS (Fig. 6A). These data indicate that Syk functions downstream from both Dectin-1 and Dectin-2.
FIGURE 6.
CARD9 and Syk function independently regulates IKK phosphorylation and ubiquitination. A, wild-type (WT) BMDMs were stimulated with C. albicans hyphae (Hyphae), zymosan (50 μg/ml), heat-inactivated C. albicans (Yeast), or LPS (100 ng/ml) for 1 h with or without pretreatment of the Syk inhibitor piceatanol for 0.5 h. The nuclear extracts were prepared and subjected to EMSA using 32P-labeled NF-κB or Oct-1 probe. The cell lysates were subjected to immunoblotting analysis using the indicated antibodies. B, wild-type and _Card9_−/− (KO) BMDMs, as well as wild-type BMDMs pretreated with piceatanol, were stimulated with C. albicans hyphae (MOI = 1) or LPS (L) for the indicated time points. The cell lysates from these samples were prepared and subjected to immunoblotting analysis using the indicated antibodies. C, wild-type and _Card9_−/− (KO) BMDMs and wild-type BMDMs pretreated with piceatanol were stimulated with C. albicans hyphae (MOI = 1). The cell lysates from these samples were subjected to immunoprecipitation using NEMO/IKKγ antibodies. The immunoprecipitates and cell lysates were subjected to immunoblot analysis using ubiquitin or NEMO antibodies. D, wild-type BMDMs were treated with or without piceatanol and then challenged with C. albicans hyphae at the indicated time points. The cell lysates were collected and subjected to the immunoprecipitation using CARD9 antibodies. The precipitates and lysates were subjected to immunoblotting analysis using the indicated antibodies. Unsti., unstimulated; Pic., piceatanol.
Our above data demonstrate that CARD9 is selectively involved in the Dectin-2 pathway, whereas Syk is required for NF-κB activation induced by Dectin-1 and Dectin-2. Therefore, we hypothesized that CARD9 and Syk might function independently. To test this hypothesis, we examined the different effect of CARD9 deficiency and inhibition of Syk on Hyphae-induced signaling. Interestingly, we found that although both CARD9 deficiency and inhibiting Syk resulted in the blocking of IκBα phosphorylation and degradation induced by Hyphae (Fig. 6B), inhibiting Syk also blocked Hyphae-induced IKKα/β phosphorylation, whereas CARD9 deficiency did not affect this signal-induced phosphorylation (Fig. 6B). In addition, inhibiting Syk but not CARD9 deficiency also resulted in inhibition of downstream ERK activation (Fig. 6B).
It has been proposed that NEMO, the regulatory subunit of the IKK complex, undergoes a signal-induced ubiquitination (6). We next examined the effect of CARD9 deficiency or Syk inhibition on NEMO-associated ubiquitination. Interestingly, we found that CARD9 deficiency blocked NEMO-associated ubiquitination, but Syk inhibition had no effect on this ubiquitination (Fig. 6C), suggesting that Syk and CARD9 do not function in a linear arrangement in a signaling pathway. Consistent with this hypothesis, we found that inhibition of Syk did not significantly affect the inducible association of CARD9 with Bcl10 but inhibited IKK phosphorylation following hyphal stimulation (Fig. 6D). Together, these results challenge the existing model of Syk function upstream of CARD9 in the Dectin-1 signaling pathway (1, 2, 8). Instead, our studies suggest a new model for _C. albicans_-induced NF-κB activation (Fig. 7) in which CARD9 and Syk mediate ubiquitination and phosphorylation of the IKK complex, respectively.
FIGURE 7.
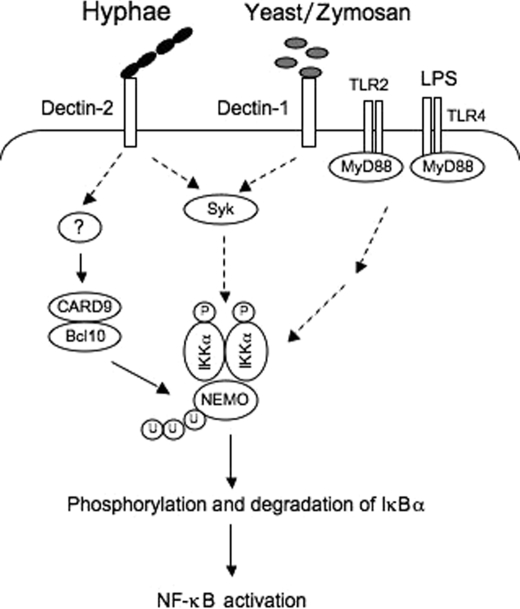
The model for _C. albicans_-induced signaling pathways leading to NF-κB activation. The hyphal form of C. albicans recognizes Dectin-2, whereas yeast and β-glucans, such as zymosan, activate Dectin-1. Although both Dectin-1 and Dectin-2 pathways are required for Syk that regulates IKK phosphorylation, CARD9 and Bcl10 are selectively involved in Dectin-2-induced signaling cascades leading to regulation of NEMO ubiquitination. The phosphorylation and ubiquitination of IKK in the Dectin-2-induced signaling pathway are required for activation of the IKK complex for NF-κB activation. LPS and other TLR ligands activate NF-κB through a MyD88-dependent but CARD9-independent pathway.
DISCUSSION
In this study, we have demonstrated that C. albicans can induce NF-κB activation through two independent pathways. One pathway is induced by the yeast-like, unicellular form of C. albicans through recognizing Dectin-1 to activate downstream signaling cascades, whereas the other pathway is induced by the hyphal form of C. albicans through recognizing Dectin-2 to activate downstream signaling cascades. Although Syk is required for both the Dectin-1 and Dectin-2 pathways, CARD9 is mainly involved in the Dectin-2 pathway. Our data further demonstrate that Syk and CARD9 function independently with Syk regulating IKK phosphorylation and CARD9 regulating IKK ubiquitination (Fig. 7). Therefore, our study proposes a new model for fungi-induced NF-κB activation in which C. albicans infection mainly induces NF-κB through a Dectin-2-CARD9 signaling cascade, instead of the previously proposed model that C. albicans infection induces NF-κB through the Dectin-1-Syk-CARD9 signaling cascade.
Fungi such as C. albicans are rapidly transformed from their yeast forms to the hyphal forms under the vegetative growth condition, such as in condition for mammalian cell culture or after infecting into mammalian bodies. After they are transformed to hyphae, the β-glucan in the surface of yeast cell wall is mainly buried by other components (19), such as monosylated glycoproteins (Mannan) (1). Therefore, it is likely that macrophages recognize the Mannan moiety on their surface of Hyphae and lead to activation of inflammatory signaling cascades. Our findings that Dectin-2 is required for Hyphae-induced NF-κB activation and cytokine production are consistent with previous observations that Dectin-2 recognizes Man9GlcNAc2 with a high affinity (22) and that C. albicans contains Man9GlcNAc2 on its cell wall (34). Although our preliminary studies indicate that Man9GlcNAc2 is not sufficient to activate NF-κB (data not shown), it will be interesting to determine whether Man9GlcNAc2 structure is required for hyphae-mediated NF-κB.
Our data showing that CARD9 is required for the inflammatory response induced by the live or hyphal form of C. albicans but not by heat-inactive C. albicans suggest that the CARD9-dependent inflammatory response induced by C. albicans is not induced by β-glucan in the cell wall of C. albicans. Consistent with this notion, we have found that β-glucans, such as zymosan and curdlan, induce NF-κB through a CARD9-independent pathway. This result appears to directly contradict a previous report that zymosan-induced NF-κB is dependent on CARD9 (27). In our studies, we have used both EMSA and immunoblotting analysis and demonstrated that the nuclear translocation of NF-κB and IκB phosphorylation/degradation is not defective in CARD9-deficient macrophages. Although we cannot explain the discrepancy regarding the role of CARD9 in zymosan-induced NF-κB activation between our results and those reported by Gross et al. (27), one possible explanation is that Gross et al. used bone marrow-derived dendritic cells, whereas we used bone marrow-derived macrophages. However, in our studies, we observed only a slight difference of NF-κB activation in wild-type and CARD9 knock-out dendritic cells (supplemental Fig. S1).
Recently, Robinson et al. (24) reported that Dectin-1 and Dectin-2 appear to function in a redundant manner for anti-fungi immunity (24). Their data also suggest that Syk functions downstream of both Dectin-1 and Dectin-2 but upstream of CARD9. Our data support the notion that Syk functions downstream of both Dectin-1 and Dectin-2. However, our results argue that Dectin-1 and Dectin-2 function independently in response to different ligands from C. albicans. In particular, we find that inhibiting Dectin-2 expression completely blocks hyphae-induced NF-κB and cytokine production. The difference between our findings versus those of Robinson et al. is likely due to the fact that Robinson et al. used Dectin-2 antibodies to trigger the endocytosis of surface Dectin-2, whereas we used Dectin-2 shRNA to block Dectin-2 expression. During the reviewing process of our manuscript, Saijo et al. (35) published a paper demonstrating that Dectin-2-deficient mice but not Dectin-1-deficient mice are highly susceptible to C. albicans infection.
Our results also show that CARD9 is inducibly associated with Bcl10 following stimulation of the Dectin-2-dependent pathway, which provides direct evidence that CARD9 functions downstream of Dectin-2. However, it remains to be determined what molecule(s) link CARD9 to Dectin-2 (Fig. 7). In addition, we find that CARD9 deficiency significantly affects signal-induced IKK ubiquitination, whereas inhibition of Syk activity blocks the signal-induced IKK phosphorylation, suggesting that CARD9 and Syk may not work in a linear arrangement in the signaling pathway following C. albicans stimulation.
Although our data showing that suppressing Dectin-2 expression blocks hyphae-induced NF-κB activation clearly demonstrate that Dectin-2 is an essential component of the receptor for mediating C. albicans hyphae-induced NF-κB activation, we cannot rule out the possibility that other receptors, such as TLRs, may also participate in mediating C. albicans hyphae-induced NF-κB activation. Thus, future studies will need to determine whether Dectin-2 is the only component or whether it also forms a complex with other receptors to mediate hyphae-induced NF-κB activation.
In summary, several important conclusions can be drawn from our study. First, C. albicans induces the host-inflammatory response mainly through its hyphal form. Second, unknown components other than β-glucan on the hyphal surface are recognized by Dectin-2 on host cells. The stimulation of Dectin-2 by hyphae induces at least two independent signaling cascades in which the Syk-dependent cascade regulates IKK phosphorylation, whereas the CARD9-dependent cascade controls IKK ubiquitination. Thus, phosphorylation and ubiquitination of the IKK complex induce its kinase activity and leads to phosphorylation and degradation of IκBα and activation of NF-κB (Fig. 7). Together, our study reveals a more detailed molecular mechanism of fungal infection-induced inflammatory response and provides potential targets for designing therapeutic agents against fungus infection.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Drs. D. Underhill, S. Akira, and M. Lorenz for reagents.
*
This work was supported, in whole or in part, by National Institutes of Health Grants RO1AI050848, RO1GM079451, and R01GM065899 (to X. L.). This work was also supported in part by a Graduate Student Fellowship from the China Scholarship Council (to L. B.).
2
The abbreviations used are:
IKK
IκBα kinase
TLR
Toll-like receptor
LPS
lipopolysaccharide
ERK
extracellular signal-regulated kinase
BMDM
bone marrow-derived macrophage
shRNA
small hairpin RNA
IL
interleukin
EMSA
electrophoretic mobility shift assay
MOI
multiplicity of infection.
REFERENCES
- 1.Netea M. G., Brown G. D., Kullberg B. J., Gow N. A. (2008) Nat. Rev. Microbiol. 6, 67–78 [DOI] [PubMed] [Google Scholar]
- 2.Robinson M. J., Sancho D., Slack E. C., LeibundGut-Landmann S., Reis e Sousa C. (2006) Nat. Immunol. 7, 1258–1265 [DOI] [PubMed] [Google Scholar]
- 3.Willment J. A., Brown G. D. (2008) Trends Microbiol. 16, 27–32 [DOI] [PubMed] [Google Scholar]
- 4.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 5.Karin M., Ben-Neriah Y. (2000) Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 6.Chen Z. J. (2005) Nat. Cell Biol. 7, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ariizumi K., Shen G. L., Shikano S., Xu S., Ritter R., 3rd, Kumamoto T., Edelbaum D., Morita A., Bergstresser P. R., Takashima A. (2000) J. Biol. Chem. 275, 20157–20167 [DOI] [PubMed] [Google Scholar]
- 8.Reid D. M., Gow N. A., Brown G. D. (2009) Curr. Opin. Immunol. 21, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown G. D., Taylor P. R., Reid D. M., Willment J. A., Williams D. L., Martinez-Pomares L., Wong S. Y., Gordon S. (2002) J. Exp. Med. 196, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown G. D., Herre J., Williams D. L., Willment J. A., Marshall A. S., Gordon S. (2003) J. Exp. Med. 197, 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Underhill D. M., Rossnagle E., Lowell C. A., Simmons R. M. (2005) Blood 106, 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers N. C., Slack E. C., Edwards A. D., Nolte M. A., Schulz O., Schweighoffer E., Williams D. L., Gordon S., Tybulewicz V. L., Brown G. D., Reis e Sousa C. (2005) Immunity 22, 507–517 [DOI] [PubMed] [Google Scholar]
- 13.Gantner B. N., Simmons R. M., Canavera S. J., Akira S., Underhill D. M. (2003) J. Exp. Med. 197, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gantner B. N., Simmons R. M., Underhill D. M. (2005) EMBO J. 24, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown G. D. (2006) Nat. Rev. 6, 33–43 [DOI] [PubMed] [Google Scholar]
- 16.Saijo S., Fujikado N., Furuta T., Chung S. H., Kotaki H., Seki K., Sudo K., Akira S., Adachi Y., Ohno N., Kinjo T., Nakamura K., Kawakami K., Iwakura Y. (2007) Nat. Immunol. 8, 39–46 [DOI] [PubMed] [Google Scholar]
- 17.Taylor P. R., Tsoni S. V., Willment J. A., Dennehy K. M., Rosas M., Findon H., Haynes K., Steele C., Botto M., Gordon S., Brown G. D. (2007) Nat. Immunol. 8, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferwerda B., Ferwerda G., Plantinga T. S., Willment J. A., van Spriel A. B., Venselaar H., Elbers C. C., Johnson M. D., Cambi A., Huysamen C., Jacobs L., Jansen T., Verheijen K., Masthoff L., Morré S. A., Vriend G., Williams D. L., Perfect J. R., Joosten L. A., Wijmenga C., van der Meer J. W., Adema G. J., Kullberg B. J., Brown G. D., Netea M. G. (2009) New Engl. J. Med. 361, 1760–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler R. T., Fink G. R. (2006) PLoS Pathog. 2, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ariizumi K., Shen G. L., Shikano S., Ritter R., 3rd, Zukas P., Edelbaum D., Morita A., Takashima A. (2000) J. Biol. Chem. 275, 11957–11963 [DOI] [PubMed] [Google Scholar]
- 21.Graham L. M., Brown G. D. (2009) Cytokine 48, 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGreal E. P., Rosas M., Brown G. D., Zamze S., Wong S. Y., Gordon S., Martinez-Pomares L., Taylor P. R. (2006) Glycobiology 16, 422–430 [DOI] [PubMed] [Google Scholar]
- 23.Sato K., Yang X. L., Yudate T., Chung J. S., Wu J., Luby-Phelps K., Kimberly R. P., Underhill D., Cruz P. D., Jr., Ariizumi K. (2006) J. Biol. Chem. 281, 38854–38866 [DOI] [PubMed] [Google Scholar]
- 24.Robinson M. J., Osorio F., Rosas M., Freitas R. P., Schweighoffer E., Gross O., Verbeek J. S., Ruland J., Tybulewicz V., Brown G. D., Moita L. F., Taylor P. R., Reis e Sousa C. (2009) J. Exp. Med. 206, 2037–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertin J., Guo Y., Wang L., Srinivasula S. M., Jacobson M. D., Poyet J. L., Merriam S., Du M. Q., Dyer M. J., Robison K. E., DiStefano P. S., Alnemri E. S. (2000) J. Biol. Chem. 275, 41082–41086 [DOI] [PubMed] [Google Scholar]
- 26.Hsu Y. M., Zhang Y., You Y., Wang D., Li H., Duramad O., Qin X. F., Dong C., Lin X. (2007) Nat. Immunol. 8, 198–205 [DOI] [PubMed] [Google Scholar]
- 27.Gross O., Gewies A., Finger K., Schäfer M., Sparwasser T., Peschel C., Förster I., Ruland J. (2006) Nature 442, 651–656 [DOI] [PubMed] [Google Scholar]
- 28.Hara H., Ishihara C., Takeuchi A., Imanishi T., Xue L., Morris S. W., Inui M., Takai T., Shibuya A., Saijo S., Iwakura Y., Ohno N., Koseki H., Yoshida H., Penninger J. M., Saito T. (2007) Nat. Immunol. 8, 619–629 [DOI] [PubMed] [Google Scholar]
- 29.Glocker E. O., Hennigs A., Nabavi M., Schäffer A. A., Woellner C., Salzer U., Pfeifer D., Veelken H., Warnatz K., Tahami F., Jamal S., Manguiat A., Rezaei N., Amirzargar A. A., Plebani A., Hannesschläger N., Gross O., Ruland J., Grimbacher B. (2009) New Engl. J. Med. 361, 1727–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeibundGut-Landmann S., Gross O., Robinson M. J., Osorio F., Slack E. C., Tsoni S. V., Schweighoffer E., Tybulewicz V., Brown G. D., Ruland J., Reis e Sousa C. (2007) Nat. Immunol. 8, 630–638 [DOI] [PubMed] [Google Scholar]
- 31.Xue L., Morris S. W., Orihuela C., Tuomanen E., Cui X., Wen R., Wang D. (2003) Nat. Immunol. 4, 857–865 [DOI] [PubMed] [Google Scholar]
- 32.Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 115–122 [DOI] [PubMed] [Google Scholar]
- 33.Goodridge H. S., Shimada T., Wolf A. J., Hsu Y. M., Becker C. A., Lin X., Underhill D. M. (2009) J. Immunol. 182, 1146–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mora-Montes H. M., López-Romero E., Zinker S., Ponce-Noyola P., Flores-Carreón A. (2004) Glycobiology 14, 593–598 [DOI] [PubMed] [Google Scholar]
- 35.Saijo S., Ikeda S., Yamabe K., Kakuta S., Ishigame H., Akitsu A., Fujikado N., Kusaka T., Kubo S., Chung S. H., Komatsu R., Miura N., Adachi Y., Ohno N., Shibuya K., Yamamoto N., Kawakami K., Yamasaki S., Saito T., Akira S., Iwakura Y. (2010) Immunity 32, 681–691 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data