Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells (original) (raw)
Abstract
Human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) are promising candidate cell sources for regenerative medicine. However, despite the common ability of hiPSCs and hESCs to differentiate into all 3 germ layers, their functional equivalence at the single cell level remains to be demonstrated. Moreover, single cell heterogeneity amongst stem cell populations may underlie important cell fate decisions. Here, we used single cell analysis to resolve the gene expression profiles of 362 hiPSCs and hESCs for an array of 42 genes that characterize the pluripotent and differentiated states. Comparison between single hESCs and single hiPSCs revealed markedly more heterogeneity in gene expression levels in the hiPSCs, suggesting that hiPSCs occupy an alternate, less stable pluripotent state. hiPSCs also displayed slower growth kinetics and impaired directed differentiation as compared with hESCs. Our results suggest that caution should be exercised before assuming that hiPSCs occupy a pluripotent state equivalent to that of hESCs, particularly when producing differentiated cells for regenerative medicine aims.
Introduction
Human induced pluripotent stem cells (hiPSCs) can be derived from differentiated somatic cells by a reprogramming process involving overexpression of a key set of transcription factors (1, 2). hiPSCs behave similarly to human embryonic stem cells (hESCs), with respect to their self-renewal and pluripotent potential in vitro and in vivo, and can therefore be used to generate large quantities of differentiated cells needed for regenerative medicine applications (3). Comparison of the gene expression profiles of hiPSCs and hESCs has revealed a globally similar pattern, with significant upregulation of key pluripotency maintenance network genes, such as Oct4 (also known as Pou5f1), Nanog, Sox2, and DNMT3B.
A growing body of evidence suggests that cell populations, particularly stem cells, do not comprise a homogenous cellular entity either in vitro or in vivo (4). Rather, stem cells display an inherent heterogeneity at the molecular level, which underlies the probabilistic element of their fate determination (5–13). Single cell analysis has aided our ability to scrutinize the dynamic fluctuations in gene expression changes among seemingly homogenous cell populations in other settings (7, 12, 14), but single cell analysis of hiPSCs has not been previously reported (see Supplemental Discussion; supplemental material available online with this article; doi:10.1172/JCI44635DS1). Here, we use a microfluidic platform to resolve the gene expression profiles of single hiPSCs for an array of 28 genes known to characterize the pluripotent state of hESCs as well as 14 genes associated with differentiated lineages. Such comparisons between hiPSCs and hESCs will be critical prior to deciding which cell type is the most promising for various regenerative medicine applications.
Results
Heterogeneity in gene expression levels is much greater among hiPSCs than amongst hESCs.
Three well-established hESC lines, namely H7, H9, and HES2, were selected to form a reference standard against which to compare hiPSCs. Four hiPSC lines were used, originating from either IMR90 dermal fibroblasts or human adipose stromal cells (hASCs), using lentiviral transgenesis or nonviral minicircle-based reprogramming (Supplemental Table 1). Consistent with previous reports (2, 15, 16), the hiPSC populations used in this study meet criteria typically used to define pluripotency (Supplemental Figure 1). They expressed high levels of pluripotency-related transcripts and cell surface antigens, generated teratomas containing all 3 germ layers, and expressed globally similar microarray profiles. These analyses were performed using cell populations, like most currently available gene expression data analysis, and mask important differences between individual cells.
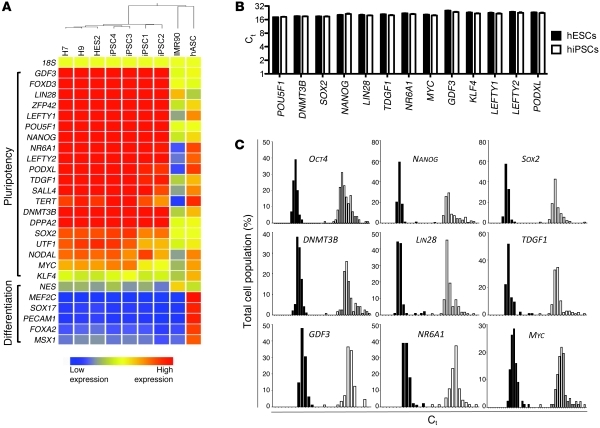
After validating the ability of our microfluidic platform to detect transcript levels in single cells (Supplemental Figure 2), we isolated a total of 282 single, viable hESCs and hiPSCs using flow cytometry and analyzed the mRNA levels of a key set of 28 pluripotency-related transcripts (see Supplemental Discussion). Pluripotency-related transcripts at the single cell level were highly expressed in the pluripotent stem cells relative to the differentiated cells (Figure 1A). The Ct values of key pluripotency-related transcripts were nearly identical between hESCs and hiPSCs when assessed in aggregate by calculation of the geometric mean, indicating equivalent gene expression levels in the cell types when regarded as a population (Figure 1B). However, an analysis of the frequency distribution allowed us to visualize the heterogeneity in the single cells, as reflected by the horizontal spread of a histogram plot. Interestingly, the variation in transcript levels was demonstrably higher in hiPSCs than in hESCs for a number of key pluripotency-related transcripts (Figure 1C and Supplemental Figure 3). We computed the variance in our sample populations and statistically determined that the difference in variance between hESC and hiPSC transcript levels was significant for a number of key regulators of the pluripotency maintenance network, such as OCT4, SOX2, NANOG, and GDF3 (Supplemental Figure 4 and Supplemental Table 2). Interestingly, single hiPSCs did not segregate according to donor cell line during any of our analyses (Supplemental Figure 5), indicating that the variation in gene expression levels was not dependent on derivation technique (viral vs. nonviral) or starting cell source (IMR90 vs. hASCs). Heterogeneity in transcript expression levels could not be attributed to differences in passage number, variable expression of Dlk1-Dio3 locus (Supplemental Figure 7), or altered cell cycle regulation (Supplemental Figure 8 and Supplemental Discussion).
Figure 1. Gene expression profiling of 282 single hESCs and hiPSCs.
(A) An array showing that pluripotency (top) and lineage (bottom) marker transcripts are differentially expressed in pluripotent stem cell types (H7, H9, HES2, hiPSC1–4) versus somatic cell types (IMR90 and hASCs). (B) Comparison of the Ct values obtained by single cell qRT-PCR shows that pluripotency transcript levels are equivalent when considering the cell populations as a whole (geometric mean ± SD). (C) Population distribution plots (horizontal axis represents Ct value; vertical axis represents percentage of total cell population) reveal that single hiPSCs (white bars) display considerable heterogeneity in transcript expression levels in comparison with that of single hESCs (black bars).
We also assayed hESCs and hiPSCs for 14 transcripts associated with the differentiated state. Most transcriptional markers of differentiation, such as PAX6, AFP, and T (also called Brachyury), were rarely detected amongst undifferentiated hESCs and hiPSCs (Supplemental Figure 8). Coexpression of lineage-related transcripts alongside pluripotency markers suggests reversible lineage priming toward a particular cell fate, as has been observed in multipotent hematopoietic progenitors (17–19). Coexpression of ectodermal lineage markers, such as nestin (NES) and microtubule associated protein-2 (MAP2), was observed relatively often in undifferentiated hESCs and hiPSCs, suggesting lineage priming toward the ectodermal fate under feeder-free culture conditions. Priming toward the ectodermal fate was most likely due to the high concentrations of bFGF, a known neurotrophic factor, in the culture media (20).
Gene expression profiling of single cells isolated on the basis of surface antigen expression or colony position.
The cell surface antigens Tra-1-60 and SSEA-4 have been previously reported as potential markers of bona fide hiPSCs (21). We sought to better define this subtype of pluripotent stem cell using FACS for high Tra-1-60 and SSEA-4 antigen expression levels (Supplemental Figure 9). We selected a total of 84 single, viable Tra-1-60+/SSEA-4+ hESCs and hiPSCs for gene expression profiling of an expanded array of pluripotency-related genes. After selection of cells based on this particular immunophenotype, lack of homogeneity in transcript levels among hESCs was still evident, but an even greater gradient of expression in pluripotency-related transcripts was present in the hiPSCs (Figure 2A). After applying principal components analysis to the data (see Supplemental Discussion), we observed that hESCs occupied a discrete focus with a small percentage of outliers within principal component space, while hiPSCs spanned a large area of the space with no discrete concentration (Supplemental Figure 10), demonstrating that even Tra-1-60+/SSEA-4+ hiPSCs exhibit considerable cell-to-cell variability in gene expression levels.
Figure 2. Immunophenotypic and positional variation in single cell gene expression.
(A) Gene expression profiling of single hiPSCs expressing the Tra-1-60+/SSEA-4+ immunophenotype. Heat map representations of gene expression levels in immunophenotyped hESCs and hiPSCs versus differentiated fibroblast cells (IMR90), with each column representing a single cell. Single hESCs (left) show minimal heterogeneity, while single hiPSCs (middle) show increased variability in comparison with 4 somatic IMR90 cells (right), with no segregation of cells according to cell line when subjected to a hierarchical clustering algorithm. A gradient of hiPSC expression is evident, with cells expressing low levels of pluripotency transcripts enriched to the right. (B and C) Positional variation in transcript expression levels within pluripotent stem cell colonies. (B) A positional gradient of expression is evident in both hESCs and hiPSCs, with lower expression of pluripotency transcripts observed in the periphery of the colony. (C) Expression levels of ectoderm (PAX6 and NES), early mesoderm (GATA4), and endoderm (SOX17) transcripts are uniform across hESC colonies. However, the periphery of hiPSC colonies has undergone relative downregulation of endoderm marker SOX17 and relative upregulation of ectoderm marker PAX6.
The hiPSCs exhibiting large variations in gene expression levels had been randomly chosen from dissociated colonies. We next sought to determine whether transcript levels were dependent on the cell’s positional context within the colony under feeder-free conditions. We isolated cells from the peripheral and central regions of hESC and hiPSC colonies grown under feeder-free conditions using 0.2-mm glass capillary micropipettes and observed downregulation of pluripotency-related transcripts toward the periphery of colonies (Figure 2B). Among detectable lineage-specific genes, such as NES, PAX6, SOX17, and GATA4, no significant positional variation in their expression was observed in the hESC colonies, although the hiPSC colonies did display a pattern suggestive of increased peripheral ectodermal lineage priming (Figure 2C). As expected, expression of housekeeping genes (GAPDH and 18S rRNA) did not vary significantly between the central and peripheral regions of either cell type.
Differentiation of hiPSCs is slow and inconsistent.
The in vivo teratoma formation assay is a commonly used test to definitively assay the pluripotent capacity of hESCs and hiPSCs. In order to track their growth in vivo, we stably transduced hESCs (H7) and hiPSCs (iPSC1) with a double fusion reporter gene construct expressing firefly luciferase and enhanced green fluorescent protein. After selection by FACS, the hESCs and hiPSCs stably expressed firefly luciferase, such that the bioluminescent signal intensity correlated linearly with the number of surviving cells (Supplemental Figure 11). We then analyzed the relative growth rate and time course of hiPSC-derived versus hESC-derived teratoma growth. When immunodeficient mice were injected with equal numbers of stably transduced hESCs and hiPSCs, both had well-formed teratomas, exhibiting all 3 germ layers, as shown by histology, at week 6 after implantation (Supplemental Figure 12). However, hiPSC-derived teratomas had much slower growth kinetics than hESC-derived teratomas (Figure 3, A and B), suggesting a relative deficit in their in vivo self-renewing capability (Supplemental Figure 13 and see Supplemental Discussion). By day 21 after transplantation, the bioluminescent signal intensity was significantly different between the 2 groups (5.73 × 106 ± 0.3 × 106 vs. 3.00 × 107 ± 1.2 × 107 photons/s/cm2/steradian for hiPSCs vs. hESCs, respectively; P < 0.05).
Figure 3. Limited growth and differentiation potential of hiPSC-derived cells.
(A) Representative bioluminescence images of immunocompromised SCID beige mice implanted with 106 hESCs or 106 hiPSCs stably expressing firefly luciferase reporter construct. (B) Quantitative analysis of bioluminescence imaging data shows slower teratoma growth kinetics of hiPSC-derived teratomas (mean ± SEM for n = 10). sr, steradian. (C) Immunostaining for cardiac troponin T (cTnT) of hESC- and hiPSC-derived cardiomyocytes (original magnification, ×20). (D) Assessment of the percentage of cell aggregates containing beating cardiomyocytes after 14 days shows substantial variation in the yield of hiPSC-CMs, while the yield of hESC-CMs is stable (mean ± SEM for n = 8). (E) Immunostaining for CD31 (left) and CD144 (top right) EC markers and robust LDL uptake (bottom right) for hESC-derived ECs after 14 days of differentiation before (bottom left) and after (top left and top right) FACS enrichment (original magnification, ×20). (F) Cell proliferation and CD31 expression over 2–3 weeks after isolation of EC populations show limited stability and proliferative capacity of hiPSC-ECs in comparison with hESC-ECs (mean ± SD for n = 4).
Next, we compared the response of hESCs and hiPSCs to directed differentiation protocols for the production of cells that may be of use in cardiac and vascular repair, namely beating cardiomyocytes and ECs. Using a well-established protocol, we found that hESCs from 3 lines differentiated robustly into hESC-derived cardiomyocytes (hESC-CMs), with 27.0% ± 3.6% of the cell aggregates containing beating cardiomyocytes (Supplemental Video 1). However, hiPSCs exhibited a variable response to identical differentiation cues, despite changing the cell line and inductive cytokine concentrations, with 11.2% ± 11.5% of the hiPSC-derived cell aggregates containing beating cardiomyocytes (hiPSC-CMs) (Figure 3, C and D). We also sought to assess the capability of hiPSCs to produce ECs, since hESC-derived ECs (hESC-ECs) have been previously demonstrated as being capable of in vitro expansion and in vivo vasculogenesis (22). However, hiPSC-derived ECs (hiPSC-ECs) had limited expansion capacity in vitro, suggesting early senescence, and exhibited a gradual loss of CD31 marker expression over a period of 2 weeks (Figure 3, E and F). In contrast, hESC-ECs were capable of robust in vitro expansion, with only minimal loss of CD31 marker expression (85.3% ± 6.32% for hESC-ECs vs. 28.2% ± 8.77% for hiPSC-ECs at day 21; P < 0.05).
In this study, we compared hESCs to hiPSCs that had been stringently validated in a variety of assays, including microarrays, quantitative PCR (qPCR), immunocytochemistry, karyotyping, and teratoma formation (Supplemental Figures 1, 7–9, 12, 14, and 15). Taken together, our data indicate that if hESCs are to be thought of as the “gold standard” pluripotent cells derived from the blastocyst, then hiPSCs are in vitro constructs that only partially recapitulate hESC biology. Consistent with previous reports, we show that hiPSC populations collectively maintain a similar gene expression profile to hESCs, express high levels of pluripotency-related transcripts and cell surface antigens, and are capable of generating endothelial progenitor cells, beating cardiomyocytes, and teratomas containing all 3 germ layers. However, hiPSC-derived cells also manifest important differences compared with their hESC-derived counterparts, which may limit their ultimate clinical applicability (Figure 3 and refs. 23, 24). The molecular mechanisms underlying the dissimilar growth and differentiation potential of hiPSCs and hESCs have remained elusive (see Supplemental Discussion).
Although mouse iPSCs can be tested in tetraploid blastocyst complementation assays, hiPSCs need to be evaluated according to molecular surrogates of their pluripotent and self-renewing capabilities. Robust quality control standards must be instituted to assign hiPSCs to “pluripotent grades” according to quantitative measurements of transcript and protein abundance. High-throughput microfluidic devices can enable rapid, efficient assessment of complex stem cell populations, such as hiPSCs. Quantitative single cell gene expression profiling of hESCs and comparison of hiPSCs to this reference standard are the initial steps toward achieving this goal. Here, we demonstrate that microfluidic single cell gene expression profiling can uncover instability in hiPSC gene expression profiles, which may significantly affect the ultimate clinical translatability of these exciting sources for cellular repair.
Methods
Cell culture.
hESC and hiPSC lines were cultured on Matrigel-coated tissue culture dishes (Growth Factor Reduced; BD Biosciences) with mTeSR-1 growth medium (Stem Cell Technologies). See Supplemental Methods for details.
Single cell gene expression profiling.
hESCs and hiPSCs were dissociated into single cells using Accutase (Sigma-Aldrich) and resuspended in PBS-based FACS buffer supplemented with 2% FBS (Gibco) before being passed through a cell strainer. Propidium iodide was added to the single cell suspension just prior to sorting using a FACS Aria (BD Biosciences) into 96-well 0.2-ml PCR plates containing buffers and enzymes for reverse transcription per manufacturer’s instructions (Fluidigm; see Supplemental Methods). All reactions were performed in duplicates or triplicates, and Ct values were directly used in data analysis after normalization to the 18S endogenous control gene. Any cells with an 18S endogenous control Ct value below 15 were discarded as cell debris (Supplemental Figure 2).
FACS.
See Supplemental Methods for details.
Teratoma formation and bioluminescence imaging.
See Supplemental Methods for details. All animal procedures were approved by the Administrative Panel on Laboratory Animal Care at Stanford.
Cardiomyocyte and EC differentiation.
See Supplemental Methods for details.
Statistics.
Gene expression data analysis was carried out using GeneSpring GX 11.0 software (Agilent). Test of homogeneity of variances was performed using an Ansari-Bradley 2-sample test. Data are shown as mean ± SD, except as indicated. Differences were considered significant at P < 0.05.
Supplementary Material
Supplemental data
Supplemental video 1
Acknowledgments
We would like to acknowledge funding support from NIH grants DP2OD004437, HL091453, HL 089027, AI085575, and AG036142; Edward Mallinckrodt Jr. Foundation (to J.C. Wu); NIH U01 HL099776 (to R.C. Robbins); the Howard Hughes Medical Institute (to K.H. Narsinh); and SNF grant PBBEP3 129803 (to V. Sanchez-Freire).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(3):1217–1221. doi:10.1172/JCI44635.
References
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Boheler KR. Pluripotency of human embryonic and induced pluripotent stem cells for cardiac and vascular regeneration. Thromb Haemost. 2010;104(1):23–29. doi: 10.1160/TH09-07-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T, Pera M, Peterson C, Andrews PW. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4(5):387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Franco CB, Chen C-C, Drukker M, Weissman IL, Galli SJ. Distinguishing mast cell and granulocyte differentiation at the single-cell level. Cell Stem Cell. 2010;6(4):361–368. doi: 10.1016/j.stem.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Wang CC, Holmberg KJ, Cabral K, Brugge JS. Identifying single-cell molecular programs by stochastic profiling. Nat Methods. 2010;7(4):311–317. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canham MA, Sharov AA, Ko MSH, Brickman JM. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8(5):e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T, et al. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7(7):e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135(5):909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103(47):17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods. 2006;3(10):807–815. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- Foygel K, et al. A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS One. 2008;3(12):e4109. doi: 10.1371/journal.pone.0004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A. 2009;106(37):15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7(3):197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11(6):774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Huang S, Guo Y-P, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007;305(2):695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Papapetrou EP, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A. 2009;106(31):12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, et al. Generation of dopaminergic neurons in vitro from human embryonic stem cells treated with neurotrophic factors. Neurosci Lett. 2004;359(1–2):99–103. doi: 10.1016/j.neulet.2004.01.073. [DOI] [PubMed] [Google Scholar]
- Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27(11):1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. Functional and transcriptional characterization of human embryonic stem cell-derived endothelial cells for treatment of myocardial infarction. PLoS One. 2009;4(12):e8443. doi: 10.1371/journal.pone.0008443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107(9):4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28(4):704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data
Supplemental video 1