A practical guide to evaluating colocalization in biological microscopy (original) (raw)
Abstract
Fluorescence microscopy is one of the most powerful tools for elucidating the cellular functions of proteins and other molecules. In many cases, the function of a molecule can be inferred from its association with specific intracellular compartments or molecular complexes, which is typically determined by comparing the distribution of a fluorescently labeled version of the molecule with that of a second, complementarily labeled probe. Although arguably the most common application of fluorescence microscopy in biomedical research, studies evaluating the “colocalization” of two probes are seldom quantified, despite a diversity of image analysis tools that have been specifically developed for that purpose. Here we provide a guide to analyzing colocalization in cell biology studies, emphasizing practical application of quantitative tools that are now widely available in commercial and free image analysis software.
Keywords: digital image analysis, fluorescence microscopy, confocal microscopy
fueled by developments in molecular biology, electronics, and chemistry, fluorescence microscopy has flourished in the past 30 years. One of the most common applications of fluorescence microscopy is to compare the subcellular distributions of two fluorescently labeled molecules. Such comparisons can be used to understand the function of a protein, as when the protein is found to colocalize with a marker of a particular organelle, or to understand intracellular transport, as when the protein is found to colocalize with a marker of a particular pathway. However, the data collected in these studies are seldom rigorously evaluated. Rather, cell biologists frequently treat colocalization as a subjective feature, using something like Potter Stewart's criterion for defining obscenity “I know it when I see it.” This practice persists despite a relatively large literature in methods of quantitative colocalization analysis.
Given the number of excellent reviews that have been published on the topic of colocalization analysis (7, 38, 50) and the absence of much that is fundamentally new for the past 20 years, a reader could justifiably wonder about the need for yet another review. (In fact, the reduced appetite for articles on the topic of colocalization is quantifiable; a literature search shows that while the number of biomedical review articles including the term “fluorescence microscopy” has increased exponentially since 1980, the number including the term “colocalization” started saturating at the turn of the millennium.) This article is predicated upon the proliferation of software tools for analyzing colocalization that have been implemented in image analysis software packages available to cell biologists (see partial listing in appendix). While it is laudable that these tools are now widely available to biomedical researchers, they are frequently only vaguely described. The goal of this article is to provide cell biologists with some guidance on how to select the most appropriate method for evaluating colocalization in their research and an appreciation of the factors that need to be considered for meaningful interpretation of colocalization studies. By using examples from our own research, this guide also emphasizes practical aspects of colocalization analysis that are seldom represented in the simulated data typically used to characterize colocalization methods.
For the purposes of this guide, we will assume that the investigator has conducted image collection appropriately for colocalization analysis so that the distribution of signal in each image is a reliable representation of the distribution of each probe in the sample. Thus the signal in each image is sufficient to distinguish from noise and background, uncontaminated by autofluorescence arising from the sample itself, and free of signal bleed-through between the two images. That said, noise is an inevitable feature of fluorescence microscopy, and image quality is necessarily limited for many interesting studies. Investigators should not be intimidated by methods that “require” high-quality images. Excellent descriptions of the various aspects of proper image collection are presented in Refs. 7, 38, 43, and 50.
What Do You Mean By Colocalization?
We will focus on how the analysis of the distribution of signals in fluorescence microscopy images can be used to determine whether two probes codistribute with one another. This approach is not appropriate for detecting molecular interactions; the resolution of the light microscope, even a “super-resolution” system, is simply insufficient to identify the physical apposition of two molecules through a comparison of their distributions in fluorescence images. Such studies require higher resolution techniques such as fluorescence resonance energy transfer or electron microscopy. Fluorescence colocalization analysis is more appropriately used to determine whether two molecules associate with the same structures; for example, to determine whether a particular protein associates with endosomes (4, 25, 26, 36, 42, 46, 49), mitochondria (31), or microtubules (6, 37) or whether two proteins associate with the same subnuclear structures (19, 33, 45) or with the same plasma membrane domains (27, 39). Evaluating colocalization at this scale is still susceptible to the limits of resolution; an overlap in fluorescence does not necessarily indicate colocalization of two probes in the same cellular structure. However, the observation of repeated coincidence of two probes in multiple structures throughout a cell increases the confidence that the two occupy the same structures. As described below, the codistribution of two probes in fluorescence microscope images may be evaluated visually, quantitatively, and statistically.
Colocalization can be thought of as consisting of two components: co-occurrence, the simple spatial overlap of two probes, and correlation, in which two probes not only overlap with one another but codistribute in proportion to one another within and between structures. In some cases, the distribution of two probes might be expected to overlap but not proportionally. For example, the fluorescently labeled cargo of an endosome would be expected to co-occur in the same vesicles labeled with a green fluorescent protein (GFP) chimera of an endocytic Rab protein, but there is no necessary reason that the amount of cargo should scale with the amount of the Rab protein. In other cases, the two probes might be expected to codistribute proportionally with one another so that the fluorescence levels of probes labeling each would be spatially correlated. An example of this would be two molecules that interact with the same molecular complexes. Throughout this guide, we will emphasize how the various methods used to measure colocalization differ in their sensitivity to these two components, and how this factor should be considered in choosing a colocalization metric.
Visual Methods For Evaluating Colocalization
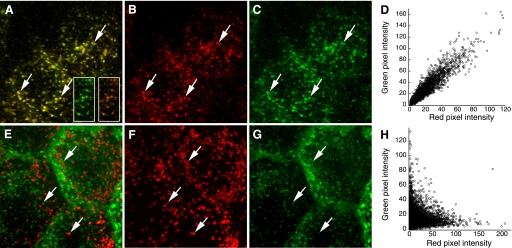
Colocalization of two probes may be subjectively identified by the appearance of structures whose color reflects the combined contribution of both probes when the images of each probe are superimposed (or “merged”). So, for example, colocalization of fluorescein and rhodamine can be apparent in structures that appear yellow, because of the combined contributions of green and red fluorescence, respectively. Figure 1_A_ shows a projected image volume of Madin Darby Canine Kidney (MDCK) cells incubated in a combination of Texas Red-labeled transferrin and Alexa 488-labeled transferrin. Since both probes are internalized via the same transferrin receptors, they would be expected to codistribute in endosomes following internalization, as is apparent in the constant yellow color of endosomes. In contrast, internalized Texas Red-dextran and Alexa 488-transferrin distribute to two distinct compartments, which appear red and green, respectively, in Fig. 1_E_.
Fig. 1.
Colocalization analysis of endocytic probes. A: maximum projection of an image volume of Madin Darby Canine Kidney (MDCK) cells following incubation with transferrin conjugated to Texas Red (B) and Alexa 488 (C) Arrows indicate endosomes containing both probes. D: scatterplot of red and green pixel intensities of the two cells shown in A. E: maximum projection of an image volume of MDCK cells following incubation in Texas Red-dextran (F) and Alexa 488-transferrin (G). Arrows indicate examples of lysosomes containing Texas Red-dextran. H: scatterplot of red and green pixel intensities of the image shown in E.
Superposition of fluorescence images is certainly the most prevalent method for evaluating colocalization, and tools for displaying multiple-channel fluorescence images as merged color images are implemented in all biological image analysis software. However, results can be ambiguous. The problem is that an intermediate color, indicating colocalization, is obtained only if the intensities of the two probes are similar. The insets in Fig. 1_A_ show how small changes in the relative intensity of two probes can completely alter the combined color of the endosomes and thus the perception of probe colocalization. For this reason, the overall degree of colocalization throughout a sample may be visually apparent only under very specific labeling conditions, when the fluorescence of the two probes occurs in a fixed and nearly equal proportion. In general, the most reliable method for visually comparing the relative distribution of two probes is a side-by-side comparison of the two images, with arrows provided as landmarks (compare the colocalization of the probes in Fig. 1, B and C, with that in Fig. 1, F and G).
The results of fluorescence colocalization studies can also be represented graphically in scatterplots where the intensity of one color is plotted against the intensity of the second color for each pixel, similar to the output provided for flow cytometry data. Under the conditions of proportional codistribution, such as in the data shown in Fig. 1_A_, the points of the scatterplot cluster around a straight line, whose slope reflects the ratio of the fluorescence of the two probes (Fig. 1_D_). In contrast, the lack of colocalization of dextran and transferrin in the image shown in Fig. 1_E_ is reflected by the distribution of points into two separate groups, each showing varying signal levels of one probe with little or no signal from the other probe (Fig. 1_H_). The ability to produce and export scatterplots is common to nearly all biological image analysis software packages.
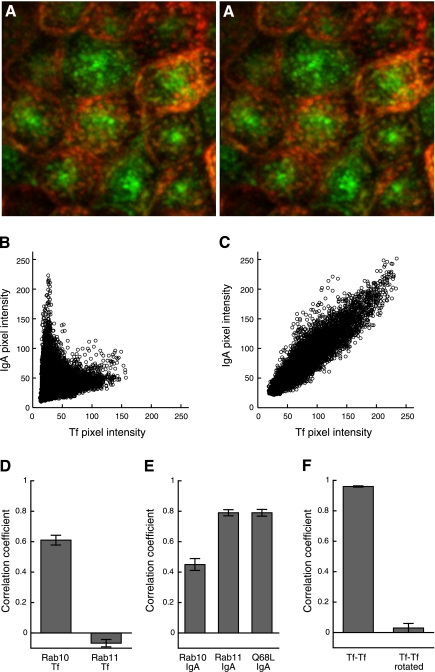
Scatterplots can provide additional insights into colocalization studies. First, they can be used to identify populations of distinct compartments. Our laboratory has used scatterplots to identify two populations of endosomes in MDCK epithelial cells, one at the apex that is enriched in internalized IgA and lacking internalized transferrin, and the other in lower portions of the cell that contains both IgA and transferrin (Fig. 2_A_) (9, 46). These two compartments are readily distinguished in scatterplots in which two different linear relationships are obtained, with the slopes reflecting the distinctive ratios of internalized IgA and transferrin in each type of compartment (Fig. 2_B_). The scatterplot obtained from images of cells treated with brefeldinA was used to support the observation that brefeldinA induced a fusion of these different compartments, resulting in a population of endosomes with a single, intermediate ratio of IgA to transferrin (Fig. 2_C_).
Fig. 2.
Colocalization analysis of endocytic probes. A: three-dimensional stereopair image of a maximum projected image volume MDCK cells following incubation with Oregon Green-IgA and Texas Red-transferrin (Tf). B: scatterplot of red and green pixel intensities of MDCK cells following incubation with Oregon Green-IgA and Texas Red-transferrin. C: scatterplot of individual red and green pixel intensities of brefeldinA-treated MDCK cells following incubation with Oregon Green-IgA and Texas Red-transferrin. D: Pearson's correlation coefficients (PCCs) of images of internalized Texas Red-transferrin and green fluorescent protein (GFP)-Rab10 or GFP-Rab11a in MDCK cells. E: PCCs of images of internalized Cy5-IgA and GFP-Rab10, GFP-Rab11a, or GFP-Rab10-Q68L in MDCK cells. F: PCCs of images of Texas Red-transferrin and Cy5-transferrin internalized by MDCK cells before and after rotating Cy5-Tf image by 90 degrees. A_–_C were reproduced with permission from the Company of Biologists (http://jcs.biologists.org), Wang EX et al. “Brefeldin A rapidly disrupts plasma membrane polarity by blocking polar sorting in common endosomes of MDCK cells.” J Cell Sci 114: 3309–3321, 2001. D_–_F were reproduced and adapted with permission from The American Society for Cell Biology, from Babbey CM et al. “Rab10 regulates membrane transport through early endosomes of polarized Madin-Darby Canine Kidney cells.” Molec Biol Cell 17: 3156–3175, 2006. (www.molbiolcell.org) (4).
The visual techniques described above are useful for exploring the relative distribution of different molecules in cells. Superposition of images is useful for providing a spatial sense of colocalization, identifying regions of the cell or compartments where molecules colocalize. Scatterplots are useful for detecting the presence of different populations of compartments. They also provide a qualitative indication of the degree of colocalization. However, these representations are generally not helpful for comparing the degree of colocalization in different experimental conditions nor for determining whether the amount of colocalization exceeds random coincidence. In the next sections we will describe several approaches that can be used to quantify colocalization. These methods are simple to employ and have been implemented in a variety of image processing software packages. However, there are numerous subtleties and assumptions in each that must be understood before they can be productively applied to biological images.
Quantifying Colocalization
Pearson's correlation coefficient.
The discussion of scatterplots above suggests the use of Pearson's correlation coefficient (PCC) as a statistic for quantifying colocalization. The formula for PCC is given below for a typical image consisting of red and green channels.PCC=∑i(Ri−R¯)×(Gi−G¯)∑i(Ri−R¯)2×∑i(Gi−G¯)2where R i and G i refer to the intensity values of the red and green channels, respectively, of pixel i, and R̄ and Ḡ refer to the mean intensities of the red and green channels, respectively, across the entire image. PCC values range from 1 for two images whose fluorescence intensities are perfectly, linearly related, to −1 for two images whose fluorescence intensities are perfectly, but inversely, related to one another. Values near zero reflect distributions of probes that are uncorrelated with one another. So, for example, PCC measures 0.944 in the image of two different colors of transferrin internalized into the top cell shown in Fig. 1_A_, whereas PCC measures only −0.045 for the image of internalized transferrin and dextran in the top cell in Fig. 1_E_. The square of PCC (generally denoted as _R_2) is also known as the “coefficient of determination,” a statistic that estimates the fraction of variability in G that can be explained by its linear regression with R. Thus, for the two colors of transferrin, 89% of the variability of Alexa 488-transferrin fluorescence can be explained by the variability of Texas Red transferrin fluorescence. In contrast, only 0.2% of the variability in Alexa 488-transferrin fluorescence is explained by variability in Texas Red dextran fluorescence.
Formulated in 1896 by mathematician/eugenicist Karl Pearson (40) and characterized for use in fluorescence microscopy nearly 100 years later by Manders (33), PCC is a statistic of which its primary advantage for colocalization analysis is its elegant simplicity. PCC measures the pixel-by-pixel covariance in the signal levels of two images. Because it subtracts the mean intensity from each pixel's intensity value, PCC is independent of signal levels and signal offset (background). Thus PCC can be measured in two-color images without any form of preprocessing, making it both simple and relatively safe from user bias. Tools for quantifying PCC are provided in nearly all image analysis software packages.
Whereas the meaning of extreme values of PCC is generally clear, intermediate values are more difficult to interpret, except when used in comparative studies. Our laboratory has used PCC in a number of studies as a means of characterizing different endocytic pathways. For example, quantitative comparisons of the distributions of Rab10, Rab11, and internalized transferrin and immunoglobulin A (IgA) were used to identify two types of endocytic compartments in polarized MDCK cells: one predominantly containing internalized transferrin and associated with Rab10 (Fig. 2_D_) and one containing internalized IgA and associated with Rab11 (Fig. 2_E_) (4). The same analysis showed that a single point mutation in Rab10 altered its distribution such that it matched Rab11. Whenever possible, the appropriate performance of a colocalization analysis should be demonstrated with positive and negative controls. In the studies described above, the positive control of the colocalization analysis was provided by evaluation of cells that had internalized two different colors of transferrin. The negative control was provided by quantifying PCC for the same images, but after rotation of one by 90 degrees, a condition in which only random colocalization is observed (Fig. 2_F_).
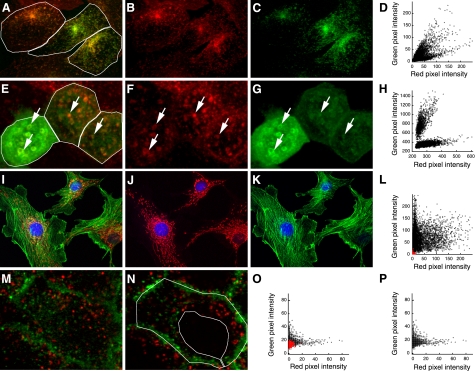
In many image analysis software packages, PCC is measured for entire images by default. However, PCC should generally be measured for individual cells, which can be accomplished by hand-drawing a “region of interest” (ROI) over the image. The issue here is that, since PCC values depend upon a simple linear relationship, they will be depressed if measured over a field of cells with heterogeneous expression or uptake of the target molecules, thus under-representing the degree of correlation. For example, we find that transferrin and IgA are both internalized into the same early endosomes of Chinese hamster ovary (CHO) cells transfected with transferrin receptor and polymeric Ig receptor (Fig. 3, A_–_C). Indeed, the two are internalized in a constant proportion that reflects the relative number of transferrin and polymeric Ig receptors expressed by the cell. However, because the numbers of transferrin and polymeric Ig receptors expressed varies between cells, the ratio of transferrin to IgA internalized likewise varies between cells, an effect that is apparent in the different color of the endosomes in the merged color image. The different ratios of transferrin to IgA are even clearer in the combined scatterplot of the three cells, which shows three different linear relationships (Fig. 3_D_). Accordingly, whereas the PCC for each cell in the field is relatively high (0.88, 0.85, and 0.89 in the indicated ROIs), the scatter in the combined data is such that PCC is reduced to 0.66 when measured over the whole image, under-representing the high degree of correlation between the two probes.
Fig. 3.
Importance of the region of interest (ROI) for measuring PCC. A: maximum projection of an image volume of transfected Chinese hamster ovary (CHO) cells following incubation with Texas Red-transferrin (B) and Oregon Green-IgA (C). D: combined scatterplot of red and green pixel intensities of the three cells in the indicated ROIs of A, showing 3 different linear relations. E: image of a single plane of polarized MDCK cells incubated in Texas Red-transferrin (F), 3 of which express GFP-Rab10 at different levels (G). Arrows indicate endosomes labeled for both transferrin and Rab10. H: combined scatterplot of red and green pixel intensities of the three ROIs shown in E, showing 2 different linear relations. I: single image plane of fluorescently labeled actin (J) and mitochondria in bovine endothelia (K). L: scatterplot of red and green pixel intensities of the image shown in I. Values from pixels outside the cells are shown in red. M: image of a single plane from an MDCK cell following incubation in Texas Red-dextran (red) and Alexa 488-transferrin (green). N: as in M but an image plane passing through the nucleus. O: scatterplot of red and green pixel intensities of the middle cell shown in N. Values from pixels from the region of the nucleus are shown in red. P: scatterplot of red and green pixel intensities from the cytosolic region of the middle cell, delineated with the ROI shown in N.
The problem of cell-cell variability is particularly pervasive in studies of cells transiently expressing GFP chimeras, since expression of the transfected protein can vary widely between cells. Figure 3_E_ shows a field of MDCK cells transiently transfected with GFP-Rab10 following internalization of fluorescently labeled transferrin. The internalized transferrin (Fig. 3_F_) colocalizes extensively with GFP-Rab10 (Fig. 3_G_), but the high expression of GFP-Rab10 in the cell at the left of the field results in a high ratio of green to red fluorescence when compared with the two cells on the right. As with the example above, the differences are more apparent in the scatterplot of the combined pixel data, which shows two different linear relationships (Fig. 3_H_). As a consequence, while measurement of the PCC in each cell indicates a reasonably strong correlation (0.69, 0.56, and 0.57 in the indicated ROIs), the PCC of the entire image measures only 0.07.
Although it may not seem intuitive, unlabeled extracellular regions (sometimes confusingly called “background pixels”) can artificially inflate PCC values if included in the region of measurement. This effect results from the fact that these empty areas contain pixels for which both the red and green signals are significantly below their average levels. This point is demonstrated by an analysis of the image shown in Fig. 3_I_, an immunofluorescence image of mitochondria (red, Fig. 3_J_) and filamentous actin (green, Fig. 3_K_). Quantification of PCC values over the region occupied by the cells indicates a poor association between the distribution of actin and mitochondria (PCC = 0.16). However, when quantified over the entire image, including empty areas between the cells, PCC increases to 0.39. By simply including pixels from extracellular regions that lack significant amounts of either red or green signal, the PCC of two essentially uncorrelated probes is significantly increased. The influence of extracellular pixels obviously depends on their number. For the example described here, more than one-third of the pixels used in this correlation arise from outside the cell (depicted in red in the scatterplot shown in Fig. 3_L_). This error, while seemingly obvious, is deceptively easy to commit. For the incautious investigator, measuring PCC over an entire field of cells labeled in this way would result in PCC values that varied inversely with cell plating density.
The examples above demonstrate that carefully outlining the region in which two probes may potentially distribute is critical to accurate measurement of PCC. However, it may not be enough to simply outline the cell; there may be regions within the cell that exclude the structures of interest. Consider the example of two probes that associate with vesicles. Insofar as intracellular vesicles are excluded from the nucleus, their mutual exclusion from the nucleus is no more meaningful than their mutual exclusion from the extracellular space. If pixels from these regions are included in analysis, they will artificially inflate PCC measurements, in the same way that pixels from the extracellular space artificially inflate PCC measurements. Figure 3_M_ shows an image of an MDCK cell in which lysosomes have been labeled with fluorescent dextran and endosomes with fluorescent transferrin. The two probes label distinct compartments, as reflected in a PCC of 0.04 for the cell at the center of this image. However, if we examine an image collected in a focal plane that includes a cross-section of the nucleus (Fig. 3_N_), from which both probes are excluded, PCC increases to 0.16. The scatterplot shown in Fig. 3_O_ shows that the points measured from the region of the nucleus (shown in red) cluster near the origin, artificially increasing the linear relationship in the data. If we measure PCC in an ROI that excludes the nuclear region, PCC returns to a measurement similar to that obtained in non-nuclear planes (0.06).
An alternative approach for excluding irrelevant pixels is to restrict analysis to pixels whose intensity falls above a threshold value. This technique may involve a process in which a region of the scatterplot (also, confusingly called the ROI) is identified as reflecting “background” and the pixels whose intensities fall in this range are omitted from analysis. This widely implemented method can be much simpler than the laborious process of manually outlining an ROI on the original image. However, eliminating low-intensity pixels runs the risk of eliminating regions of mutual exclusion within the cells which, if they represent areas in which the probes could potentially distribute, are meaningful to quantifications of probe distributions. For the example shown in Fig. 3_N_, pixel intensities of the nuclear region (shown in red in Fig. 3_O_) superimpose over a population of pixel intensities from the cytosol (shown in Fig. 3_P_). Thus an intensity-based procedure that eliminated the pixels of the nuclear region would have the undesired effect of eliminating pixels from the relevant cytosolic region as well. Thus, when using an intensity-based approach for eliminating irrelevant pixels, it is crucial to evaluate its effect on relevant pixels.
Interpreting PCC measurements: should your colocalization data be measured by their fit to a linear relationship?
To productively use PCC to measure colocalization, it is important that investigators understand exactly what PCC measures. PCC quantifies the degree to which the variability in red and green pixel intensities can be explained with a simple, linear relationship between the two. Thus it is sensitive to both signal co-occurrence (the degree to which, for each pixel, red and green intensity values are either both above background or both below background) and the more rigorous condition of signal correlation (pixel-for-pixel proportionality in the signal levels of the two channels). To the degree that the biology of the system is such that can be modeled by a linear relationship in the levels of two probes, PCC is an appropriate measure of association. However, there are many biological conditions in which this simple model is inadequate, in which cases PCC measurements only indirectly reflect probe colocalization.
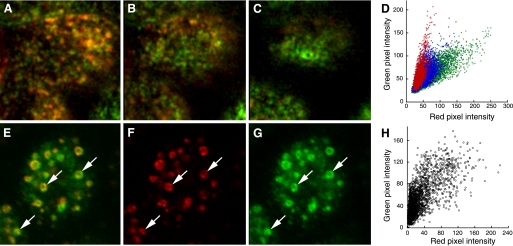
First, insofar as PCC measures fit to a single linear relationship, it provides a poor measure of colocalization in more complex situations, such as when probes co-occur in different proportions in different compartments of the cell. Much like the examples of PCC quantification in heterogeneous cells, PCC values will be depressed if measured in a population of heterogeneous intracellular compartments. For example, as mentioned previously, we have found that transfected MDCK cells internalize transferrin and IgA into the same set of endosomes from which IgA is then sorted to an apical recycling endosome. As a consequence, transferrin and IgA occur at similar concentrations in early endosomes, but the concentration of transferrin is much reduced relative to IgA in the downstream compartment. These compartments are arrayed along an apical-basal axis such that early endosomes captured in medial planes of the cell (Fig. 4_A_) are gradually replaced by apical recycling endosomes as one shifts the focus to the tops of the cells (Fig. 4, B and C). The change in the ratio of IgA to transferrin is shown not only in the change in the color of the overlaid images but also in the scatterplots of the three regions, which show the relationship of the two probes in the apical planes in red, that of the medial planes in green and the intermediate planes in blue (Fig. 4_D_). The presence of two kinds of compartments, with a ninefold difference in the proportion of IgA to transferrin, generates scatter in a two-dimensional scatterplot that cannot be explained with a simple linear model, resulting in a PCC value that under represents the degree of colocalization (in this case, 0.66 for the entire volume of the top cell in the field). In situations such as this, where the data are more complex than modeled by linear regression, PCC measurements are ambiguous, if not misleading.
Fig. 4.
Colocalization without a simple linear relationship. A: medial focal plane of MDCK cells incubated with Texas Red-transferrin (red) and Oregon Green IgA (green). B: as in A but collected 1.2 μm higher. C: as in A but collected 2.4 μm higher. D: scatterplots of red and green pixel intensities of the top cell collected from the focal plane shown in A (green), B (blue), or C (red). E: single image plane of an MDCK cell immunolabeled for EEA1 (F), expressing GFP-RhoB (G). Arrows indicate a few examples of compartments labeled with both probes. H: scatterplot of red and green pixel intensities of the cell shown in E.
As discussed previously, even if two probes co-occur on the same cellular structures, there may be no reason that they should co-occur in fixed proportion to one another. For the situation shown in Fig. 1_A_, in which two transferrin conjugates are internalized and trafficked in proportion to their concentrations, PCC provides an excellent metric of colocalization. However, for studies in which proportional codistribution is not necessarily expected, PCC can provide a poor measure of colocalization. An example of such a case is shown in the image of immunolocalized EEA1 (red) in MDCK cells expressing GFP-RhoB (green), shown in Fig. 4_E_. Although the two probes distribute to the same intracellular compartments (compare Fig. 4, F and G), the ratio of the two probes varies considerably within and between structures, as evidenced by the varying color of the combined color image and the extensive spread in the scatterplot (Fig. 4_H_). Thus, despite the extensive overlap between the two, PCC is relatively low (0.73).
Thus for investigators primarily interested in quantifying probe overlap, PCC is an equivocal measure of colocalization; depending upon proportionality, extensively overlapping probes may yield either high or low PCC values. The influence of proportionality on PCC can be directly demonstrated with the following example. Reducing the background levels in the image shown in Fig. 1_A_ to zero results in an image with a PCC of 0.92. If we then convert all of the non-zero values to a constant value, thus removing the correlation of signal levels within and between labeled structures, but not reducing overlap, PCC is reduced to 0.70. Thus two images with the same amount of signal overlap can have dramatically different PCC values; in one case explaining 85% of the variability, in the other explaining less than half.
As with the visual inspection of color-merged images, the researcher needs to be clear as to whether probe colocalization in a particular study is expected to be accurately modeled by a single linear relationship. To the degree that the signal levels of two probes are not predicted to be linearly related, and thus that the investigator is interested in probe occurrence alone, PCC provides an indirect and sometimes poor metric of colocalization.
At the opposite extreme, it has also been argued that colocalization should only quantify pixel-by-pixel correlation in the subset of pixels that contain both fluorophores (2). Limiting measurements to pixels that contain both probes profoundly changes the parameter that is measured by PCC. If one is interested in evaluating the distribution of a probe, the regions from which the probe is excluded are as meaningful as those in which the probe is found (as long as these regions are potentially accessible to the probe). In the case of analyzing cellular distributions, if one limits colocalization analysis to pixels that contain both probes, the PCC is changed from a measure of probe codistribution within the cell to one addressing probe codistribution within the structures cohabited by the probes.
As expected, this procedure yields high values of PCC for probes with proportional codistributions. For the image of two colors of transferrin internalized into endosomes of MDCK cells, shown in Fig. 1_A_, PCC measures 0.94 if measured for all pixels in the image and 0.88 if measured only for pixels that contain both red and green signals above background levels. In contrast, this approach yields low PCC values under conditions in which probes overlap but not in a fixed proportion. For the image of immunolocalized EEA1 and GFP-RhoB shown in Fig. 4_E_, PCC is reduced from 0.73 to 0.45 if measured only in pixels containing signal from both probes.
One significant complication that arises in applying this technique is that it requires methods to classify pixels as either containing or lacking a fluorescent probe. While this might seem simple, the process of distinguishing signal from background is frequently a complex problem, as described later.
Manders overlap coefficient.
In response to the perceived difficulty of interpreting negative PCC values, an alternative but closely related metric, the Manders Overlap Coefficient (MOC) (34), was developed. MOC is described by the equation:MOC=∑i(Ri×Gi)∑iRi2×∑iGi2
MOC is implemented in image analysis software packages, such as Colocalizer Pro, Image-Pro, Imaris, and Volocity and can be implemented in ImageJ via the JACoP plugin.
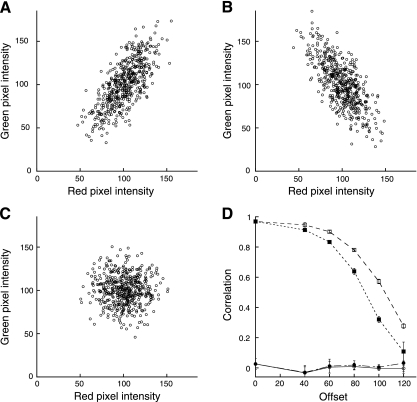
Eliminating the subtraction of mean signals from the equation has the effect of preventing negative MOC values, which may be reassuring to investigators confused by negative measurements of correlation. However, it has a variety of other consequences that are arguably more confusing. Figure 5, A_–_C, shows examples of data that are positively correlated, negatively correlated, and uncorrelated, with PCC values of 0.73, −0.71, and −0.03, respectively. Whereas an MOC value of 0.99 is obtained for the positively correlated data of Fig. 5_A_, a similar value is obtained for the negatively correlated data of Fig. 5_B_ (0.92) and even for the uncorrelated data of Fig. 5_C_ (0.97). While this is at first confusing (if not troubling), this behavior reflects the fact that, unlike PCC, MOC is almost independent of signal proportionality, instead it is primarily sensitive to co-occurrence, the fraction of pixels with positive values for both channels, regardless of signal levels. This is demonstrated in Fig. 5_D_, which shows MOC and PCC measurements for sets of random data like those shown in Fig. 5_C_ but with different constant values subtracted from each. Whereas PCC values are essentially unaffected by the downward shift in the distributions, MOC values decline as the fraction of pixels with positive values decreases. MOC reaches 0 only when the two probes are completely mutually exclusive; i.e., there are no pixels with positive values for both channels (not shown).
Fig. 5.
Manders' Overlap Coefficient (MOC) as a measure of colocalization. A: scatterplot of positively related red and green pixel values, PCC = 0.73, MOC = 0.99. B: scatterplot of negatively related red and green pixel values, PCC = −0.71, MOC = 0.92. C: scatterplot of uncorrelated red and green pixel values, PCC = −0.03, MOC = 0.97. D: PCC (circles) and MOC (squares) measurements for uncorrelated data similar to that shown in C, as a function of offset. Solid symbols reflect offset in both red and green channels. Open symbols reflect offset in the red channel alone.
In the context of the argument made above, that colocalization need not imply proportional codistribution, the insensitivity of MOC to signal proportionality might suggest MOC as a better indicator of colocalization than PCC for some analyses. However, MOC only indirectly and somewhat unpredictably measures co-occurrence. [A very complete dissection of this and several other problems with MOC as a measure of colocalization has recently been provided by Adler et al. (2).] If the goal is to measure the co-occurrence of two probes, it can be better measured directly as the fraction of one probe that is coincident with the second probe, as described below.
Fractional overlap.
Whereas PCC provides an effective statistic for measuring overall association of two probes in an image, it has the major shortcoming that it indirectly (and sometimes poorly) measures the quantity that is typically at the heart of most analyses of colocalization in cell biology: the fraction of one protein that colocalizes with a second protein. This quantity can be measured via Manders' Colocalization Coefficients (MCC) (34), metrics that are widely used in biological microscopy and have been implemented in all biological image analysis software packages. For two probes, denoted as R and G, two different MCC values are derived, _M_1, the fraction of R in compartments containing G and _M_2, the fraction of G in compartments containing R. These coefficients are simply calculated as:M1=∑iRi,colocal∑iRiwhere R i,colocal = R i if G i > 0 and R i,colocal = 0 if G i = 0 andM2=∑iGi,colocal∑iGiwhere G i,colocal = G i if R i > 0 and G i,colocal = 0 if R i = 0
By providing measures of the fraction of total probe fluorescence that colocalizes with the fluorescence of a second probe, MCCs provide an intuitive and direct metric of the quantity of interest for most biological colocalization studies. Unlike PCC, MCC strictly measures co-occurrence independent of signal proportionality.
While simple in principle, measuring MCC is complicated by the fact that the input values used to measure MCC can almost never be taken directly from the original images. The problem is in the numerator of each expression, where pixel values are included in the sum if they occur in pixels in which the signal from the second probe exceeds zero. While such pixels might seem to be self-evident, pixel values of zero are seldom obtained in fluorescence images, which typically contain “background,” low signal levels in the image derived from light leakage into the system, autofluorescence, “nonspecific” labeling, and probe fluorescence arising from out-of-focus image planes. Although background could be eliminated in the collection process by adjusting detector settings, microscopists generally maintain a positive offset in detector settings to ensure that weak signals are detected; thus the detection process additionally contributes to background. Since MCC depends utterly upon the ability to distinguish pixels with signal derived from a labeled structure in the focal plane from pixels whose signal results strictly from background sources, a necessary preliminary step is thus to eliminate the component of pixel intensity derived from background. The bulk of the thought and effort of applying MCCs generally goes into identifying and subtracting background values from images, so that the image contains meaningful “zero” values.
The most obvious method for eliminating background is to subtract a global threshold value from the pixel intensities of the image. However, for many images, estimating the appropriate threshold value representing background is challenging. More importantly, for images in which much of the fluorescence occurs in structures whose intensity is close to that of the background, MCC is very sensitive to the value chosen for the threshold. Taking the example of the image shown in Fig. 1_A_, increasing the threshold of the green channel by only five gray levels decreases the estimated overlap of red signal with green signal from 92% to 82%. The sensitivity of MCC to the estimate of background is disquieting and indicates the importance of an automatic, or at least nonsubjective, reproducible method for determining background.
Costes et al. (14) developed a unique approach for automatically identifying the threshold value to be used to identify background based on an analysis that determines the range of pixel values for which a positive PCC is obtained. In this approach, PCC is measured for all pixels in the image and then again for pixels for the next lower red and green intensity values on the regression line. This process is repeated until pixel values are reached for which PCC drops to or below zero. The red and green intensity values on the regression line at this point are then used as the threshold values for identifying background levels in each channel. Only those pixels whose red and green intensity values are both above their respective thresholds are considered to be pixels with colocalized probes. MCC is then calculated as the fraction of total fluorescence in the region of interest that occurs in these “colocal” pixels.
The Costes method for estimating thresholds is a robust and reproducible method that can be easily automated, both speeding processing and eliminating user bias. The method has been implemented in the Imaris, Slidebook, and Volocity software and in ImageJ plugins (JACoP, WCIF) and has been widely applied to studies in cell biology (5, 10, 11, 18, 44, 47, 49).
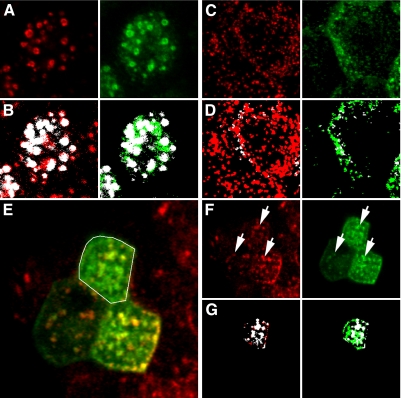
In many cases, the Costes method provides a quick and effective method for distinguishing labeled structures from background, thus supporting accurate measurement of MCC. For example, for the images of immunolocalized EEA1 and GFP-RhoB, shown in Fig. 6_A_, the Costes method effectively distinguishes labeled endosomes from background, as shown in the binarized versions of the images after applying the calculated thresholds (Fig. 6_B_). MCC calculations, based on these thresholds, indicate that 85% of the EEA1 is found in compartments associated with GFP-RhoB and 78% of the GFP-RhoB associates with compartments containing EEA1 (shown in white).
Fig. 6.
The Costes automatic thresholding method. A: single image plane of a polarized MDCK cell immunolabeled for EEA1 (red) and expressing GFP-RhoB (green). B: binary versions of images shown in A, after applying Costes threshold. Pixels with positive signals for both probes are shown in white. C: maximum projection of an image volume of Texas Red-dextran (red) and Alexa 488-transferrin (green) internalized by an MDCK cell. D: binary versions of images shown in C, after applying Costes threshold. Pixels with positive signals for both probes are shown in white. E: image of a single focal plane image of MDCK cells expressing GFP-EHD1 (green) after incubation with Texas Red-transferrin (red). F: individual images from E, with arrows indicating compartments containing both probes. G: binary versions of the cell within the ROI shown in E, after applying Costes threshold. Pixels with positive signals for both probes are shown in white.
Application of Costes thresholds to images of internalized transferrin and dextran in MDCK cells (Fig. 6_C_) likewise effectively distinguishes labeled structures from background (Fig. 6_D_). MCC calculations from these thresholds indicate that 5% of the internalized dextran localizes with transferrin and 19% of the transferrin localizes with dextran (shown in white).
The Costes approach also effectively distinguishes intracellular compartments labeled with GFP-EDH1. Figure 6_E_ shows an image of a single focal plane of fluorescent transferrin (red) internalized into MDCK cells expressing GFP-EHD1 (green). Comparison of the individual images (Fig. 6_F_) suggests that the majority of the internalized transferrin is found in compartments associated with GFP-EHD1. This impression is supported by MCC analysis of the cell delineated with the ROI in Fig. 6_E_ following application of Costes thresholds (Fig. 6_G_), indicating that 71% of the transferrin is found in compartments associated with GFP-EHD1.
As with any image analysis technique, the results of the Costes thresholding method should always be checked visually. Under some circumstances, the Costes procedure can fail to identify a useful threshold. For example, previous studies have shown that Costes thresholding struggles with images that have very high labeling density or large differences in the number of structures labeled with each probe (13). In our experience, the Costes method is effective for images with high signal-to-background ratios, but in images with low signal levels it frequently identifies a threshold value that is so low that it fails to discriminate labeled structures from background. For example, in the image of fluorescent IgA and transferrin internalized into CHO cells (shown in Fig. 7, A and B), the Costes threshold for the cell in the indicated ROI is so low that more than 81% of the cellular region is scored positive for transferrin and 92% of the cellular region is scored positive for IgA (Fig. 7_C_). Not surprisingly, MCC analysis indicates an astonishingly high overlap between the two probes: 97% of the transferrin is identified as colocalizing with IgA and 100% of the IgA is identified as colocal with transferrin (shown in white). Similar results were obtained in an analysis of the distribution of two transferrin probes, shown in Fig. 1_A_; the Costes method identifies 93% and 97% of the cell as positive for red and green probes, respectively, resulting in MCC values of 100% for each. For these examples, it is clear that pixel values corresponding to the point on the regression where the correlation shifts from positive to negative are too low to adequately discriminate labeled from unlabeled cellular structures, leading to meaningless MCC measurements. Software implementations of the Costes algorithm typically accommodate this problem by including the capability to change the point on the regression used to identify thresholds. However, in doing this, one negates a primary advantage of the Costes approach: that it removes subjectivity from MCC computations. An alternative nonsubjective approach might be to identify the thresholds as the lowest points on the regression line where the correlation remains significant. This criterion will result in higher thresholds but is complicated by the problem of significance testing of correlations, a thorny problem that we discuss later.
Fig. 7.
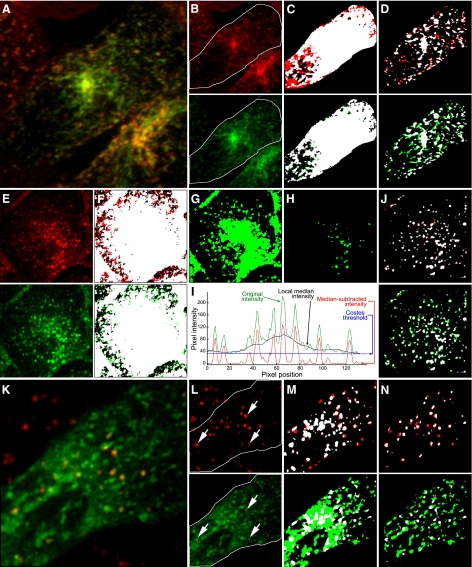
Background correction via Costes automatic thresholding method and median subtraction. A: maximum projection of an image volume of transfected CHO cells following incubation with Texas Red-transferrin (red) and Oregon Green-IgA (green). B: individual images from A. C: binary versions of images of the cell in the ROI shown in B, after applying Costes threshold. Pixels with positive signals for both probes are shown in white. D: binary versions of images of the cell in the ROI shown in C following subtraction of a 32 × 32 pixel median and thresholds of 8 and 8. E: maximum projection of an image volume of MDCK cells incubated with Texas Red-transferrin (red) and Alexa 488-transferrin (green). F: binary versions of images shown in E, after applying Costes threshold. Pixels with positive signals for both probes are shown in white. G: binary version of image of Texas Red transferrin shown in E, after applying a threshold of 57. H: binary version of image of Texas Red transferrin shown in E, after applying a threshold of 139. I: pixel intensities along a line in the image of Texas Red transferrin shown in E. Green, original intensities. Black, local median intensities. Red, intensities after subtraction of local median from original intensities. Blue, Costes threshold. J: binary versions of the images shown in E following subtraction of a 32 × 32 pixel median, and thresholds of 27 and 23. K: single image plane of an MDCK cell expressing GFP-Rab7 (green) following incubation with diI-LDL (red). L: individual images from K. Arrows indicate endosomes labeled for both probes. M: binary versions of images of the cell in the ROI shown in L, after applying Costes threshold. Pixels with positive signals for both probes are shown in white. N: binary versions of images of the cell in the ROI shown in L following subtraction of a 32 × 32 pixel median, and thresholds of 42 and 22.
Regardless of the criterion used to select the thresholds, the Costes approach fails to address a more general problem for thresholding images: background levels vary spatially in many cases so that no one background value is appropriate for the entire image. Spatial variation in background may result from spatial inhomogeneity in illumination or detection or from out-of-focus fluorescence, which may be appreciable even in confocal fluorescence images. This problem is exemplified in the image of an MDCK cell that has internalized two fluorescent conjugates of transferrin into endosomes shown in Fig. 7_E_. Costes thresholding results in classification of nearly the entire image as positive for red and green probes (Fig. 7_F_). As with the other examples described above, the Costes method identifies thresholds that are too low to distinguish labeled structures from background in this image. However, the problem is not simply one of underestimating the appropriate threshold value. Increasing the threshold distinguishes individual endosomes in the periphery of the cell, but not at the center, a thicker part of the cell (Fig. 7_G_). Increasing the threshold further eliminates background around the endosomes at the center of the cell but eliminates the peripheral endosomes altogether (Fig. 7_H_). It is clear that a single threshold value will not satisfactorily identify background throughout this image.
For studies of dispersed objects, such as endosomes, we have found that an effective measure of local background can be derived from the median intensity in a relatively large region surrounding each pixel in the image (typically between 24 × 24 and 36 × 36 pixels in size) (4, 15, 16, 35, 46). In this approach, the image is spatially filtered to remove pixel noise and a background image is constructed in which the value of each pixel in the original image is replaced with the median intensity in the region surrounding the pixel. This background image is then subtracted pixel-by-pixel from the filtered image to obtain a background-subtracted quotient image. As long as the size and density of objects is such that they occupy less than half of the region size, the median provides an accurate measure of the local background. Thus the size of the region is critical; it must be large enough to predominantly measure the region surrounding objects but small enough to reflect spatial variation. Since there are some low residual values remaining in pixels of unlabeled regions after median subtraction, a small value is subtracted from the quotient image, producing an image with zero values in unlabeled regions, making it suitable for quantifying MCCs.
The effectiveness of median subtraction is demonstrated in the line intensity profiles shown in Fig. 7_I_. The pixel intensity along a line traced over the image shown in Fig. 7_E_ is shown in green, with spikes occurring at the points along the line that pass through endosomes. It can be seen that a background level determined by the Costes threshold (indicated in blue) fails to discriminate individual endosomes, identifying the entire region of this line as above background. In contrast, the median intensity of a 32 × 32 pixel region (indicated in black), follows the low-frequency background variation in the image closely, more closely approximating the background levels between endosomes even in the center of the region. Subtracting the median intensities from the original image results in an intensity profile (shown in red), for which a single value can be subtracted to distinguish individual endosomes from background.
Figure 7_J_ shows binarized versions of the images shown in Fig. 7_E_, following background subtraction and thresholding. Visual inspection of this image shows that this process has more effectively eliminated background from the image, more clearly distinguishing individual transferrin-containing endosomes in both the periphery and perinuclear regions of the cell. MCC calculations using these background values indicate that 93% of the red transferrin occurs in compartments containing green transferrin, and 66% of the green transferrin occurs in compartments containing red transferrin (reflecting the dimmer labeling from the red transferrin). Median subtraction has likewise effectively isolated endosomes from background in the image shown in Fig. 7_A_, as shown in the binarized images shown in Fig. 7_D_. MCC calculations using these background values indicate that 76% of the internalized IgA occurs in compartments containing transferrin and 74% of the internalized transferrin occurs in compartments containing IgA.
A more complex situation is shown in Fig. 7_K_, which shows the intracellular distribution of internalized diI-LDL (red) in a living MDCK cell expressing GFP-Rab7 (green). The individual channels show that LDL localizes predominantly in compartments associated with GFP-Rab7 (see arrows in Fig. 7_L_). Whereas the Costes method effectively isolates individual endosomes in the image of diI-LDL, a much larger region of the cytosol is identified as above background in the image of GFP-Rab7 (Fig. 7_M_). Better discrimination of the intracellular compartments is obtained using backgrounds applied after median subtraction (Fig. 7_N_). MCC analysis of these images indicates that 71% of the diI-LDL occurs in compartments associated with GFP-Rab7 and 20% of the vesicular GFP-Rab7 is found in compartments containing diI-LDL.
However, one could fairly ask whether median subtraction is appropriate for analyzing the distribution of a protein like GFP-Rab7. Whereas we can confidently ignore cytosolic signals in samples labeled by probes that we know are contained within punctate vesicular compartments, a sizable fraction of peripheral membrane proteins like Rab7 will in fact be located in the cytosol. By eliminating this diffuse fluorescence from analysis, median subtraction restricts colocalization analysis to the vesicular pool of GFP-Rab7. This is appropriate for estimating the fraction of vesicular GFP-Rab7 associated with endosomes containing diI-LDL but will lead to overestimates of the fraction of total cellular GFP-Rab7 associated with diI-LDL. As with the Costes method described above, one must always evaluate the results of the method used to identify image background and choose the method that is appropriate to the biology of the system.
Comparison of Pearson's Correlation Coefficient and Manders' Colocalization Coefficient
PCC and MCC represent the two major metrics of colocalization used in biomedical research. Strictly speaking, neither is superior to the other; both have strengths and weaknesses that, depending on the situation, make one or the other the preferred metric.
The most obvious advantage of MCC is that it is a more intuitive measure of colocalization than PCC. MCC is also more useful for data that are poorly suited to the simple, linear model that underlies PCC. For example, in the image shown in Fig. 4_E_, nearly all of the immunolocalized EEA1 appears to occur in compartments associated with GFP-RhoB. This appearance is quantitatively supported by MCC analysis indicating that 85% of the EEA1 localizes to compartments associated with GFP-RhoB. However, PCC measurement indicates a relatively poor association between the probes (PCC = 0.73) due to the fact that, while colocalized, the ratio of the probes varies widely (Fig. 4_H_). For this study, MCC would be considered the better metric of colocalization because it is independent of signal proportionality. In general, cases in which probes are not proportionally codistributed will yield ambiguous, intermediate PCC values that are hard to interpret. Rather than indicating a partial colocalization, these intermediate values may indicate a mismatch between the data and the underlying model of PCC.
Another obvious advantage of MCC over PCC is that it provides two components: the fraction of A with B and the fraction of B with A. This is important when the probes distribute to different kinds of compartments, as for example, in the case in which all of A is found in compartments containing B, but B is also found in additional compartments lacking A. Consider the example of a study in which an investigator has fluorescently labeled a protein that associates with vesicles and would like to know if these vesicles associate with microtubules. If one numerically simulates this situation such that all of the vesicular fluorescence overlaps that of the microtubules but only 20% of the microtubule fluorescence overlaps that of vesicles, one obtains a depressingly low PCC of 0.2. However, this same value is obtained if only 20% of the vesicular fluorescence overlaps that of microtubules, if 100% of the microtubule fluorescence overlaps that of the vesicles. Thus, with respect to the question of whether the vesicular protein associates with microtubules, the same low PCC value is obtained for essentially opposite experimental outcomes. This problem is realized in the analysis of diI-LDL and GFP-Rab7 in the image shown in Fig. 7_K_. Although MCC analysis indicates that 71% of the internalized LDL localizes to compartments associated with GFP-Rab7, the PCC value obtained for this cell is very low (0.32) because of the large number of GFP-Rab7 compartments in excess of those labeled with diI-LDL (80% of the GFP-Rab7 occurs in regions lacking diI-LDL). This is another example of complex data in which pixel intensities of the two probes are not related by a simple, linear relationship. For these kinds of data, PCC yields ambiguous results, whereas MCC more directly measures the quantity of interest.
MCC analysis is also more appropriate for three-dimensional analysis of colocalization, which is required for studies in which probe colocalization varies spatially in a cell. While these variations may be appropriately sampled when they occur within a single focal plane, they are insidious when they occur vertically in a cell since results become completely dependent on the particular focal plane that was captured. This kind of vertical heterogeneity is apparent in the distributions of internalized IgA and transferrin shown in Figs. 2 and 4. While PCC can be quantified in three-dimensional image volumes, it is poorly suited to the kind of complexity that requires three-dimensional analysis in the first place, as discussed above. MCC analysis is also much easier to extend to three-dimensional analyses (in which case it is sometimes called an “object-based” analysis). Quantification of an overall PCC for a three-dimensional volume of the cell requires delineation of the region-of-interest for each focal plane from the volume and combination of all of the identified voxels into a single array from which PCC is calculated (46). Since MCCs are not influenced by areas from which both probes are excluded, MCC does not require this painful delineation of the region-of-interest. Measurement of three-dimensional MCCs can be accomplished either manually, by simply dividing colocal fluorescence by total fluorescence from an entire stack of images (as described in Refs. 32 and 42) or automatically in software that supports three-dimensional quantification of MCC (e.g., Volocity, as applied by Refs. 25, 26, and 44).
If circumstances require measurement of PCC throughout the volume of a cell, a simpler alternative to three-dimensional analysis that frequently yields comparable results is to measure PCC in a projected image of the volume (4). The projected image may consist either of the sum of all of the images of the volume or the maximum intensity value for each (x,y) position found throughout the volume. This approach is best limited to cells with limited depth and label density such that structures do not overlap when projected into a single image. For such cells, projection essentially results in collection of all of the structures from the volume into a single image that is much easier to evaluate. In the case of the relatively flat cells shown in Fig. 1, PCC of the projected volume of Fig. 1_A_ compares well with PCC quantified over the three-dimensional volume (0.94 vs. 0.90), and PCC of the projected volume of Fig. 1_E_ compares well with PCC measured over the three-dimensional volume (−0.045 vs. −0.008). MCC analysis is less forgiving of overlap occurring in the projection process and should seldom be applied to projected data, where it is likely to generate spuriously high estimates of overlap. For example, analysis of the three-dimensional volume of the two cells shown in Fig. 3_A_ indicates that 51–52% of the IgA colocalizes with transferrin, whereas analysis of the projected volume indicates 66–71% overlap. We emphasize that although we have included projected images as examples in this review, quantifications of projections must always be carefully validated by comparison with results obtained from three-dimensional analysis.
In summary, MCC offers many practical and theoretical advantages over PCC. The major drawback of MCC is that it is complicated by the need to be able to reliably identify background levels in an image and thereby identify labeled structures. The surprisingly difficult problem of distinguishing label from background is one that has no single answer; different images require different strategies. For some images a single threshold value derived via the Costes approach may suffice. Other images may require locally determined background levels. Still others may require more elaborate methods of object discrimination that may be daunting for many cell biologists seeking an answer to what seems like a relatively simple question. In our experience median subtraction is effective for discriminating punctate compartments but less effective for other kinds of structures. Other studies have demonstrated effective segmentation of biological structures using Laplace (41), Sobel (24), or watershed filters (27). The utility of these approaches for any given application will generally require careful evaluation and optimization. For images where background correction is challenging, PCC analysis may still be the preferred method, as it requires no image preprocessing of any kind.
An Outline of a Colocalization Analysis Workflow
Despite what may seem like a relentless emphasis on problems and complications, this review is not intended to dishearten investigators seeking a rigorous method for analyzing colocalization. The wide availability of software tools for visualizing and quantifying colocalization has made it extraordinarily easy to conduct colocalization studies, and while it is important to consider the foregoing caveats, identifying and applying the proper technique is actually straightforward. A general workflow is schematized in Fig. 8.
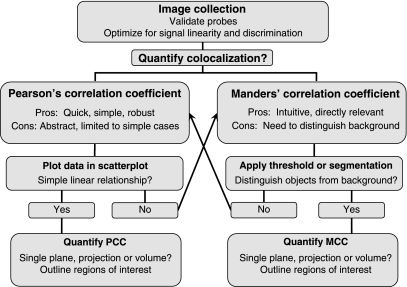
Fig. 8.
Schematic of a general colocalization analysis workflow. Upon determining that colocalization analysis is appropriate to the biological question, an in investigator first needs to validate the fluorescent probes used to identify molecular localizations. The image collection system must be optimized to detect the dimmest structures without saturating signal levels of the most heavily labeled structures. The optics of the system should be optimized to provide effective discrimination such that each detector channel sensitively detects one or the other probe without cross-contamination of signal between the two signals. Colocalization can be quantified either as Pearson's correlation coefficient (PCC) or Manders' correlation coefficient (MCC). A preliminary step in PCC analysis is to visually evaluate scatterplots of the pixel intensity data to evaluate whether the data follow a simple linear relationship. With this condition satisfied PCC can be quantified either in single images or for three-dimensional image volumes in designated ROIs that limit analysis to individual cells. If the data fail to follow a simple linear relationship, MCC may be the preferred approach for quantifying colocalization. A preliminary step in MCC analysis is to identify the threshold value necessary to distinguish labeled structures from background. With this accomplished, MCC can be quantified either in single images or for three-dimensional image volumes in designated ROIs that limit analysis to individual cells. To the degree that effective thresholding cannot be accomplished, PCC may be the preferred approach.
An investigator first needs to identify the nature of the colocalization question. If one is trying to identify a molecular interaction, this whole review is irrelevant, and the investigator should instead use a high-resolution technique such as electron microscopy or fluorescence resonance energy transfer.
The first step in colocalization analysis is to design and conduct image collection (a topic more completely described in Refs. 7, 38, 43, and 50). Investigators need to establish that probes are both specific and sensitive, ideally as determined in control studies in which the distributions of the probes are compared with those of validated probes. The parameters of image collection (illumination level, detector gain, integration time) should be adjusted such that fluorescence signals are collected in the linear range of the detector system (detecting the dimmest structures without saturating signal levels in the brightest structures). The optics of the system should be such that crosstalk of signals between channels is negligible, a condition that can be verified by collecting images of singly labeled samples, using the same settings used for colocalization studies. Finally, images of the entire three-dimensional volume of the sample should generally be acquired, by collecting a series of images using optical sectioning techniques such as confocal microscopy, multiphoton microscopy, or image deconvolution microscopy. Although three-dimensional analysis may not always be necessary, in these times of cheap digital storage it is better to collect volumetric data that you do not need than to have to repeat the study should volumetric analysis prove to be warranted.
Next, the investigator needs to determine whether quantification is necessary. Quantification is primarily useful for generating values that can be compared between different conditions. If no such comparisons are conducted, quantification may add nothing more than a veneer of legitimacy to a technique that has been traditionally been viewed as qualitative. In addition, there are cases where colocalization is so visually obvious that a number value is no more compelling than the merged image or the data scatterplot.
If quantification is desired, the choice for most studies is between some form of PCC and MCC. Both PCC and MCC have strengths and shortcomings. Despite some surprising controversy on the topic (2, 3, 7, 8), neither should be considered superior to the other; the choice of one over the other depends on the biological question and the nature of the images themselves. In contrast, MOC is a truly troubled metric (2) that we cannot recommend in any circumstance that we can imagine.
PCC is a very simple, thoroughly characterized, robust measure that can be remarkably free of the influence of the investigator's wishes. Since PCC reflects the fit of the data to a simple, linear relationship between the signals of the two probes, data should first be evaluated for linearity by plotting the values of a few representative cells in scatterplots. Pixel intensities should follow a single, linear relationship, without excessive scatter and without multiple relationships; e.g., additional compartments with one, but not the other probe. If the data indicate a simple linear relationship, one should then randomly identify cells to be quantified and outline the relevant ROI for each. One should then compare values measured from single planes or from projected images with those obtained from the entire volume to determine whether the additional effort of measuring PCC in volumes is warranted.
To the degree that probe distributions are at least approximately linearly related, and one has carefully identified an appropriate ROI, PCC provides a meaningful measure of probe colocalization for most studies. That said, PCC is a somewhat abstract measure of probe colocalization and can be misleading in some applications. In summarizing the relative distribution of two probes into a single statistic, it cannot separately measure the amount of red that co-occurs with green and the amount of green that co-occurs with red, the central question of many colocalization studies. Perhaps more significantly, this also means that PCC can be confounded by differences in the number of compartments labeled with each probe. This and other complex relationships in the data will be apparent in the scatterplots of the pixel data, which will indicate studies for which colocalization might be better measured as MCC.
MCC provides a measure of colocalization that is much more meaningful to most investigators: the fraction of each probe that is colocalized with the other. By providing two separate measures, MCC is also independent of differences in the number of structures labeled by each probe. Finally, MCC does not depend on a linear relationship between the signal levels of the two probes and is less finicky with respect to defining the ROI, making it simpler to implement for measurements of volumes. The major drawback of MCC is that measured values are very sensitive to the estimated level of background, the threshold value used to distinguish labeled structures from unlabeled background. In general, accurate estimation of background is subjectively evaluated via visual inspection of the thresholded images. Given this subjectivity, it is important to avoid bias by consistently applying the same thresholding technique to all experimental samples. For some images, effective thresholding can be accomplished by relatively standard methods. However, many images obtained in biological microscopy are challenging for standard thresholding techniques and may require more elaborate methods of image segmentation, which is itself a distinct field of research for which most cell biologists have neither the time, the training, nor the inclination.
Finally, colocalization studies will generally require some sort of statistical analysis for interpretation, a topic considered below.
Significance Testing in Colocalization Studies
One problem with quantifications of colocalization is that they are seldom supported with statistical analysis. In the absence of a statistical context for a particular value, it is frequently difficult to interpret its meaning. Significance testing in colocalization analysis can take two forms. The first form seeks to estimate the statistical significance of differences in the amount of colocalization measured between experimental groups. The second form seeks to identify the statistical significance of colocalization measurements for a single experimental group.
Before the significance testing is discussed, a short discussion of the concept of noise is in order. Sometimes confused with “background,” the term “noise” refers to the variability in a measurement, whereas background refers to the amount of offset in an image. Variability is incorporated into the calculation of PCC, such that PCC declines with noise. Noise indirectly affects MCC by complicating the process of thresholding. Noise thus results in underestimates of “true” colocalization, a somewhat slippery concept except in simulated images. The bigger issue with noise is that it confounds comparisons when it is manifest in one condition more than the other. Although noise is an inevitable characteristic of fluorescence microscopy, every effort should be made to optimize image collection techniques for maximum signal-to-noise ratios in images to be quantified. After collection, image noise can be reduced by either deconvolution (28) or low-pass filtering (but see discussion of autocorrelation later). Finally, the effect of noise can be effectively corrected via a correction factor based on an estimate of noise derived from repeated imaging of the same field (1).
As with any metric, the significance of a difference between two groups can be evaluated statistically. For example, our group and others have used Student's _t_-tests to test the significance of differences in PCCs (4, 46) and MCCs (26, 42). As long as none of the confounding variables described above differ between experimental groups (e.g., differences in signal level, noise or the relative amount of labeling between the two probes for PCC, differences in the accuracy of background estimation for MCC), statistical comparison of populations is straightforward.
A much more challenging task is to estimate the significance of a colocalization measurement made for a single group. In other words, while values of PCC approaching 0 or 1 may convincingly demonstrate the absence or presence of colocalization, respectively, what does one conclude about a PCC measurement of 0.4? Actually, the statistical question is no different from that for comparing two groups, except that rather than evaluating the magnitude of the difference between two groups, one evaluates the difference between the value measured for the experimental group and for some sort of “null” model, which reflects the measurement that would be obtained for random data.
Readers familiar with statistics will recognize that the statistical significance of a measured PCC value can be directly derived from the data; the probability that a given PCC value could be obtained by chance is a function of the deviations of the individual values from the best-fit regression line and the number of measurements. However, the large sample size of image PCC analysis, in which even small regions contain thousands of values, provides enormous statistical power, such that even subtle and biologically meaningless correlations result in statistically significant PCC values. For example, Fig. 9_A_ shows a scatterplot of 500 pairs of random numbers, for which PCC measures 0.014 (P = 0.38). Figure 9_B_ shows an equally unimpressive scatterplot but one for which PCC measures 0.177, a highly significant value (P < 0.00003). Regarded cynically, this analysis demonstrates the kind of issue that many cell biologists have with statistics, whose attitude is like that of Mark Twain, who identified statistics as the third form of lies (“lies, damned lies and statistics”). However, a more enlightened perspective is that statistical analysis is capable of identifying even subtle effects, some of which may be irrelevant to the desired comparison. For example, correlations of this magnitude can easily result from gradients in the field of illumination or lack of a flat-field correction in the microscope objective. In these cases, statistically significant PCC values may be obtained that reflect optical artifacts rather than a correlation in the distribution of the two probes.
Fig. 9.
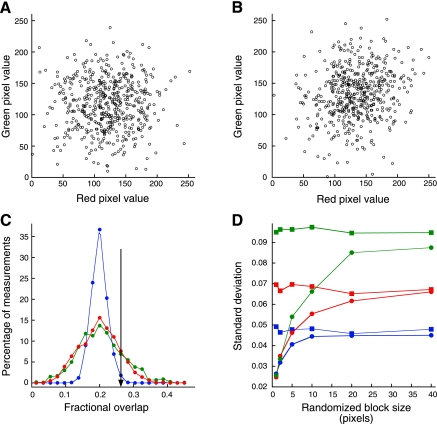
Significance testing of colocalization data. A: scatterplot of 500 red and green pixel values with PCC = 0.014, P = 0.38. B: scatterplot of 500 red and green pixel values with PCC = 0.177, P <0.00003. C: green line is distribution of 1,000 MCC values obtained after randomly distributing 20 red and 20 green pixel segments, each 10 μm in length, along a 1,000-pixel line. Blue line is distribution of MCC values obtained after scrambling individual pixels from each original line. Red line is distribution of MCC values obtained after scrambling 10-pixel blocks from each original line. Black line indicates MCC value whose magnitude was exceeded in only 1% of the MCC values obtained from scrambled pixels but was exceeded by 20% of MCC values obtained from the original lines. D: standard deviation of 100 MCCs measured for original lines (squares) and after scrambling blocks of various sizes (circles). Values were computed for line segments consisting of 40 5-pixel segments (blue), 20 10-pixel segments (red), and 10 20-pixel segments (green).
A bigger problem with statistical analysis of microscopy image data is that significance testing of correlation coefficients is confounded by spatial autocorrelation, the condition in which the value of one pixel is likely to be similar to that of its neighboring pixels. Autocorrelation is essentially ubiquitous in microscope images, resulting from two sources. First, pixel sampling is typically arranged such a point source forms an image over several adjacent pixels, according to the point spread function of the imaging system. Second, labeled structures are almost invariably larger than a point source, so that their images project onto tens or hundreds of adjacent pixels. Shortly after the development of PCC, it was demonstrated that autocorrelation can result in statistically significant correlations even for variables with no real association (22). For autocorrelated variables, calculating the significance of PCC using the usual _t_-test will yield a P value that is too low; red and green values with no real association will appear to be significantly correlated with each other far too often. Since low-pass filtering increases autocorrelation, the effect of autocorrelation on significance testing in correlation is aggravated by spatial filters used to reduce image noise.
An alternative approach for evaluating the significance of a colocalization measurement is to compare the mean of measured values to the mean obtained from pairs of images that are out of registration with one another, a condition that should yield random colocalization. This can be accomplished by rotating the image of one channel relative to the other (4), shifting the image of one channel relative to the other (19, 45) or selecting different regions of the two images (27). This situation is essentially identical to a comparison of two experimental groups, except that in this case one statistically compares the mean of a set of experimental values to the mean of a set of values obtained from misregistered data.
The most accurate method for estimating the significance of an individual measurement of colocalization would be based on comparison with a large number of measurements taken in comparable samples in which the distributions of the two probes are unrelated. The probability that the measure could be obtained by chance, the P value, is then determined by the fraction of measurements in the random distribution that are greater than the measured value. However, since such “comparable” samples seldom exist, a practical alternative is a randomization approach in which the random probability distribution is generated from measurements of images in which one channel is scrambled or translated. In the “scrambling” approach, the random probability distribution is derived from repeated measurements of colocalization between the image of one channel and a version of the second channel in which the pixels, or blocks of pixels are randomly rearranged (14), an approach subsequently applied by Refs. 7, 36, and 37. In the “frame-translation” approach, the random probability distribution is derived from a number of measurements obtained after translating the image of one channel relative to the other (19, 41).
Unfortunately, the scrambling approach is again complicated by autocorrelation. Rearranging the individual pixels of one channel eliminates the spatial autocorrelation of that channel, because the rearranged pixels are no longer adjacent to their original neighbors but instead are adjacent to pixels from other parts of the image. To the degree that the sample has autocorrelation that is not represented in the randomized data, the probability distribution of PCCs of the randomized data will be narrower than that for the original data. Consequently, deviations from the mean will appear to be more significant than they are.
Autocorrelation also affects significance testing of MCC. Figure 9_C_ shows the results of a simulation in which 20 red and 20 green line segments, each 10 pixels in length, were arrayed randomly along a line 1,000 pixels in length. This process was repeated 1,000 times, and MCC was quantified for each trial. The resulting distribution, shown in green, represents the distribution of MCC measurements that would be expected to occur for two unrelated distributions by chance alone. We next simulated a random probability distribution by randomly distributing the 200 green pixels individually throughout the 1,000 pixel line segment, repeating this process 1,000 times and calculating MCC for each trial. The resulting probability distribution (shown in blue) is much narrower than the true random probability distribution. If used to estimate random probability of a given measurement, this narrow distribution would consistently overestimate the significance of a measurement. For example, the vertical line in Fig. 9_C_ indicates a measurement of 0.26. While an overlap of this size or greater occurred in almost 20% of the random scrambling of the original objects, it occurs in only 1% of the random scramblings of the pixels. Thus an MCC measurement that occurred well within the range of chance would be judged highly significant if evaluated relative to the scrambled data.
The accuracy of the probability distribution can be improved by generating the scrambled image using blocks of pixels rather than individual pixels. This procedure retains more of the autocorrelation of the original data, resulting in a probability distribution that more closely approximates that of the original variates. If one repeats the randomization process shown in Fig. 9_C_, but scrambles 10 pixel blocks rather than single pixels, the distribution of overlap for the scrambled data nearly superimposes that of the true random probability distribution (red line). This distribution will provide a more accurate estimate of the random probability of a measurement, and thus its statistical significance. Similar results have been obtained in two-dimensional simulations, demonstrating that small block sizes yield unrealistically narrow and misleading probability distributions of MCC (41) and PCC (14, 41).
Although most descriptions of this method indicate that block size should be adjusted to match the size of the point-spread function, our simulations indicate that block size should equal or exceed the size of the objects in an image, whose area will necessarily be at least as large as that of the point-spread-function. To test the role of block size, we conducted a simulation in which MCC was quantified for each of 1,000 trials in which red and green line segments of different sizes (5, 10, or 20 pixels) were randomly arrayed along a 1,000 pixel line segment. For each random trial, the green pixels from the resulting line were divided into blocks ranging from 1 to 40 pixels in length, which were then randomly distributed to generate a scrambled line segment and MCC was calculated again. Figure 9_D_ shows that the standard deviations obtained for the scrambled line segments (circles) approach those of the original line segments (squares) only when the block size exceeds the size of the original line segments. Since autocorrelation is always reduced by the process of fragmenting the original image, the standard deviations of the randomized block data never quite reach the levels obtained with the original data.
As described above, the use of the narrow probability distributions obtained with small block sizes would result in systematic overestimates of the significance of measured MCC values. The results of this simulation suggest that to generate a representative random probability distribution, the size of pixel blocks chosen for randomization should exceed that of the size of the objects in the image.
The pixel block scrambling approach for estimating the random probability of a colocalization measurement has been implemented in the Slidebook image analysis software package and the JACoP and WCIF ImageJ plugins, which include the capability to adjust the block size used in randomization. Alternative methods for generating “randomized” images have also been implemented in which an image of randomly distributed pixels is first generated, based either on a white noise image (Imaris) or a pixel-scrambled version of one of the test images (WCIF Colocalization Test ImageJ plugin). This image is then convolved with a Gaussian filter whose size matches that of the point-spread function of the imaging system. These methods will be appropriate for analysis of structures whose images are similar in size to the point-spread-function. However, since they fail to reproduce the autocorrelation resulting from structures larger than the point-spread-function, they will yield spurious identifications of significant correlations in many samples.
The problem of spatial autocorrelation can be avoided using a frame translation approach, in which the random probability distribution is generated from measurements of colocalization obtained after shifting one image relative to the other. Since the random measurements are obtained from measurements of the unaltered structures, the autocorrelation of the original data is preserved so that accurate random probability distributions can be generated. In a study of simulated punctate objects, this approach was found to yield broader, more accurate random probability distributions than those obtained after pixel block scrambling (41).
To generate meaningful values, the frame-translation method requires that the “random” measurements be obtained from mismatched regions that can otherwise be considered equivalent. Thus the random measurements must be obtained from displaced regions that remain within the region of potential interaction. So for example, one cannot compare PCC measured within a cell to a random probability distribution that includes values measured with frames that extend outside of the cell. For many studies, this can limit the number of random scenarios that can be measured, compromising the accuracy of the random probability distribution. For example, Fay et al. (19) generated only 75 random scenarios using translations in x, y, and z. One way to increase the number of random measurements in a frame-translation method is to apply a “wrap-around” technique, in which translated regions that depart the region of interaction (e.g., extending outside the cell) are populated with pixels from the opposite side of the region. This approach, which is sometimes applied to spatial correlation studies in ecology (20), has been implemented in the SCIAN-Lab CDA ImageJ plugin, permitting measurements of random PCC and MCC values for even very small image regions (41). Because this method fragments the original image at the edges, it will decrease the autocorrelation slightly and thus increase the probability of a false positive. However, for images of reasonable size, only a small portion of the image is fragmented, so this artifact will generally be quite small. P values derived from this technique may generally be more accurate than those resulting from the scrambling approach.
Although the frame-translation approach has been widely applied for spatial correlation studies in other fields, it has received little attention in biological microscopy. Thus, whereas it appears to offer promise as a tool for significance testing in colocalization studies in cell biology, its general practicality and utility in biological microscopy remain to be demonstrated.
Both of the randomization approaches are complicated by the need to identify the region of potential interaction between the two probes. If, in the process of simulating random probe distributions, one maps the randomized probe fluorescence to regions of the cell that are not actually physically accessible to the probe, one increases the number of simulations with no overlap between the two probes. A distribution of measurements obtained from such simulations will contain an inordinate number of low colocalization values, resulting is mistaken estimates of significance. Whereas we have discussed this issue with respect to restricting frame translation to the region of the cell, the problem is equally important to the scrambling approach, in which one must identify the region in which pixel blocks should be randomly distributed. Whereas we can restrict block distribution to the area occupied by the cell, it may not be the case that all intracellular regions are actually accessible to the two probes. The nucleus is an obvious example of a structure that reduces the potential space of interaction of cytosolic molecules, but what about all of the other unlabeled intracellular structures that limit the potential space of interaction? To the degree that probes cannot actually distribute throughout what appears to be a homogeneous isotropic region, probability distributions generated with either the scrambling or frame translation approaches will be biased toward low colocalization measures, leading to systematic overestimates of significance. Productive use of the randomization approaches may require that colocalization be measured in the context of a explicitly defined region, as in some studies of the distribution of integral membrane proteins in the plasma membrane (27, 39), or the distribution of nuclear proteins (45).
In summary, whereas the idea of directly estimating the probability that a given measurement of colocalization could be obtained by chance is very attractive, it is not simple to implement in practice. The process of generating randomized data is complicated by the difficulty of reproducing the autocorrelation present in the original data and by the difficulty of identifying the region of potential interaction of the two probes. Failure to appreciate these factors will generally lead to systematic overestimation of the significance of colocalization measurements. These problems may underlie the fact that, in a survey of applications of this approach, we find an inordinately large number of studies in which the experimentally measured values fall outside the entire range of randomized values, indicating for each the statistically unlikely random probability of zero. While extreme values are possible, they should be regarded critically and ideally compared with values obtained for images of probes with unrelated distributions.
The problem of significance testing of colocalization data is one with no simple answer at this point. Potential solutions may have been developed in the field of ecology, where spatial autocorrelation is a major concern (21, 29), and where methods for testing the significance of correlations between autocorrelated variables have been developed (12, 17). Applying these methods to image analysis in cell biology may be fruitful (Dunn and McDonald, unpublished observations).
Summary
There are a number of different metrics that can be used to measure colocalization in fluorescence microscope images. Here we have described Pearson's Correlation Coefficient, Pearson's Correlation Coefficient measured in regions of probe overlap, Manders Overlap Coefficient, and Manders Colocalization Coefficient, the metrics most widely applied in biological microscopy. Additional measures not discussed here include the Intensity Correlation Quotient (30) and Spearmann's Rank Coefficient (nonparametric versions of PCC) (1), as well as various object-based measures (7, 23, 27, 38, 48). The process of measuring each of these metrics typically involves multiple decisions as to the region of interest in the image and/or the range of intensity values to be analyzed. Thus an investigator new to quantitative microscopy is presented with a dizzying range of possible approaches, each providing a range of outcomes, depending on parameter settings.
All of the colocalization measurements described here are widely available to biomedical researchers, either as ImageJ plugins or in commercial image analysis software, sometimes provided with microscope systems. The different techniques are invariably simple to apply and frequently arrayed conveniently side by side. Ironically, the simplicity with which different colocalization metrics can be tweaked and compared becomes dangerous, since the algorithms and their parameters are poorly described. This is a situation that is ripe for misuse, innocent or otherwise. In the absence of an understanding of what each metric actually measures, and of the basis and consequences of the parameter settings, it is seductively easy to uncritically apply a variety of different methods using a variety of different parameter settings. The problem is that when the user is given little or no guidance as to how to choose the correct assay or how to set parameter values, it becomes easy to evaluate the legitimacy of the assay from the results, a practice that is actually suggested by at least one review. We are reminded of a colleague who, when asked how one recognizes when digital image analysis has been properly conducted, responded “when it gives you the answer you expect.”
The single most important point of this review is that there are some very powerful and apparently simple approaches for quantifying colocalization, but none should be considered to be turn-key methods. Investigators need to understand the strengths and weaknesses of each metric, particularly in practical application. Once the researcher has identified the colocalization measurement that is appropriate to the biological question and to the nature of the samples, every step in the image analysis process should be scrutinized. Software that provides the ability to visually evaluate the effects of parameter adjustments is preferable to software that simply spits out a final numerical value.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant DK-51098 and a George M. O'Brien Award from the NIH Grant DK-61594 (to K. Dunn).
DISCLOSURES
No conflicts of interest, financial or otherwise are declared by the authors.
ACKNOWLEDGMENTS
Image collection and digital image analysis were conducted at the Indiana Center for Biological Microscopy. We thank Jeff Clendenon and Jason Byars for technical assistance.
APPENDIX
Appendix of Image Analysis Software Providing Colocalization Assays
Nearly all of the image analysis methods described here are incorporated into image analysis software provided with microscope systems, in stand-alone software designed for cell biologists, and in ImageJ plugins, some of which are listed below.
Commercially Available Software
| Colocalizer Pro: | Colocalization Research Software |
|---|---|
| Image-Pro: | Media Cybernetics, Inc. |
| Imaris: | Bitplane Scientific Software AG. |
| Metamorph: | Molecular Devices, Inc. |
| Slidebook: | Intelligent Imaging Innovations, Inc. |
| Huygens Profession: | Scientific Volume Imaging BV. |
| Volocity: | PerkinElmer, Inc. |
| Axiovision: | Carl Zeiss Microimaging, LLC. |
ImageJ Plugins
REFERENCES
- 1.Adler J, Pagakis SN, Parmryd I. Replicate-based noise corrected correlation for accurate measurements of colocalization. J Microsc 230: 121–133, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Adler J, Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A 77: 733–742 [DOI] [PubMed] [Google Scholar]
- 3.Adler J, Parmryd I. Recent review on colocalization seem to misunderstand the Pearson correlation coefficient. J Microsc 227: 83; author reply 84–85, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Babbey CM, Ahktar N, Wang E, Chen CCH, Grant BD, Dunn KW. Rab10 regulates membrane transport through early endosomes of polarized Madin-Darby Canine Kidney cells. Molec Biol Cell 17: 3156–3175, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakrac B, Kladnik A, Macek P, McHaffie G, Werner A, Lakey JH, Anderluh G. A toxin-based probe reveals cytoplasmic exposure of Golgi sphingomyelin. J Biol Chem 285: 22186–22195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci 18: 251–265, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Bolte S, Cordelieres FP. Reply to Letter to the Editor. J Microsc Oxford 227: 84–85, 2007 [Google Scholar]
- 9.Brown PS, Wang EX, Aroeti B, Chapin SJ, Mostov KE, Dunn KW. Definition of distinct compartments in polarized Madin-Darby Canine Kidney (MDCK) cells for membrane-volume sorting, polarized sorting and apical recycling. Traffic 1: 124–140, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Burette AC, Strehler EE, Weinberg RJ. “Fast” plasma membrane calcium pump PMCA2a concentrates in GABAergic terminals in the adult rat brain. J Comp Neurol 512: 500–513, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrel D, Masson J, Al Awabdh S, Capra CB, Lenkei Z, Hamon M, Emerit MB, Darmon M. Targeting of the 5-HT1A serotonin receptor to neuronal dendrites is mediated by Yif1B. J Neurosci 28: 8063–8073, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clifford P, Richardson S, Hemon D. Assessing the significance of the correlation between 2 spatial processes. Biometrics 45: 123–134, 1989 [PubMed] [Google Scholar]
- 13.Comeau JW, Costantino S, Wiseman PW. A guide to accurate fluorescence microscopy colocalization measurements. Biophys J 91: 4611–4622, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86: 3993–4003, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn KW, Maxfield FR. Delivery of ligands from sorting endosomes to late endosomes occurs by maturation of sorting endosomes. J Cell Biol 117: 301–310, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn KW, Mcgraw TE, Maxfield FR. Iterative fractionation of recycling receptors from lysosomally destined ligands in an early sorting endosome. J Cell Biol 109: 3303–3314, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutilleul P. Modifying the _t_-test for assessing the correlation between 2 spatial processes. Biometrics 49: 305–314, 1993 [Google Scholar]
- 18.Estrada-Bernal A, Gatlin JC, Sunpaweravong S, Pfenninger KH. Dynamic adhesions and MARCKS in melanoma cells. J Cell Sci 122: 2300–2310, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay FS, Taneja KL, Shenoy S, Lifshitz L, Singer RH. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A). Exp Cell Res 231: 27–37, 1997 [DOI] [PubMed] [Google Scholar]
- 20.Fortin MJ, Dale MRT. Spatial Analysis: A Guide For Ecologists. Cambridge, NY: Cambridge University Press, 2005 [Google Scholar]
- 21.Fortin MJ, Dale MRT. Spatial autocorrelation in ecological studies: a legacy of solutions and myths. Geograph Anal 41: 392–397, 2009 [Google Scholar]
- 22.Gosset WS. The elimination of spurious correlation due to position in time or space. Biometrika 10: 179–181, 1914 [Google Scholar]
- 23.Helmuth JA, Paul G, Sbalzarini IF. Beyond co-localization: inferring spatial interactions between sub-cellular structures from microscopy images. BMC Bioinform 11: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaskolski F, Mulle C, Manzoni OJ. An automated method to quantify and visualize colocalized fluorescent signals. J Neurosci Methods 146: 42–49, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Khandelwal P, Ruiz WG, Apodaca G. Compensatory endocytosis in bladder umbrella cells occurs through an integrin-regulated and RhoA- and dynamin-dependent pathway. EMBO J 29: 1961–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khandelwal P, Ruiz WG, Balestreire-Hawryluk E, Weisz OA, Goldenring JR, Apodaca G. Rab11a-dependent exocytosis of discoidal/fusiform vesicles in bladder umbrella cells. Proc Natl Acad Sci USA 105: 15773–15778, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lachmanovich E, Shvartsman DE, Malka Y, Botvin C, Henis YI, Weiss AM. Co-localization analysis of complex formation among membrane proteins by computerized fluorescence microscopy: application to immunofluorescence co-patching studies. J Microsc 212: 122–131, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Landmann L, Marbet P. Colocalization analysis yields superior results after image restoration. Microsc Res Tech 64: 103–112, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Legendre P, Dale MRT, Fortin MJ, Gurevitch J, Hohn M, Myers D. The consequences of spatial structure for the design and analysis of ecological field surveys. Ecography 25: 601–615, 2002 [Google Scholar]
- 30.Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci 24: 4070–4081, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch RM, Carrington W, Fogarty KE, Fay FS. Metabolic modulation of hexokinase association with mitochondria in living smooth muscle cells. Am J Physiol Cell Physiol 270: C488–C499, 1996 [DOI] [PubMed] [Google Scholar]
- 32.Lynch RM, Fogarty KE, Fay FS. Modulation of hexokinase association with mitochondria analyzed with quantitative three-dimensional confocal microscopy. J Cell Biol 112: 385–395, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manders EMM, Stap J, Brakenhoff GJ, Vandriel R, Aten JA. Dynamics of 3-dimensional replication patterns during the S-phase, analyzed by double labeling of DNA and confocal microscopy. J Cell Sci 103: 857–862, 1992 [DOI] [PubMed] [Google Scholar]
- 34.Manders EMM, Verbeek FJ, Aten JA. Measurement of colocalization of objects in dual-color confocal images. J Microsc Oxford 169: 375–382, 1993 [DOI] [PubMed] [Google Scholar]
- 35.Maxfield FR, Dunn KW. Studies of endocytosis using image intensification fluorescence microscopy and digital image analysis. In: Optical Microscopy for Biology, edited by Herman BaKJ: Wiley-Liss, 1990, p. 357–371 [Google Scholar]
- 36.Mukhopadhyay S, Shah M, Xu F, Patel K, Tuder RM, Sehgal PB. Cytoplasmic provenance of STAT3 and PY-STAT3 in the endolysosomal compartments in pulmonary arterial endothelial and smooth muscle cells: implications in pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol 294: L449–L468, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Nicolas CS, Park KH, El Harchi A, Camonis J, Kass RS, Escande D, Merot J, Loussouarn G, Le Bouffant F, Baro I. IKs response to protein kinase A-dependent KCNQ1 phosphorylation requires direct interaction with microtubules. Cardiovasc Res 79: 427–435, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oheim M, Li D. Quantitative colocalisation imaging: concepts, measurements, and pitfalls. In: Imaging Cellular And Molecular Biological Functions, edited by Shorte SL, Frischknecht F. Berlin: Springer, 2007, p. xxi, 449 p. [Google Scholar]
- 39.Parmryd I, Adler J, Patel R, Magee AI. Imaging metabolism of phosphatidylinositol 4,5-bisphosphate in T-cell GM1-enriched domains containing Ras proteins. Exp Cell Res 285: 27–38, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Pearson K. Mathematical contributions to the theory of evolution III. Regression, heredity and panmixia. Philos Trans R Soc Lond B Biol Sci 187: 253–318, 1896 [Google Scholar]
- 41.Ramirez O, Garcia A, Rojas R, Couve A, Hartel S. Confined displacement algorithm determines true and random colocalization in fluorescence microscopy. J Microsc 239: 173–183 [DOI] [PubMed] [Google Scholar]
- 42.Rondanino C, Rojas R, Ruiz WG, Wang E, Hughey RP, Dunn KW, Apodaca G. RhoB-Dependent modulation of postendocytic traffic in polarized madin-darby canine kidney cells. Traffic 8: 932–949, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Scriven DR, Lynch RM, Moore ED. Image acquisition for colocalization using optical microscopy. Am J Physiol Cell Physiol 294: C1119–C1122, 2008 [DOI] [PubMed] [Google Scholar]
- 44.Tzaban S, Massol RH, Yen E, Hamman W, Frank SR, Lapierre LA, Hansen SH, Goldenring JR, Blumberg RS, Lencer WI. The recycling and transcytotic pathways for IgG transport by FcRn are distinct and display an inherent polarity. J Cell Biol 185: 673–684, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Steensel B, van Binnendijk EP, Hornsby CD, van der Voort HT, Krozowski ZS, de Kloet ER, van Driel R. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J Cell Sci 109: 787–792, 1996 [DOI] [PubMed] [Google Scholar]
- 46.Wang EX, Pennington JG, Goldenring JR, Hunziker W, Dunn KW. Brefeldin A rapidly disrupts plasma membrane polarity by blocking polar sorting in common endosomes of MDCK cells. J Cell Sci 114: 3309–3321, 2001 [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Azuma Y, Moore D, Osheroff N, Neufeld KL. Interaction between tumor suppressor adenomatous polyposis coli and topoisomerase IIalpha: implication for the G2/M transition. Mol Biol Cell 19: 4076–4085, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodcroft BJ, Hammond L, Stow JL, Hamilton NA. Automated organelle-based colocalization in whole-cell imaging. Cytometry A 75: 941–950, 2009 [DOI] [PubMed] [Google Scholar]
- 49.Zhang D, Manna M, Wohland T, Kraut R. Alternate raft pathways cooperate to mediate slow diffusion and efficient uptake of a sphingolipid tracer to degradative and recycling compartments. J Cell Sci 122: 3715–3728, 2009 [DOI] [PubMed] [Google Scholar]
- 50.Zinchuk V, Zinchuk O. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr Protoc Cell Biol Chapter 4: Unit 4: 19, 2008 [DOI] [PubMed] [Google Scholar]