TDP-43 neurotoxicity and protein aggregation modulated by heat shock factor and insulin/IGF-1 signaling (original) (raw)
Abstract
TAR DNA-binding protein 43 (TDP-43) plays a key role in the neurodegenerative diseases including amyotrophic lateral sclerosis and frontotemporal lobar degeneration. The nature of the TDP-43-mediated neurotoxicity associated with these diseases is not yet understood. Here, we have established transgenic Caenorhabditis elegans models that express human TDP-43 variants in the nervous system, including the full-length wild-type (WT) and mutant proteins and a pathologic C-terminal fragment. The C. elegans models developed severe locomotor defects associated with the aggregation of TDP-43 in neurons. In comparison to parallel Cu/Zn superoxide dismutase worm models, transgenic full-length TDP-43, including the WT protein, was highly neurotoxic. In addition, TDP-43 demonstrated an unusually high tendency to aggregate, a property intrinsic to the WT protein. The C-terminal 25 kDa fragment of TDP-43 was unstable but remarkably aggregation-prone. Distinct disulfide-linked TDP-43 dimers and oligomers were detected. In C. elegans, the neurotoxicity and the protein aggregation of TDP-43 were regulated by environmental temperature and heat shock transcriptional factor 1, indicating that a deficiency in protein quality control is a risk factor for TDP-43 proteinopathy. Furthermore, the neurotoxicity and the protein aggregation of TDP-43 can be significantly attenuated by a deficiency in the insulin/insulin-like growth factor 1 (IGF-1) signaling in C. elegans and mammalian cells. These results suggest that protein misfolding underlies the aging-dependent neurodegeneration associated with TDP-43 and that the insulin/IGF-1 signaling may be a target for therapies.
INTRODUCTION
TAR DNA-binding protein 43 (TDP-43) is an evolutionarily conserved DNA/RNA-binding protein that has been associated with neurodegenerative diseases including amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). ALS is characterized by degeneration of motor neurons that leads to motor dysfunction (1,2), and FTLD by degeneration of the frontal and temporal lobes of the brain that leads to dementia (3). Wild-type (WT) TDP-43 protein and its proteolytic fragments have been found as characteristic components of the ubiquitinated proteinaceous inclusions in the brain and spinal cord of ALS and FTLD patients (4). Furthermore, more than 30 dominant mutations in TDP-43 have been identified in ∼4% of familial ALS (5,6), and rare mutations have been found in several FTLD patients (7–9). The common TDP-43 proteinopathy suggests that ALS and FTLD might represent different manifestations of a spectrum of related neurodegenerative disorders (10).
The exact function of TDP-43 remains unclear, although it has been implicated in transcription regulation and RNA metabolism (11,12). TDP-43 regulates the processing of various RNA species (12–18), and its function is essential for early mouse embryogenesis (19,20). Expression of both WT and mutant human TDP-43 proteins in transgenic mice and rats leads to neurodegeneration (21–26). The cellular pathologies include ubiquitinated inclusions (21,23), TDP-43 nuclear and cytoplasmic aggregates (22,24,26), mitochondrial accumulation (24,25) and disruption of Gemini of coiled bodies (25). Studies in other animal models, such as Drosophila and zebrafish, have suggested that both loss of function and gained toxicity of TDP-43 could lead to the neurodegeneration (27–29). Recent Caenorhabditis elegans models suggested that RNA binding and phosphorylation of TDP-43 influences its toxicity (30,31). However, the exact mechanism of TDP-43-associated neurodegeneration remains largely unknown (6). TDP-43 is primarily a nuclear protein and forms both nuclear and cytoplasmic complexes (18,32). It is degraded through both the ubiquitin-proteasome system and autophagy (33,34). It is recruited to cytoplasmic stress granules under oxidative insults (35). Cytoplasmic mislocalization of mutant TDP-43 has been associated with its cytotoxicity (36). A common feature shared by many neurodegeneration-associated proteins, including ALS-linked Cu/Zn superoxide dismutase (SOD1), is the heightened propensity to form non-native high-molecular-weight species, including soluble oligomers and insoluble aggregates (37–39). Although TDP-43 has been shown to be aggregation-prone in vitro (40), it remains unclear whether the misfolding and the aggregation of TDP-43 play an important role in its neurotoxicity.
Aging is a risk factor for adult-onset neurodegenerative diseases. One molecular pathway that regulates aging is insulin/insulin-like growth factor 1 (IGF-1) signaling, which is evolutionarily conserved from nematode to mammals (41,42). daf-2 encodes the sole insulin/IGF-1 receptor (IGF1R) in C. elegans, and a loss-of-function daf-2 mutation doubles the lifespan in C. elegans (41). DAF-2-mediated longevity is partially mediated by downstream DAF-16, a forkhead-related transcriptional factor that DAF-2 negatively regulates (43). Deficiency in insulin/IGF-1 signaling has previously been shown to suppress the aggregation of polyglutamine peptides and an amyloid peptide in C. elegans body wall muscles (44,45). The heat shock transcriptional factor 1 (HSF-1), a master transcriptional factor that positively regulates the expression of many heat shock proteins, is also involved in aging (43).
To establish tractable models for efficient probing of the age-dependent human neurodegenerative diseases, we have created transgenic C. elegans that express various forms of human TDP-43 and develop profound movement defects associated with protein misfolding and aggregation. A comparison of TDP-43 and SOD1 in human cells revealed that TDP-43 had an unusually high propensity to form protein aggregates, and a C-terminal fragment of TDP-43 found in FTLD patients was particularly aggregation-prone. We found that the neurotoxicity and the protein aggregation of TDP-43 in C. elegans were increased by thermal stress and were dependent on HSF-1. In addition, the DAF-2-mediated insulin signaling that regulates the aging program significantly modulated the neurotoxicity and the protein aggregation of TDP-43 in neurons. We further extended these observations to mammalian systems using a human cell model of TDP-43 protein aggregation.
RESULTS
Transgenic C. elegans expressing neuronal full-length and truncated human TDP-43 develop locomotor defects and protein aggregation
To explore the potential use of C. elegans as a model organism to study TDP-43-linked ALS and FTLD, we generated transgenic C. elegans expressing the human TDP-43 gene via a pan-neuronal promoter of the C. elegans gene synaptobrevin (snb-1) (46). Both WT TDP-43 and ALS-linked mutant forms of this protein, Q331K and M337V, were expressed in the nervous system of transgenic animals. An enhanced yellow fluorescent protein (YFP) tag at the C-terminus of TDP-43 was used to visualize the protein in the intact transparent animals.
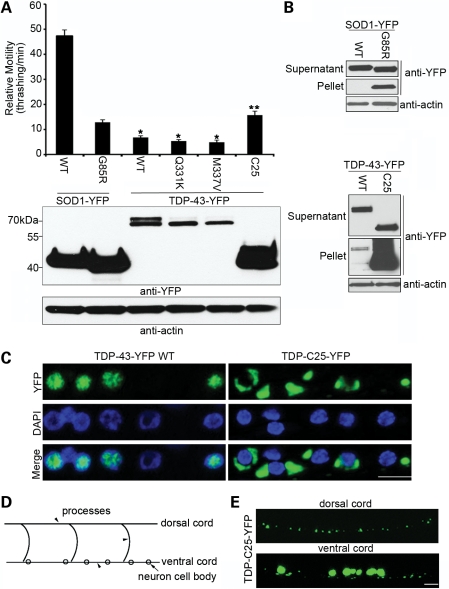
Transgenic C. elegans expressing both the WT and the ALS-linked mutant forms of TDP-43-YFP developed profound locomotor defects, as measured by the crawling speed of the worms on solid medium and the thrashing rate in liquid medium (Fig. 1A; Supplementary Material, Videos S1–6), providing a direct readout of the neurotoxicity of the protein. The expression of YFP alone using the same expression vector, as a negative control, did not produce any significant defect. Next, we compared the phenotypes of the transgenic strains expressing TDP-43-YFP with those of SOD1-YFP strains, using transgenic lines exhibiting comparable intensity of YFP fluorescence. The WT and the G85R mutant SOD1 were significantly different in their neurotoxic properties: whereas WT SOD1-YFP had only a mild effect on the behavior of the transgenic animals, the G85R SOD1-YFP mutant exhibited severe locomotor defects (Fig. 1A) (46). In comparison, both the WT and the mutant forms of TDP-43-YFP exhibited particularly strong locomotor defects that were consistently more severe than those of the mutant SOD1-YFP, even when the total protein levels of TDP-43 were less than that of mutant SOD1 (Fig. 1A). In addition, the locomotor defect in the transgenic C. elegans expressing the ALS-linked TDP-43 mutants, Q331K and M337V, appeared to be slightly more severe than that associated with WT TDP-43 (Fig. 1A). These phenotypes suggested an intrinsic toxicity of the TDP-43 protein when it accumulated at pathological conditions.
Figure 1.
Transgenic C. elegans expressing neuronal TDP-43 develops pronounced movement defects associated with protein aggregation. (A) Relative motility, as measured by the thrashing rate in liquid medium, was compared among 1-day adult C. elegans expressing variants of human SOD1-YFP or TDP-43-YFP under the control of the snb-1 promoter. n = 32, error bars represent the standard error of means (SEM). The TDP-43 strains had significantly worse locomotion than the mutant SOD1 strain (*P < 0.05). The TDP-C25 strain had better movement than the other TDP-43 strains (**P < 0.05). The expression levels of total SOD1 or TDP-43 proteins were shown by immunoblotting against YFP. (B) The insolubility assay as a measure of the aggregation of SOD1-YFP or TDP-43-YFP variants. From detergent-extracted C. elegans homogenates, 10 µg (∼1/20) of soluble supernatant protein and 5 µg (∼1/4) of insoluble pellet protein were analyzed by immunoblotting. (C) Nuclear localization of full-length TDP-43-YFP and cytoplasmic localization of the fragment TDP-C25-YFP. Representative images of motor neurons in the ventral cord stained with DAPI are shown. Note the well-demarked aggregates of TDP-C25-YFP of various sizes in the cytoplasm. (D) Schematic drawing of the ventral and dorsal cords in the C. elegans nervous system. The neuronal cell bodies (arrow) are located in the ventral cord, and the neuronal processes (arrowheads) are projected circumferentially and located in both ventral and dorsal cords. (E) In addition to the large aggregates of TDP-C25-YFP in the neuronal cell bodies, smaller aggregates were observed in the neuronal processes of both ventral and dorsal cords. Scale bars: 5 µm.
The C-terminal fragments of TDP-43 are signature components of the ubiquitinated inclusions in the affected brain or spinal cord of human patients (4). We then generated transgenic C. elegans expressing a C-terminal 25 kDa fragment (TDP-C25, residues 219–414) that has been identified in FTLD brains (47). Interestingly, although the TDP-C25 transgenic C. elegans exhibited a significantly worse locomotor defect than WT SOD1, the neurotoxicity of the truncated protein is less than that of the full-length TDP-43 proteins (Fig. 1A).
To understand the basis of the locomotor defect of the TDP-43 transgenic C. elegans, we used Nomarski optics and YFP fluorescence microscopy to examine the ventral cord motor neurons that innervate body wall muscles. The TDP-43 transgenic C. elegans did not exhibit significant loss of neurons or gross abnormality of neuronal morphology. To assess the function of the motor neurons, we used a cholinergic agonist, aldicarb, which is an established agent for probing neurotransmission in nematodes (48). Like the mutant SOD1 transgenic C. elegans (46), the TDP-43 strains were more resistant than the non-transgenic or YFP-only controls to the paralytic effect of aldicarb (data not shown), suggesting that the locomotor defects in the TDP-43 transgenic C. elegans were likely mediated by a reduced efficiency in synaptic transmissions.
In addition to their locomotor defects, the TDP-43-YFP transgenic C. elegans strains exhibited growth defects similar to those observed in G85R SOD1-YFP transgenic C. elegans (46). When compared with the non-transgenic and YFP-only controls, the TDP-43-YFP transgenic C. elegans had slower growth rates during larval developmental stages from L1 to L4. Overall, the growth defects in the transgenic C. elegans expressing TDP-43-YFP and mutant SOD1-YFP were correlated with the locomotor defects in these animals, suggesting a common origin related to the neurotoxicity induced by these proteins.
To assess the aggregation of TDP-43 in C. elegans neurons, we applied a biochemical assay for detecting protein aggregates to the transgenic C. elegans (Fig. 1B). The TDP-43 and SOD1 transgenic C. elegans were homogenized and extracted to separate supernatant and pellet fractions. In this process, detergent-insoluble protein aggregates were enriched in the pellets. WT SOD1-YFP completely segregated with the supernatant, whereas G85R SOD-YFP had readily detectable aggregates in the pellet. In comparison, a fraction of the full-length WT TDP-43-YFP was detected in the pellet, suggesting its aggregation in C. elegans neurons. In the case of TDP-C25-YFP, it was predominantly enriched in the pellet, suggesting its remarkably high propensity for aggregation (Fig. 1B).
We then examined the distribution of the TDP-43-YFP in the live neurons of intact C. elegans. The WT TDP-43 (Fig. 1C) and full-length mutant proteins, Q331K and M337V (Supplementary Material, Fig. S1), were mostly localized to the nucleus in C. elegans neurons. In contrast, the TDP-C25-YFP were localized exclusively to the cytoplasm and formed discrete aggregates (Supplementary Material, Fig. 1C). The TDP-C25-YFP aggregates started as relatively small globular structures, but some grew into larger structures filling the cytoplasmic space. In addition, smaller puncta of TDP-C25-YFP were also observed throughout the neuronal processes (Fig. 1D and E). The puncta were evenly distributed along the long processes, and no fast movements were observed for these structures, suggesting that small aggregates could form locally in the neuronal processes. Such puncta were absent in transgenic C. elegans expressing full-length TDP-43 proteins.
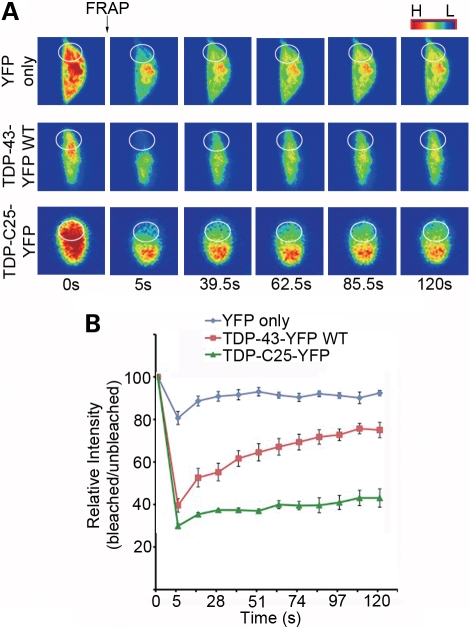
The nature of the aggregates was further confirmed by fluorescence recovery after photobleaching (FRAP) analysis. When neurons expressing YFP only were partially bleached in the cytoplasm, the fluorescent protein was immediately redistributed within the cytoplasm, suggesting that YFP was completely soluble (Fig. 2). FRAP in the nuclei of YFP-expressing neurons indicated that the nuclear pool of YFP was equally mobile (Supplementary Material, Fig. S2). However, the recovery of TDP-43-YFP, which was predominantly localized to the nucleus, occurred at a slower rate, suggesting that a subset of the protein molecules was either in an aggregated state or associated with less mobile macromolecular structures (Fig. 2). The nature of the cytoplasmic aggregates of TDP-C25 was clearly demonstrated by FRAP. After a portion of TDP-C25 fluorescent aggregates was bleached, the fluorescence in the bleached region did not recover after several minutes, indicating that the TDP-C25-YFP present in the structure were highly immobile (Fig. 2).
Figure 2.
FRAP analysis of TDP-43 aggregates in C. elegans neurons. (A) Representative images from the FRAP analysis of neurons expressing YFP only, WT TDP-43-YFP localized to the nucleus and TDP-C25-YFP, which form aggregates in the cytoplasm. The circles mark the photobleached region. Colors indicate high (H) or low (L) intensity of fluorescence. (B) The ratio of the intensities in bleached and adjacent unbleached regions is used to assess the diffusion rate of the fluorescent proteins. n = 5, error bars represent the SEM.
The high aggregation propensity of both full-length and truncated TDP-43 in mammalian cells
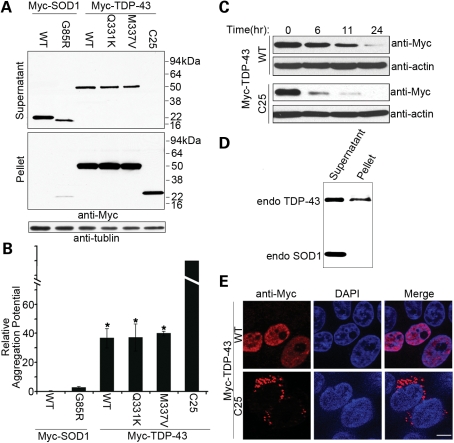
To further analyze the aggregation propensity of the TDP-43 variants in mammalian cells, we tagged these proteins with an N-terminal Myc and compared them with SOD1 variants in human embryonic kidney 293T (HEK293T) cells (Fig. 3). The cells were lysed at 48 h after cell transfection, and the lysates were extracted and separated into soluble supernatants and insoluble pellets. The relative ratio of insoluble to soluble proteins was used as a measure of the aggregation propensity of TDP-43 and SOD1. The representative of ALS-linked SOD1 mutant, G85R, showed a heightened aggregation propensity when compared with the WT SOD1 protein. Interestingly, all the TDP-43 variants, including WT, Q331K and M337V, exhibited a higher aggregation propensity than any form of SOD1, even mutant SOD1 (Fig. 3A and B). The particularly aggregation-prone TDP-C25 fragment fractionated predominantly into the insoluble pellet, consistent with the observation that it formed discrete aggregates in C. elegans neurons. The low steady-state level of soluble TDP-C25 is likely due to its rapid turnover. The instability of the TDP-C25 protein was confirmed by its estimated half-life of 3.3 h, when compared with 11.3 h for the WT TDP-43 protein (Fig. 3C). Given that G85R represents a protein of relatively high aggregation propensity among ALS-linked SOD1 mutants (49), these results revealed unusually high aggregation potential for TDP-43, which appeared to be inherent in the WT protein. Furthermore, endogenous TDP-43 in HEK293T cells fractionated significantly into the insoluble pellet, when compared with SOD1, suggesting that TDP-43 exists in large sedimentable structures (Fig. 3D). In addition, immunostaining for Myc-tagged TDP-43 in HEK293T or U2OS cells showed a cellular distribution pattern similar to that observed in C. elegans neurons. The WT TDP-43 proteins and the full-length mutants, Q331K and M337V, showed a predominantly nuclear localization (Supplementary Material, Fig. S3). In contrast, the TDP-C25 fragment formed discrete and exclusively cytoplasmic aggregates (Fig. 3E).
Figure 3.
High aggregation potential of TDP-43 proteins in human cells. (A) N-terminal Myc-tagged TDP-43 and SOD1 were transiently overexpressed in HEK293T cells, and the lysates were extracted with detergents. Two micrograms (∼1/500) of soluble supernatant protein and 8 µg (∼1/50) of insoluble pellet protein were analyzed by denaturing and reducing SDS–PAGE followed by immunoblotting with anti-Myc antibodies. (B) The relative aggregation potential, as measured by the ratio of insoluble pellet proteins to soluble supernatant proteins, is compared among variants of SOD1 and TDP-43. TDP-43 is more aggregation-prone than mutant G85R SOD1 (*P < 0.05). A nominal aggregation potential for TDP-C25 is given because of high degree of instability of the soluble protein. n = 3, error bars represent the SEM. (C) Cycloheximide chase experiments for TDP-43 proteins. Twenty-four hours after the transfection of the Myc-tagged proteins, HeLa cells were treated with 200 µg/ml of cycloheximide. The cells were harvested at various time points during the subsequent 24 h. The cells were lysed with the extraction buffer and centrifuged, and 8 µg of soluble supernatant protein was analyzed by immunoblotting with the anti-Myc antibody (long exposure for the TDP-C25 immunoblot). The TDP-C25 protein was estimated to have a half-life of 3.3 h, when compared with 11.3 h for the WT TDP-43 protein. (D) Endogenous TDP-43 in HEK293T cells is relatively enriched in insoluble pellets, when compared with the endogenous SOD1. (E) Nuclear localization of full-length Myc-TDP-43 and cytoplasmic localization of aggregated Myc-TDP-C25 are indicated by immunofluorescence staining in transfected HEK293T cells. Scale bar: 5 µm.
TDP-43 protein dimers and oligomers linked by disulfide bonds in vivo
There are six cysteine residues in TDP-43 (C38, C49, C172, C174, C197 and C243), with one (C243) included in the TDP-C25 fragment. Because aberrant disulfide cross-linking of SOD1 has been implicated in protein aggregation in cell and mouse models (50–53), we asked whether intermolecular disulfide bonds played any role in the aggregation of TDP-43. As described above, the Myc-tagged proteins were expressed in the HEK293T cells, detergent-extracted, and then electrophoresed under denaturing but non-reducing conditions with sample buffer containing sodium dodecyl sulfate (SDS) but no reducing agents (Fig. 4). Because the protein extraction buffer contained iodoacetamide, which binds covalently with cysteine and prevents its ex vivo oxidation, only disulfide bonds formed in vivo were detected by this method.
Figure 4.
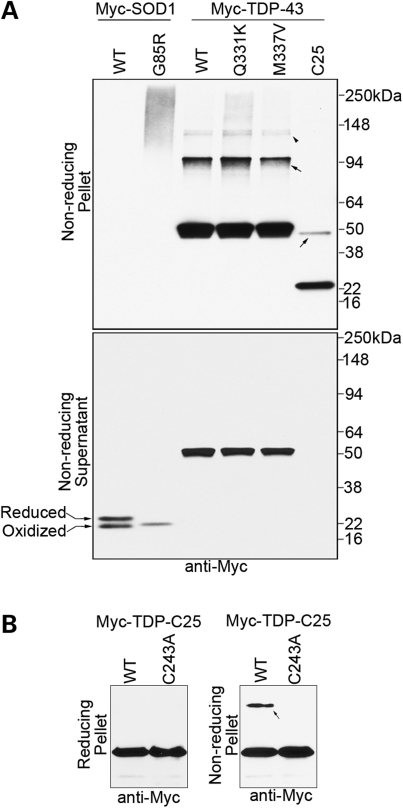
TDP-43 protein dimers and oligomers linked by disulfide bonds are enriched in aggregates in vivo. (A) N-terminal Myc-tagged TDP-43 and SOD1 variants were transfected into HEK293T cells, and the lysates were detergent extracted in the presence of the thiol blocker iodoacetamide. The insoluble pellet and the soluble supernatant fractions were both analyzed with denaturing and non-reducing SDS–PAGE without β-mercaptoethanol. The top panel shows the pellet fraction in which the aggregated protein was enriched. In contrast to mutant SOD1 G85R, which is ‘glued’ together by disulfide bonds into large aggregates, distinct TDP-43 dimers (arrows) and oligomers (arrowhead) dependent on β-mercaptoethanol are observed. The bottom panel shows the supernatant fraction, in which the majority of soluble TDP-43 or SOD1 proteins are not disulfide cross-linked. The doublet bands of WT SOD1 are due to its intramolecular disulfide bond. A fraction of soluble WT SOD1 protein was disulfide-reduced and subsequently modified by iodoacetamide, resulting in a slower migration, as described previously (68). The soluble SOD1 G85R protein has a characteristic faster migration than WT SOD1 and is presumably all in a reduced state. (B) Mutating the only cysteine in TDP-C25 (C243A) blocked the formation of the insoluble dimers (arrow), but the aggregation of the majority of the protein was not significantly affected.
Under the non-reducing conditions, the insoluble G85R SOD1 proteins in the pellet completely shifted to high-molecular-weight species in denaturing SDS–polyacrylamide gel electrophoresis (PAGE), suggesting that the aggregated SOD1 proteins were all conjugated to themselves or to other proteins via disulfide bonds. In comparison, a significant fraction of the insoluble WT and mutant TDP-43 proteins in the pellets was not disulfide-linked and could be disassociated by the SDS. However, a fraction of the insoluble TDP-43 protein appeared to form distinct dimers and oligomers via disulfide bonds, a result observed for all the tested full-length variants and TDP-C25 (Fig. 4A, top panel). When the soluble fractions were treated under the same conditions, no disulfide-linked SOD1 G85R or TDP-43 high-molecular-weight species were detected (Fig. 4A, lower panel), indicating that these non-native species enriched in the insoluble fractions were not a product of treatments ex vivo. To test the role of the disulfide bond in the aggregate formation, we generated a cysteine-free TDP-C25 mutant (C243A) and performed aggregation analysis. C243A blocked the formation of the disulfide-linked dimer, but the aggregation of the majority of the TDP-C25 protein was not affected significantly (Fig. 4B). Taken together, these results suggest that disulfide-linked TDP-43 dimers and oligomers can be formed in vivo but they are not required for the aggregate formation.
Transgenic C. elegans expressing TDP-43 are sensitive to elevated environmental temperature
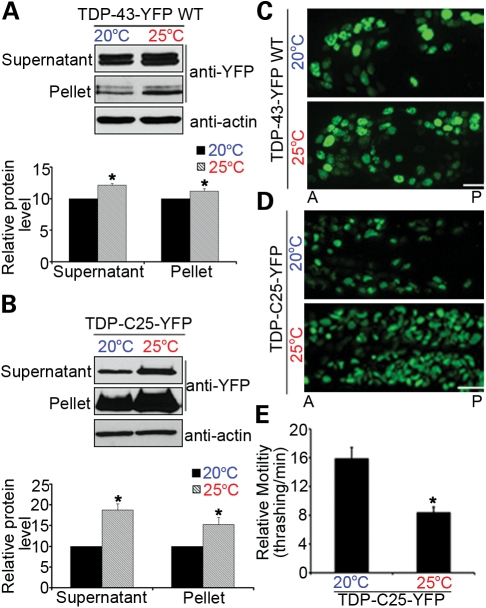
Since protein misfolding is sensitive to the thermal environment, we asked whether the locomotor defects and protein aggregation in the transgenic C. elegans models expressing TDP-43 are dependent on the environmental temperatures. The C. elegans models expressing full-length TDP-43 variants and the TDP-C25 fragment were grown at the preferred temperature of 20°C or at an elevated temperature, 25°C. Compared with those grown at 20°C, the TDP-43 transgenic C. elegans grown at 25°C exhibited more pronounced phenotypes, including higher levels of soluble and insoluble proteins (Fig. 5A and B), more accumulation of fluorescently tagged proteins in neurons (Fig. 5C and D) and worsened locomotor defects (Fig. 5E). The C. elegans strains expressing the highly aggregation-prone protein, TDP-C25, appeared to be particularly sensitive to the elevated temperature. Quantitative reverse transcription polymerase chain reaction (RT–PCR) was used to measure the mRNA levels of the TDP-43 variant, and the temperature change did not change the gene expression levels (Supplementary Material, Fig. S5). These data suggested that elevated temperature could increase TDP-43 protein misfolding and the overall burden on protein quality control, resulting in the accumulation of soluble and insoluble TDP-43 and a more severe neurotoxicity.
Figure 5.
Elevated environmental temperature increases protein aggregation and neurotoxicity in transgenic TDP-43 C. elegans. Both WT TDP-43-YFP (A) and TDP-C25-YFP (B) proteins show increased accumulation in soluble and insoluble fractions at 25°C when compared with those at 20°C (*P < 0.05; n = 3). Ten micrograms (∼1/20) of soluble supernatant protein and 5 µg (∼1/4) of insoluble pellet protein were analyzed by immunoblotting. (C) Elevated temperature increases the accumulation of WT TDP-43-YFP in C. elegans neurons. Anterior (A) and posterior (P) regions of heads of L4 animals are shown. (D) Aggregates of TDP-C25-YFP are markedly increased in number and size in response to the elevated temperature. Head neurons of L1 animals are shown. (E) Quantitation of the effects of temperature elevation on the locomotor behavior of 1-day adult TDP-C25-YFP animals (*P < 0.05). n = 32, error bars represent the SEM. Scale bars: 5 µm.
HSF-1 is a major factor required for protection against the neurotoxicity and protein aggregation of TDP-43
Next, we asked whether TDP-43-mediated neurotoxicity and protein aggregation were directly regulated by components of the protein quality-control system. We first used RNAi to knockdown the expression of candidate genes and test how the RNAi knockdown affected TDP-43-induced neurodegeneration. To enhance the effect of the RNAi treatments on neurons, transgenic C. elegans stably expressing WT TDP-43-YFP or TDP-C25-YFP were crossed into a background of eri-1(mg366);lin-15B(n744) (54). The sensitized C. elegans strains were then fed with bacteria expressing double-stranded RNA against target genes.
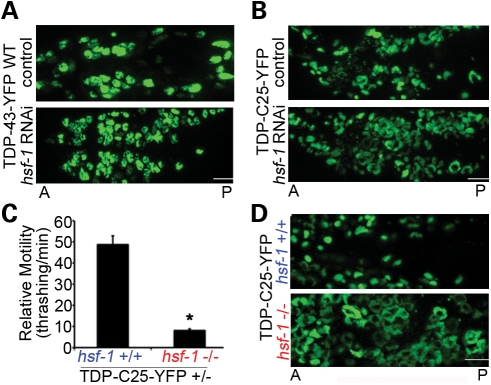
Among the candidate genes, we identified HSF-1 as a major protective factor because its deficiency resulted in a strong aggravation of the neurodegenerative and protein aggregation phenotypes associated with TDP-43 (Fig. 6A and B). The C. elegans strains expressing TDP-C25-YFP, which is highly unstable and aggregation-prone, were more sensitive to the HSF-1 knockdown than the strains expressing WT TDP-43-YFP. Moreover, we validated the RNAi results by crossing the TDP-43 transgenic strains to a loss-of-function allele of hsf-1, sy441. The hsf-1(sy441) mutant itself has largely normal movement behavior at 20°C (Supplementary Material, Fig. S6); however, it significantly worsened the locomotor phenotypes of TDP-C25-YFP strains (Fig. 6C, the hemizygous strain was tested due to the developmental arrest of the homozygous transgeneic animals in the hsf-1 background). The phenotype of the full-length TDP-43 transgenic worms was so severe that a worsening effect of hsf-1(sy441) was difficult to measure by the thrashing assay. hsf-1(sy441) also worsened the growth defect of TDP-C25-YFP strains. In fact, the TDP-C25 homozygous animals for hsf-1(sy441) had such retarded growth at 20°C that they failed to develop and pass the L2 developmental stage. There was a concomitant pronounced increase in the aggregation of TDP-C25-YFP in C. elegans neurons; the aggregates grew significantly in size, filling the cytoplasmic space of the majority of neurons (Fig. 6D). The effect of loss of function of hsf-1 on the aggregation of TDP-43 was not a result of altered gene expression, as confirmed by quantitative RT–PCR (Supplementary Material, Fig. S5), suggesting that HSF-1 acted through protein quality control.
Figure 6.
HSF-1 robustly modulates protein aggregation and neurotoxicity in transgenic TDP-43 C. elegans. (A and B) Reduction in hsf-1 by RNAi increases the accumulation of WT TDP-43-YFP and TDP-C25-YFP in C. elegans neurons. Integrated TDP-43 transgenic C. elegans lines were sensitized for RNAi treatment by crossing into a background of eri-1(mg366);lin-15B(n744) and fed with bacteria expressing double-stranded RNA targeting hsf-1. The hsf-1 RNAi increases the accumulation of TDP-43-YFP in the neuronal nuclei and the aggregates of TDP-C25-YFP in the cytoplasm. Anterior (A) and posterior (P) regions of heads of L4 animals are shown. (C) Since homozygous TDP-C25-YFP transgenic animals fail to thrive in the presence of hsf-1(sy441), quantitation of the effects on the locomotor behavior was carried out on TDP-C25-YFP hemizygous transgenic animals (TDP-C25-YFP +/−; 1-day adult) (*P < 0.05). n = 32, error bars represent the SEM. (D) In the presence of loss-of-function mutant hsf-1(sy441), aggregates of TDP-C25-YFP are markedly increased in number and size in response to the elevated temperature. Head neurons of L1 animals are shown. Scale bars: 5 µm.
Insulin/IGF-1 signaling modulates the neurotoxicity and protein aggregation of TDP-43
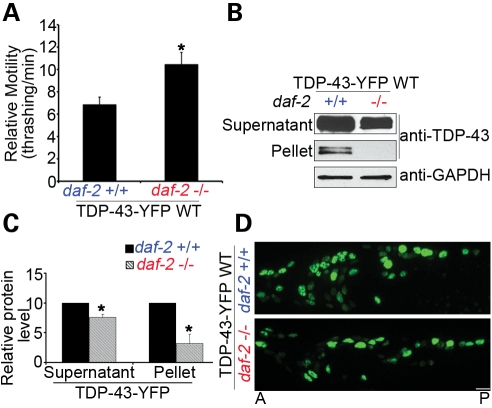
Aging is associated with the TDP-43 proteinopathy in humans. In our relatively short-lived C. elegans models, the locomotor defects deteriorated over time significantly faster than WT N2 animals (Supplementary Material, Fig. S4). We asked whether the C. elegans genes involved in longevity could modulate the neurotoxicity of TDP-43. We crossed a loss-of-function allele of the gene encoding the insulin/IGF1R daf-2, e1370, which doubles the lifespan in C. elegans, to the transgenic C. elegans stably expressing the full-length WT TDP-43-YFP. The daf-2(e1370) allele led to a marked improvement in the locomotor defects in the TDP-43 transgenic C. elegans (Fig. 7A) as well as a significant reduction in aggregates in the insoluble pellets of extracted worm homogenates (Fig. 7B and C). Consistent with the reduction in the insoluble protein, the accumulation of WT TDP-43-YFP in the nucleus was significantly attenuated (Fig. 7D).
Figure 7.
Loss-of-function mutant daf-2(e1370) suppresses neurotoxicity and aggregation of WT full-length TDP-43-YFP in C. elegans. (A) The neurotoxicity of WT TDP-43-YFP in the C. elegans models, as measured by the thrashing rate in liquid medium, is significantly suppressed by a loss-of-function allele of the insulin/IGF1R daf-2, e1370. n = 32, error bars represent the SEM. (B) Loss of function of daf-2 decreases the soluble and insoluble aggregated TDP-43 proteins in C. elegans. The C. elegans homogenates were detergent extracted, and 10 µg (∼1/20) of soluble supernatant protein and 5 µg (∼1/4) of insoluble pellet protein were analyzed by immunoblotting. (C) Quantification of the TDP-43 proteins levels (*P < 0.05, n = 3). (D) Loss of function of daf-2 decreases the accumulation of full-length TDP-43-YFP in the nuclei of C. elegans neurons. Representative 1-day adult images are shown. Scale bars: 5 µm.
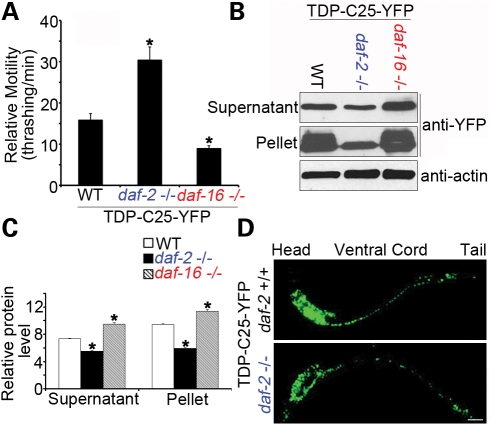
To further assess the effect of insulin/IGF-1 signaling on the protein aggregation phenotypes displayed by the TDP-43 transgenic C. elegans models, we crossed the daf-2(e1370) allele to the transgenic C. elegans stably expressing the highly aggregation-prone variant TDP-C25-YFP. daf-2(e1370) robustly protected against the neurotoxicity and protein aggregation of TDP-C25-YFP, as the number and size of the fluorescent aggregates were significantly decreased (Fig. 8A–D). To further study the role of insulin/IGF-1, we crossed the TDP-43 transgenic animals to a daf-16 loss-of-function allele, mu86, that blocks the lifespan extension effect of daf-2(e1370) (55,56). The phenotype of the full-length TDP-43 transgenic worms was so pronounced that a worsening effect of daf-16(mu86) was difficult to measure by the thrashing assay. However, an increase in the protein aggregation of TDP-C25-YFP was observed in live C. elegans carrying daf-16(mu86). Indeed, in sensitive behavioral and biochemical assays, daf-2(e1370) and daf-16(mu86) showed opposing effects on the neurotoxicity and aggregation of TDP-C25-YFP (Fig. 8A–C). The IGF-1 pathway did not significantly alter the behaviorsly of the YFP-only animals (Supplementary Material, Fig. S6), suggesting that the genetic modifiers acted independently of the YFP tag. For the TDP-43 variants, both the supernatant and the pellet protein fractions were significantly decreased by daf-2(e1370 ) and increased by daf-16(mu86) (Fig. 8C). No evidence of gene expression changes was found (Supplementary Material, Fig. S5), suggesting that the deficiency in insulin/IGF-1 signaling exerted its protection against the neurotoxicity by clearing misfolded or aggregated proteins. Since the centrifugation sedimented only large aggregates, the alterations of the TDP-43 protein levels in the supernatant fractions could be due to the misfolded and aggregated proteins retained in the supernatants.
Figure 8.
daf-2(e1370) and daf-16(mu86) exert opposing effects on neurotoxicity and aggregation of TDP-C25-YFP in C. elegans. (A) The neurotoxicity of TDP-C25-YFP in the C. elegans models, as measured by the thrashing rate in liquid medium, is significantly suppressed by the loss-of-function mutant daf-2(e1370) but increased by the loss-of-function allele daf-16(mu86). n = 32, error bars represent the SEM. (B) daf-2(e1370) and daf-16(mu86) show opposing effects on the aggregation of TDP-C25-YFP. Ten micrograms (∼1/20) of soluble supernatant protein and 5 µg (∼1/4) of insoluble pellet protein were analyzed by immunoblotting. (C) Quantification of the TDP-C25 proteins levels (*P < 0.05, n = 3). (D) Loss of function of daf-2 decreases the number and size of neuronal aggregates of TDP-C25. Representative L2 worm images are shown. Scale bar: 15 µm.
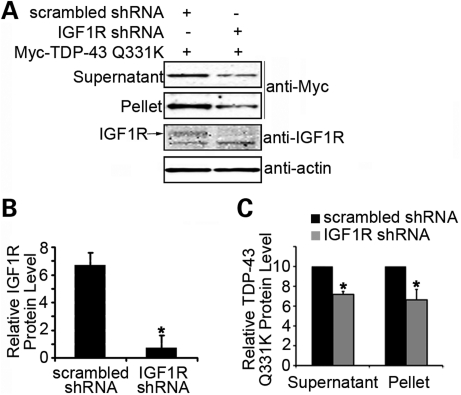
To determine whether the insulin/IGF-1 signaling also modulates the aggregation of TDP-43 in a mammalian system, we used the aforementioned cell-based model of TDP-43 aggregation. Human HEK293T cells were transiently transfected with Myc-tagged human TDP-43 carrying Q331K, a mutation linked to familial ALS, with or without an small hairpin RNA (shRNA)-mediated knockdown of the IGF1R. Multiple shRNA against different regions of IGF1R were used, and the knockdown was verified by anti-IGF1R immunoblotting (Fig. 9A and B). The cell lysates were fractionated into supernatants and pellets and analyzed by anti-Myc immunoblotting. As was observed for C. elegans, a deficiency of the mammalian IGF1R led to a significant reduction in both soluble and aggregated TDP-43 (Fig. 9A and C), suggesting that the insulin/IGF-1 pathway regulates the clearance of misfolded and aggregated TDP-43 through an evolutionarily conserved mechanism.
Figure 9.
Reduction in the function of insulin/IGF1R suppresses the aggregation of TDP-43 in human cells. (A) Knockdown of IGF1R by shRNA in HEK293T cells decreases the accumulation of soluble and insoluble aggregated Myc-TDP-43 Q331K. Twelve micrograms (∼1/50) of soluble supernatant protein and 15 µg (∼1/10) of insoluble pellet protein were analyzed by immunoblotting with the anti-Myc antibody. (B) Quantitation of the knockdown of the IGF1R protein expression is shown. (C) Quantitation of soluble and insoluble Myc-TDP-43 Q331K protein levels after the knockdown of IGF1R. *P < 0.05, n = 3, error bars represent the SEM.
DISCUSSION
In the present study, we have established C. elegans models expressing full-length and truncated human TDP-43 variants that are associated with neurodegenerative diseases ALS and FTLD. These C. elegans models developed robust behavioral phenotypes including the locomotor defects that provide a sensitive readout for studying the neurotoxicity of TDP-43. We further demonstrated in C. elegans and human cells that TDP-43 has a high aggregation propensity that is intrinsic to its WT form. A C-terminal 25 kDa fragment of TDP-43 implicated in human patients was found to be particularly aggregation-prone. An elevation in environmental temperature was associated with increased aggregation and neurotoxicity of the TDP-43 proteins in the C. elegans models. Thus, the results of this study suggest that TDP-43 misfolding and aggregation contribute to its neurotoxicity. Consistent with this conclusion was the observation that HSF-1, a key mediator of protein quality control, was a potent regulator of the TDP-43-induced neurotoxicity in C. elegans. Furthermore, we found that alterations in the insulin/IGF-1 signaling pathway, an evolutionarily conserved pathway that regulates lifespan, significantly ameliorated the neurotoxicity and protein aggregation associated with TDP-43, providing a mechanistic link between the aging program and the TDP-43 proteinopathy.
Neurotoxicity and protein aggregation of TDP-43
The nature of the toxicity of TDP-43 that leads to neurodegeneration in ALS and FTLD is poorly understood. The results of this study indicate that TDP-43 is prone to misfold and aggregate, a property tightly associated with its neurotoxicity. Although the phenotype of a loss-of-function mutant of the C. elegans TDP-43 ortholog, tdp-1, is largely normal (unpublished observation), transgenic animals expressing human TDP-43 in neurons developed pronounced behavioral phenotypes. In C. elegans and human cells, accumulated TDP-43 has a high propensity to form insoluble protein aggregates. When compared with SOD1, TDP-43 has several distinct features with regard to its aggregation and neurotoxicity. First, unlike SOD1, in which a single point mutation can transform a highly stable protein into an aggregation-prone and toxic mutant, the WT form of TDP-43 is already highly aggregation-prone and potentially toxic, suggesting that neurons are sensitive to the abnormal accumulation of this protein. This feature is in accord with the recent reports of the toxicity of WT TDP-43 in rodent models (22,24,25), and the observation that inclusion bodies immunopositive for TDP-43 have been found in the central nervous system of a subset of ALS and FTLD patients without TDP-43 mutations (4). Second, the neurotoxicity induced by TDP-43 in the C. elegans model is more severe than that induced by mutant SOD1 under the same experimental conditions. The neurotoxicity of TDP-43 may be related to its high propensity to aggregate. In addition to its unusually high aggregation propensity, there could be other factors that contribute to the severe toxicity of TDP-43, such as those related to its functions in transcription regulation and RNA processing. Disease-associated mutations or elevation of expression levels may perturb the normal functions of TDP-43 and lead to neurotoxicity. Recent C. elegans studies reported that RNA-binding functions or phosphorylation of TDP-43 promoted the toxicity of TDP-43 (30,31). TDP-43 may carry out its normal functions in large protein complexes, because a significant fraction of the endogenous protein exists in sedimentable structures (Fig. 3D). We propose that the misfolding of TDP-43 causes disruption of its normal binding complexes as well as gain of aberrant interaction, leading to cellular dysfunction. Moreover, in contrast to SOD1, which is mainly cytoplasmic, full-length TDP-43 is predominantly localized to the nucleus. Although tags such as YFP may influence protein behavior, we observed that full-length TDP-43-YFP was localized to the C. elegans neuronal nucleus as expected. Furthermore, the high aggregation propensity of TDP-43 was observed when studied with different tags in both C. elegans and mammalian cells. Indeed, the endogenous TDP-43 appeared to be much more insoluble than SOD1 in human cells. Further studies are needed to clarify the toxic effects of TDP-43 misfolding and aggregation in the nucleus.
Several cleavage sites have been reported to generate C-terminal 25 kDa fragments of TDP-43 in cells and patients (47,57,58). The pathologic TDP-C25 fragment (residues 219–414) tested in this study showed a remarkably high propensity to aggregate. Compared with the full-length proteins, soluble TDP-C25 is rapidly degraded, perhaps as a result of misfolding and disassociation from its protective partners. Lacking the predicted nuclear localization signal (57), TDP-C25 aggregated selectively into the cytoplasm. Consistent with the report that TDP-C25 is enriched in the insoluble aggregates in human patients (4), our identification of the high aggregation potential of TDP-C25 suggests that it could nucleate the aggregate formation despite of its low steady-state levels in neurons. Interestingly, TDP-C25 is less toxic to C. elegans neurons than full-length TDP-43 under the same experimental conditions. In comparison, TDP-C25 is much more insoluble than its full-length counterparts. This observation supports the notion that although the insoluble aggregates are potentially toxic to the cell, the toxicity is not linearly proportional to the protein mass in the aggregates, as the proteins trapped inside the aggregates are probably functionally inert or species other than large aggregates are more toxic. In addition to the insoluble aggregates, soluble non-native species such as oligomers or misfolded protein itself could also contribute to the toxicity. Moreover, the detection of distinct dimers and oligomers of TDP-43 linked by disulfide bonds suggests that specific oligomeric species exist in the cell. However, the disulfide bond is not required for the formation of the TDP-C25 aggregates, suggesting that the highly aggregation-prone protein can form polymeric structures through other forces such as hydrophobic interactions. Further studies are needed to understand the TDP-43 protein complexes under native and disease conditions.
TDP-43 toxicity modulated by HSF-1 and insulin/IGF-1 signaling
The abnormal accumulation of TDP-43 proteins is the hallmark in a subset of cases of ALS and FTLD (4,10). Our demonstration that the neurotoxicity of TDP-43 is modulated by HSF-1 and the observation that deficiency in insulin/IGF-1 signaling suppresses the toxicity support the concept that the toxicity is dependent on protein misfolding (Fig. 10). TDP-43 is implicated in RNA processing and DNA transcription (11,12). Nuclear and cytoplasmic inclusions of TDP-43 were found in neurons of ALS and FTLD patients (4). The truncated variant, TDP-C25, is a component of the human pathology and has been observed to form cytoplasmic aggregates (Figs 1 and 3). TDP-43 misfolding could disrupt its native complexes or cause aberrant protein interactions and thus perturb the cellular functions. HSF-1 was found to be a particularly strong regulator of the TDP-43 toxicity. In response to protein misfolding, HSF-1 is activated and acts as a master transcriptional regulator of molecular chaperones that are critical facilitators for protein quality control (59). A loss of function of HSF-1 could compromise this stress response and therefore worsen the misfolding-dependent toxicity of TDP-43. A related observation is that elevated environmental temperature exacerbated the TDP-43 toxicity, probably by promoting the unfolding of TDP-43 and burdening the protein quality-control systems. We also identified the insulin/IGF1R DAF-2 as a novel suppressor of the TDP-43 toxicity. Modulation of the TDP-43 neurotoxicity by DAF-2 and the downstream FOXO transcriptional factor DAF-16 are consistent with the notion that TDP-43 toxicity is mediated by stress related to protein misfolding, since this signaling has been shown to regulate protein folding-related stress responses (60). At least part of the DAF-16-induced stress responses is mediated by the activation of molecular chaperone networks (60). Indeed, we consistently observed that the modulation of the TDP-43 neurotoxicity was correlated with the changes in its protein aggregation, underscoring the role of protein misfolding in the neurotoxicity.
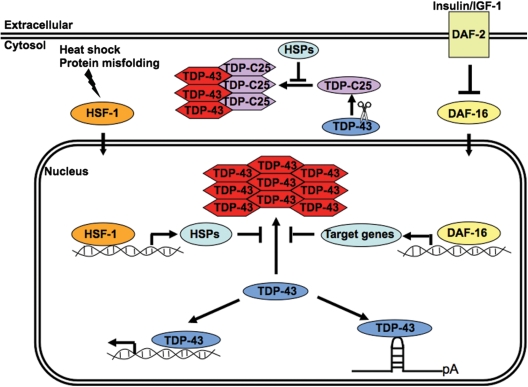
Figure 10.
A model of TDP-43 misfolding and aggregation as a modulator of its toxicity. A schematic representation of the mechanisms through which the HSF-1 and IGF-1 pathways may regulate the folding and aggregation of TDP-43 proteins. The normal DNA/RNA-binding functions of TDP-43 could be disrupted by the misfolding and aggregation of the protein. The nuclear and cytoplasmic aggregates formed by misfolded TDP-43 and its fragments, such as TDP-C25, may further perturb cellular functions by burdening the protein cellular control. The HSF-1 and IGF-1 signaling may regulate the misfolding and toxicity of TDP-43 through modulation of the protein quality-control system, such as the molecular chaperone networks. Oval and hexagon shapes represent native and misfolded TDP-43, respectively. HSP, heat shock protein; pA, polyadenylation.
Aging in C. elegans is also influenced by environmental temperature (61), HSF-1 (43) and insulin/IGF-1 signaling (41). There is an emerging notion that aging is associated with the decline in protein homeostasis (43,44). We recently observed that the molecular chaperones are up-regulated in normally aged brains (62). The results of the current study suggest that protein misfolding could be the mechanistic basis for the age dependence of the TDP-43 proteinopathy. Reduced IGF-1 signaling was recently shown to delay age-dependent Aβ toxicity in mice by modulating its oligomers and dense plaques formed outside the neurons in the brain (63). The results of our study demonstrated that reduced insulin/IGF-1 signaling could ameliorate TDP-43 toxicity by decreasing its aggregation in neurons. It was also recently shown that nuclear targeting of FOXO3a, the mammalian homolog of DAF-16, protected against various insults implicated in motor neuron diseases such as mutant SOD1 and mutant p150glued (64). In the present study, we found that, in mammalian cells, a deficiency of the insulin/IGF1R attenuated the TDP-43 protein aggregation. Future studies in rodent models would be needed to confirm the potential use of this pathway for therapeutic interventions in TDP-43-related neurodegenerative diseases.
MATERIALS AND METHODS
Deoxyribonucleic acid
The C. elegans transgene constructs were prepared by subcloning the human TDP-43 cDNAs, with or without a C-terminal YFP tag, into a modified plasmid pPD30_38 (Fire Lab Vector, Addgene), with the promoter replaced with that of the snb-1 gene. For mammalian cell transfection, the human TDP-43 and SOD1 cDNAs were subcloned into a pRK5 plasmid containing an N-terminal Myc tag (EQKLISEEDL).
Caenorhabditis elegans
The N2 Bristol C. elegans strain, cultured under standard conditions at 20°C (65), was used for all experiments unless otherwise indicated. To generate transgenic lines, a DNA solution containing 20 ng/µl of the expression construct was injected into hermaphrodite gonads (66). The expression of TDP-43-YFP was visualized by a C-terminal YFP tag. Multiple extrachromosomal lines were established based on the fluorescent markers. These lines were further treated with 30 µg/ml trimethylpsoralen (Sigma) and 300 µJ of 365 nm UV light to screen for integrated lines that stably expressed the transgenes. Each integrated line was backcrossed with the N2 strain at least four times. Integrated lines expressing only YFP were generated as controls. Integrated lines expressing the human WT SOD1-YFP and G85R variants have been described previously (2). Mutant strains obtained from the Caenorhabditis Genetics Center were the following: PS3551 [_hsf-1(sy441)_], CB1370 [_daf-2(e1370)_], CF1038 [_daf-16(mu86)_] and KP3948 [eri-1(mg366); _lin-15B(n744)_]. The transgenic strains generated in this study were the following: IW62 [_Psnb-1::YFP(iwIs25)_], IW63 [_Psnb-1::TDP-43-YFP WT(iwIs26)_], IW33 [_Psnb-1::TDP-C25-YFP(iwIs22)_], IW20 [_Psnb-1::TDP-43-YFP Q331K(iwEx20)_], IW46 [_Psnb-1::TDP-43-YFP M337V(iwEx28)_], IW31 [_Psnb-1::SOD1-YFP WT(iwIs27)_] and IW8 [_Psnb-1::SOD1-YFP G85R(iwIs8)_].
Caenorhabditis elegans behavioral assay
The C. elegans strains were observed stereoscopically, noting the duration and velocity of their spontaneous movement. Animals were also examined for their maximum motility by observing their forward and backward movements after being touched by a metal pick. To quantitate their relative motility, animals were transferred to M9 buffer (3 mg/ml KH2PO4, 6 mg/ml Na2HPO4, 5 mg/ml NaCl and 1 mm MgSO4) for the thrashing assay at room temperature. After 1 min of adaptation, the number of body bends or thrashes was counted for 1 min as an index of the locomotor phenotype. A thrash is counted when both the head and the tail bend away from the anterior–posterior axis more than 45°.
RNAi in C. elegans
To sensitize C. elegans neurons for RNAi treatments, the eri-1(mg366) and lin-15B(n744) alleles were crossed to integrated lines expressing transgenic TDP-43-YFP or TDP-C25-YFP. Selected bacterial clones from an RNAi feeding library (Source BioScience, Nottingham, England) (67) were grown and plated to feed C. elegans. L4 animals were transferred to the RNAi plates and the phenotypes of their offspring were analyzed.
4′,6′-Diamidino-2-phenylindole staining in C. elegans
For the TDP-43 co-localization studies in C. elegans, L4 worms were first washed with M9 buffer and H2O. Then, the worms were fixed in 500 µl methanol on dry ice for 5 min. Following three washes of phosphate-buffered saline with 0.1% Tween 20, the fixed worms were mounted onto microscope slides in a solution of 2.5% 1,4-diazobicyclo[2,2,2]-octane in 100 mm Tris, pH 8.8, with 50% glycerol and 0.2 µg/ml 4′,6′-diamidino-2-phenylindole (DAPI) dihydrochloride.
Microscopy and FRAP analysis
Videos of C. elegans movement were recorded using a Leica fluorescence stereoscope (MZ165) and a Leica DFC310 FX camera. For high-magnification imaging, animals were immobilized with 10 mm levamisole and examined with a Zeiss AxioObserver Z1 with Apotome imaging system. For FRAP analysis, a Zeiss LSM510 META laser confocal microscope was used.
Protein aggregation assays for C. elegans and mammalian cells
The biochemical assay used to detect insoluble aggregated proteins has been described previously (49). Caenorhabditis elegans were homogenized by sonication on ice with the extraction buffer [10 mm Tris–HCl, pH 8.0, with 1 mm ethylenediaminetetraacetic acid (EDTA),100 mm NaCl and 0.5% NP-40] supplemented with mini-EDTA protease inhibitor cocktail (Roche) and 50 mm iodoacetamide (Sigma). The lysates were then transferred to a Coulter Airfuge and centrifuged at 25 psi (∼130 000_g_) for 5 min. The supernatant was saved as the ‘S1’ sample. The remaining pellet was transferred and sonicated in extraction buffer to resuspend the pellet into solution. This resuspended pellet was ultracentrifuged again for 5 min, and the remaining pellet was transferred to 100 µl of resuspension buffer (10 mm Tris–HCl, pH 8.0, with 1 mm EDTA, 100 mm NaCl, 0.5% NP-40, 0.5% deoxycholic acid and 2% SDS) and sonicated until the pellet was resuspended in solution; this preparation was saved as the ‘P2’ sample. The S1 and P2 fractions were subjected to SDS–PAGE and transferred to nitrocellulose membranes (Bio-Rad). The immunoblotting analyses were performed using the following antibodies: 1:2000 anti-Myc (clone 9E10) conjugated to horseradish peroxidase (Roche), 1:2000 anti-YFP (BD Biosciences), 1:1000 anti-TDP-43 (Proteintech Group, Chicago, IL, USA) and 1:3000 anti-SOD1 (SOD-100, Enzo, Farmingdale, NY, USA). Proteins were visualized using enhanced chemiluminescence and quantitated using ImageJ software. The Student's _t_-tests were used for statistical analysis.
For the protein aggregation assay in mammalian cells, HEK293T cells were plated onto 60 mm tissue culture dishes at a density of 106 cells per dish. Myc-tagged TDP-43 and SOD1 variants were engineered and sequenced in their entirety. Plasmid (6 µg) was transfected using Invitrogen's Lipofectamine 2000 reagent according to the manufacturer's instructions. At 48 h following transfection, the cells were harvested in the extraction buffer and analyzed as described above.
For the denaturing and non-reducing SDS–PAGE, S1 and P2 fractions were mixed in Laemmli sample buffer (2% SDS) without β-mercaptoethanol. The samples were boiled before gel electrophoresis.
Quantitative RT–PCR
Caenorhabditis elegans was harvested in M9 buffer, and RNA was isolated using a phenol–chloroform extraction with TRIzol reagent (Invitrogen), followed by purification using RNeasy mini kit (Qiagen). A two-step RT–PCR was employed to assess relative changes in transgenic transcripts using iScript cDNA Synthesis Kit and SYBR Green Supermix (Bio-Rad). Standard curves were generated for the worm gdh-1 and human TDP-43 primer pairs. The primers for the worm gdh-1 were CGGTATCATCGAAGGACTCAT and GTCCATCTCTCCACAGCTTT. The primers for the human TDP-43 were GCCTTCGGTTCTGGAAATAACTC and CCCGACCCTGCATTGGAT (sequences included in TDP-C25). Samples were processed in triplicates, and the relative ratio between the two genes was calculated using the standard curve equation to measure the expression level of the TDP-43 or TDP-C25 transgenes.
shRNA knockdown in mammalian cells
Small hairpin oligonucleotides matching the following IGF1R RNA sequences were subcloned into the pRFP-C-RS plasmid (Origene, Rockville, MD, USA) using _Bam_HI/_Hin_dIII restriction sites: (i) GGATGCGGTGTCCAATAACTACATTGTGG, (ii) AACGGCAACCTGAGTTACTACATTGTGCG and (iii) TGGCATACCTCAACGCCAATAAGTTCGTC. HEK293T cells at a density of 3.2 × 105 per 60 mm dish were transfected with a 4 µg (total) mixture of the three shRNAs and 20 ng TDP-43 Q331K and harvested 72 h later for analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Institutes of Health (NS062089) to J.W., the Robert Packard Center for ALS Research at Johns Hopkins, the Muscular Dystrophy Association and the Johns Hopkins Claude D. Pepper Older Americans Independence Center.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
We would like to thank Katherine H. Reiter for her technical assistance and Wang laboratory members for helpful discussion. We thank Dr David Borchelt for reading the manuscript. Some nematode strains used in this work were provided by Caenorhabditis Genetics Center, an NIH supported National Center for Research Resources.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Cleveland D.W., Rothstein J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2001;2:806–819. doi: 10.1038/35097565. doi:10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 2.Bruijn L.I., Miller T.M., Cleveland D.W. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. doi:10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 3.Sleegers K., Cruts M., Van Broeckhoven C. Molecular pathways of frontotemporal lobar degeneration. Annu. Rev. Neurosci. 2010;33:71–88. doi: 10.1146/annurev-neuro-060909-153144. doi:10.1146/annurev-neuro-060909-153144. [DOI] [PubMed] [Google Scholar]
- 4.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. doi:10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 5.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. doi:10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. doi:10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benajiba L., Le Ber I., Camuzat A., Lacoste M., Thomas-Anterion C., Couratier P., Legallic S., Salachas F., Hannequin D., Decousus M., et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann. Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. doi:10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- 8.Gitcho M.A., Bigio E.H., Mishra M., Johnson N., Weintraub S., Mesulam M., Rademakers R., Chakraverty S., Cruchaga C., Morris J.C., et al. TARDBP 3'-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 2009;118:633–645. doi: 10.1007/s00401-009-0571-7. doi:10.1007/s00401-009-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacs G.G., Murrell J.R., Horvath S., Haraszti L., Majtenyi K., Molnar M.J., Budka H., Ghetti B., Spina S. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov. Disord. 2009;24:1843–1847. doi: 10.1002/mds.22697. doi:10.1002/mds.22701. [DOI] [PubMed] [Google Scholar]
- 10.Mackenzie I.R., Feldman H.H. Ubiquitin immunohistochemistry suggests classic motor neuron disease, motor neuron disease with dementia, and frontotemporal dementia of the motor neuron disease type represent a clinicopathologic spectrum. J. Neuropathol. Exp. Neurol. 2005;64:730–739. doi: 10.1097/01.jnen.0000174335.27708.0a. doi:10.1097/01.jnen.0000174335.27708.0a. [DOI] [PubMed] [Google Scholar]
- 11.Ou S.H., Wu F., Harrich D., Garcia-Martinez L.F., Gaynor R.B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y.M., Baralle F.E. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. doi:10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 13.Strong M.J., Volkening K., Hammond R., Yang W., Strong W., Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. doi:10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Bose J.K., Wang I.F., Hung L., Tarn W.Y., Shen C.K. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J. Biol. Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. doi:10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiesel F.C., Voigt A., Weber S.S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M.L., Kern J.V., Rasse T.M., et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. doi:10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S.H., Shanware N., Bowler M.J., Tibbetts R.S. ALS-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to coregulate HDAC6 mRNA. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.154831. doi:10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang P.M., Ling J., Jeong Y.H., Price D.L., Aja S.M., Wong P.C. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl Acad. Sci. USA. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. doi:10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling S.C., Albuquerque C.P., Han J.S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D.W. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl Acad. Sci. USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. doi:10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L.S., Cheng W.C., Hou S.C., Yan Y.T., Jiang S.T., Shen C.K. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 20.Sephton C.F., Good S.K., Atkin S., Dewey C.M., Mayer P., III, Herz J., Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. doi:10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegorzewska I., Bell S., Cairns N.J., Miller T.M., Baloh R.H. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl Acad. Sci. USA. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. doi:10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wils H., Kleinberger G., Janssens J., Pereson S., Joris G. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc. Natl Acad. Sci. USA. 2010;107:3858–3863. doi: 10.1073/pnas.0912417107. doi:10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stallings N.R., Puttaparthi K., Luther C.M., Burns D.K., Elliott J.L. Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol. Dis. 2010;40:404–414. doi: 10.1016/j.nbd.2010.06.017. doi:10.1016/j.nbd.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y.F., Gendron T.F., Zhang Y.J., Lin W.L., D'Alton S., Sheng H., Casey M.C., Tong J., Knight J., Yu X., et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. doi:10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan X., Chiang P.M., Price D.L., Wong P.C. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl Acad. Sci. USA. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. doi:10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H., Huang C., Chen H., Wang D., Landel C.P., Xia P.Y., Bowser R., Liu Y.J., Xia X.G. Transgenic rat model of neurodegeneration caused by mutation in the TDP gene. PLoS Genet. 2010;6:e1000887. doi: 10.1371/journal.pgen.1000887. doi:10.1371/journal.pgen.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Ray P., Rao E.J., Shi C., Guo W., Chen X., Woodruff E.A., III, Fushimi K., Wu J.Y. A Drosophila model for TDP-43 proteinopathy. Proc. Natl Acad. Sci. USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. doi:10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson K.A., Kim S.H., Wassarman D.A., Tibbetts R.S. Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS) J. Biol. Chem. 2010;285:11068–11072. doi: 10.1074/jbc.C109.078527. doi:10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabashi E., Lin L., Tradewell M.L., Dion P.A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H.D., Vande Velde C., et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. doi:10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 30.Ash P.E., Zhang Y.J., Roberts C.M., Saldi T., Hutter H., Buratti E., Petrucelli L., Link C.D. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum. Mol. Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. doi:10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liachko N.F., Guthrie C.R., Kraemer B.C. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J. Neurosci. 2010;30:16208–16219. doi: 10.1523/JNEUROSCI.2911-10.2010. doi:10.1523/JNEUROSCI.2911-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2009;9:1104–1120. doi: 10.1021/pr901076y. doi:10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Fan H., Ying Z., Li B., Wang H., Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci. Lett. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. doi:10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 34.Caccamo A., Majumder S., Deng J.J., Bai Y., Thornton F.B., Oddo S. Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J. Biol. Chem. 2009;284:27416–27424. doi: 10.1074/jbc.M109.031278. doi:10.1074/jbc.M109.031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. doi:10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 36.Barmada S.J., Skibinski G., Korb E., Rao E.J., Wu J.Y., Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. doi:10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston J.A., Dalton M.J., Gurney M.E., Kopito R.R. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. doi:10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Xu G., Borchelt D.R. High molecular weight complexes of mutant superoxide dismutase 1: age-dependent and tissue-specific accumulation. Neurobiol. Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. doi:10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- 39.Wang J., Farr G.W., Zeiss C.J., Rodriguez-Gil D.J., Wilson J.H., Furtak K., Rutkowski D.T., Kaufman R.J., Ruse C.I., Yates J.R., III, et al. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc. Natl Acad. Sci. USA. 2009;106:1392–1397. doi: 10.1073/pnas.0813045106. doi:10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson B.S., Snead D., Lee J.J., McCaffery J.M., Shorter J., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. doi:10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. doi:10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 42.Berryman D.E., Christiansen J.S., Johannsson G., Thorner M.O., Kopchick J.J. Role of the GH/IGF-1 axis in lifespan and healthspan: lessons from animal models. Growth Horm. IGF Res. 2008;18:455–471. doi: 10.1016/j.ghir.2008.05.005. doi:10.1016/j.ghir.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu A.L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. doi:10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 44.Morley J.F., Brignull H.R., Weyers J.J., Morimoto R.I. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. doi:10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen E., Bieschke J., Perciavalle R.M., Kelly J.W., Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. doi:10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 46.Wang J., Farr G.W., Hall D.H., Li F., Furtak K., Dreier L., Horwich A.L. An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000350. doi: 10.1371/journal.pgen.1000350. doi:10.1371/journal.pgen.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 2009;18:3353–3364. doi: 10.1093/hmg/ddp275. doi:10.1093/hmg/ddp275. [DOI] [PubMed] [Google Scholar]
- 48.Mahoney T.R., Luo S., Nonet M.L. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protocols. 2006;1:1772–1777. doi: 10.1038/nprot.2006.281. doi:10.1038/nprot.2006.281. [DOI] [PubMed] [Google Scholar]
- 49.Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N.G., Jenkins N.A., Borchelt D.R. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. doi:10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 50.Wang J., Xu G., Borchelt D.R. Mapping superoxide dismutase 1 domains of non-native interaction: roles of intra- and intermolecular disulfide bonding in aggregation. J. Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. doi:10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng H.X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G.H., Khan M.S., Hung W.Y., Bigio E.H., et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. doi:10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furukawa Y., Fu R., Deng H.X., Siddique T., O'Halloran T.V. Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu, Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc. Natl Acad. Sci. USA. 2006;103:7148–7153. doi: 10.1073/pnas.0602048103. doi:10.1073/pnas.0602048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karch C.M., Prudencio M., Winkler D.D., Hart P.J., Borchelt D.R. Role of mutant SOD1 disulfide oxidation and aggregation in the pathogenesis of familial ALS. Proc. Natl Acad. Sci. USA. 2009;106:7774–7779. doi: 10.1073/pnas.0902505106. doi:10.1073/pnas.0902505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang D., Kennedy S., Conte D., Kim J.K., Gabel H.W., Kamath R.S., Mello C.C., Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. doi:10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- 55.Lin K., Dorman J.B., Rodan A., Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. doi:10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 56.Ogg S., Paradis S., Gottlieb S., Patterson G.I., Lee L., Tissenbaum H.A., Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. doi:10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 57.Igaz L.M., Kwong L.K., Chen-Plotkin A., Winton M.J., Unger T.L., Xu Y., Neumann M., Trojanowski J.Q., Lee V.M. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. doi:10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y.J., Xu Y.F., Cook C., Gendron T.F., Roettges P., Link C.D., Lin W.L., Tong J., Castanedes-Casey M., Ash P., et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl Acad. Sci. USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. doi:10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorger P.K. Heat shock factor and the heat shock response. Cell. 1991;65:363–366. doi: 10.1016/0092-8674(91)90452-5. doi:10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- 60.Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. doi:10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 61.Klass M.R. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech. Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. doi:10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 62.Wang J., Martin E., Gonzales V., Borchelt D.R., Lee M.K. Differential regulation of small heat shock proteins in transgenic mouse models of neurodegenerative diseases. Neurobiol. Aging. 2008;29:586–597. doi: 10.1016/j.neurobiolaging.2006.11.009. doi:10.1016/j.neurobiolaging.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohen E., Paulsson J.F., Blinder P., Burstyn-Cohen T., Du D., Estepa G., Adame A., Pham H.M., Holzenberger M., Kelly J.W., et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. doi:10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mojsilovic-Petrovic J., Nedelsky N., Boccitto M., Mano I., Georgiades S.N., Zhou W., Liu Y., Neve R.L., Taylor J.P., Driscoll M., et al. FOXO3a is broadly neuroprotective in vitro and in vivo against insults implicated in motor neuron diseases. J. Neurosci. 2009;29:8236–8247. doi: 10.1523/JNEUROSCI.1805-09.2009. doi:10.1523/JNEUROSCI.1805-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brenner S. The genetics of C. elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. doi:10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 68.Jonsson P.A., Graffmo K.S., Andersen P.M., Brannstrom T., Lindberg M., Oliveberg M., Marklund S.L. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. doi:10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data