Molecular Nature of Spemann’s Organizer: the Role of the Xenopus Homeobox Gene goosecoid (original) (raw)
. Author manuscript; available in PMC: 2011 May 26.
Summary
This study analyzes the function of the homeobox gene goosecoid in Xenopus development. First, we find that goosecoid mRNA distribution closely mimics the expected localization of organizer tissue in normal embryos as well as in those treated with LiCI and UV light. Second, goosecoid mRNA accumulation is induced by activin, even in the absence of protein synthesis. It is not affected by bFGF and is repressed by retinoic acid. Lastly, microinjection of goosecoid mRNA into the ventral side of Xenopus embryos, where goosecoid is normally absent, leads to the formation of an additional complete body axis, including head structures and abundant notochordal tissue. The results suggest that the goosecoid homeodomain protein plays a central role in executing Spemann’s organizer phenomenon.
Introduction
The “organizer” experiment (Spemann and Mangold, 1924) is one of the best known in biology, but its molecular basis remains largely unknown. If the region where gastrulation starts in an amphibian embryo, called the dorsal lip of the blastopore, is transplanted into the opposite (ventral) side of a host embryo, an entire new body axis results. The transplanted tissue, whose normal fate is to become head (“prechordal”) mesoderm and notochord, is able to recruit cells from the ventral side of the embryo, “organizing” them into axial structures such as somites and neural tube (reviewed by Spemann, 1938; Hamburger, 1988).
This ability of the organizer region to initiate a cascade of cell–cell interactions stimulated a large body of research employing the transplantation methods of experimental embryology; in recent years most of this research has been carried out with Xenopus laevis. The formation of the organizer can be traced back to the moment of fertilization. The egg is initially radially symmetrical, but sperm entry triggers a microtubule-driven rotation of the egg cortex. This intracellular movement eventually leads to the formation of the body axis. The direction of this cortical rotation determines the position of the future dorsal lip, which usually forms opposite to the sperm entry point (Gerhart et al., 1989; Elinson and Kao, 1989). Blastomere transplantation experiments suggest that at the 32-cell stage the dorsal information is located in the most vegetal (bottom) and dorsal cells of the embryo (Gimlich and Gerhart, 1984; Gimlich, 1986), the so-called Nieuwkoop center (Gerhart et al., 1989). Experiments involving coculture of tissue fragments suggest that cells from the Nieuwkoop center release diffusible signals that in turn induce Spemann’s organizer activity in the overlying marginal zone cells (Nieuwkoop, 1973).
One of the most important recent advances in Xenopus embryology is the discovery that peptide growth factors mediate mesoderm induction. Factors related to basic fibroblast growth factor (bFGF) can induce ventral mesoderm (blood, mesothelium, mesenchyme, and some muscle), while growth factors of the TGF-β family, in particular activin (also called XTC-MIF), are very potent dorsal mesoderm inducers (prechordal mesoderm, notochord, and muscle) (Slack et al., 1989; Smith et al., 1989; Green and Smith, 1990). Uncommitted cells from the animal cap (top) region of Xenopus blastulae acquire Spemann’s organizer activity when incubated with activin, inducing secondary axes including head, trunk, and tail after transplantation into host embryos (Cooke et al., 1987; Ruiz i Altaba and Melton, 1989). Furthermore, microinjection of activin mRNA into cleaving Xenopus embryos can also induce secondary axes (Thomsen et al., 1990). Thus, in molecular terms the organizer is currently thought to be induced as a result of the release of a dorsal growth factor by the Nieuwkoop center blastomeres. This growth factor would then act upon the overlying cells of the marginal zone, inducing them to become organizer tissue.
At the late blastula stage, the organizer is located in a patch of cells encompassing 60° of arc of the dorsal marginal zone, which gives rise to head and notochordal (anterodorsal) mesoderm and determines the site where gastrulation starts. Since this tissue displays great powers of regulation after experimental manipulation, it has been termed the “organizer field” (Spemann, 1938) or “primary organization field” (Cooke, 1972). Examples of these regulatory properties include the following. First, a dorsal lip can be divided into several fragments, each of which can induce a secondary axis. Second, surgical removal of half an organizer results in entirely normal embryos (Stewart and Gerhart, 1990), while removal of more than this amount results in tadpoles with progressively more severe anterior deficiencies. Third, if an organizer is transplanted closer than 60° of arc to the site of the host’s dorsal lip, a secondary axis is not induced and a normal embryo ensues (Cooke, 1972). Fourth, if uncommitted embryonic cells are transplanted into the dorsal lip region, they will become incorporated into the organizer, adopting a notochordal cell fate (Spemann, 1938). Because it is the site where the body axis is initially formed, and because it has such interesting properties, the organizer field has fascinated embryologists for almost 70 years. To understand what makes this region unique, however, it is first necessary to isolate the molecules that control its biological behavior.
The starting point for the present investigation was provided by a previous study in which a cDNA library from manually dissected Xenopus dorsal lips was screened with an oligonucleotide probe specific for homeobox genes (Blumberg et al., 1991). We chose to start by studying homeobox genes, because they have been shown to mediate axis formation in Drosophila and vertebrates (reviewed by Gehring, 1987; De Robertis et al., 1990; Kessel and Gruss, 1990; Melton, 1991). This experiment resulted in the isolation of four types of homeobox-containing cDNAs, all of which were found to be concentrated in the dorsal lip by Northern blot analysis of dissected embryos. One cDNA, Xlab, was a labial homolog, and two others, Xcad1 and Xcad2, were caudal homologs. The fourth type of cDNA was the most interesting because it is expressed earlier than the others and especially because it encodes a protein that binds DNA in vitro with a specificity similar to that of the homeodomain of the Drosophila protein bicoid (Blumberg et al., 1991). The gene bicoid occupies a high place in the regulatory hierarchy leading to anteroposterior axis formation in fruit flies (Driever and Nüsslein-Volhard, 1988; Nüsslein-Volhard, 1991). This Xenopus gene was named goosecoid (gsc) to reflect its similarity, in parts of the homeodomain, to the Drosophila genes gooseberry and bicoid.
The present study is concerned with the function of goosecoid in Xenopus development. We find that the in situ distribution of goosecoid transcripts mimics the location of the organizer field in normal and experimentally manipulated embryos, that goosecoid expression is a primary response to activin induction, and that microinjection of its mRNA into the ventral blastomeres of the 4-cell embryo is sufficient to induce the formation of secondary body axes at high frequency. The results suggest that the goosecoid homeodomain protein plays a central role in executing Spemann’s organizer function.
Results
goosecoid Expression Demarcates the Xenopus Organizer
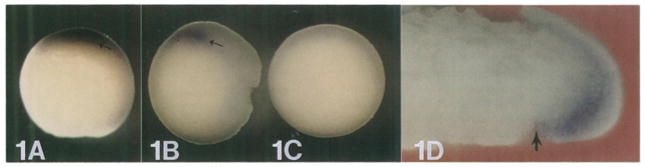
The localization of goosecoid transcripts in Xenopus embryos was analyzed by in situ hybridization (Hemmati-Brivanlou et al., 1990; Tautz and Pfeifle, 1989) with some modifications necessary for use in whole mounts of early embryos (see Experimental Procedures). At stage 10½, in which a visible dorsal lip has formed, goosecoid mRNA is found in a patch of cells of the marginal zone directly overlying the blastopore dorsal lip (Figures 1A and 1B). The specificity of the hybridization reaction was demonstrated by incubation with the sense strand of the same probe which, as expected, showed no hybridization (Figure 1C). The extent of the _goosecoid-_staining region varies somewhat from embryo to embryo, but the intense staining occupies about 60° of arc of the marginal zone (Figure 2A). The observed staining is roughly within the limits expected for the organizer region from embryological studies (Stewart and Gerhart, 1990).
Figure 1. Whole-Mount In Situ Hybridization of goosecoid Expression in Stage 10½ Gastrulae.
(A) antisense probe, vegetal view; (B) antisense probe, side view; (C) control sense probe, vegetal view; (D) sagittal section through the dorsal lip. Note that goosecoid hybridization is located in the deep layer of the upper lip of the blastopore. The fate of this region is to become head and notochord mesoderm. The arrow indicates the dorsal blastopore lip.
Figure 2. goosecoid Expression Follows the Expected Behavior of the Organizer Field in Experimentally Treated Embryos.
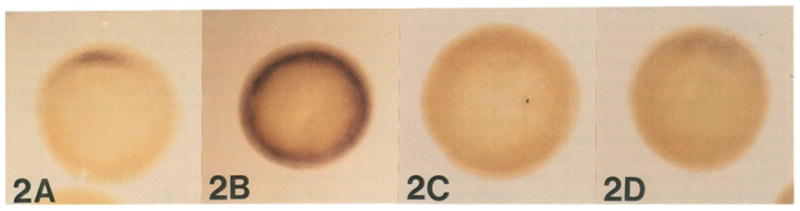
These embryos have been rendered transparent with Murray’s solution (see Experimental Procedures). (A) untreated stage 10½ gastrula; note that the goosecoid field encompasses about 60° of arc of the marginal zone. (B) LiCI-treated gastrula (0.12 M for 40 min at the 32-cell stage); goosecoid expression has become radially symmetric. (C) UV-treated gastrula (60 s, see Experimental Procedures); note that goosecoid expression is abolished. (D) RA-treated embryo (10−6 M, starting at the 2-cell stage); goosecoid expression is inhibited but still weakly detectable.
Figure 1D shows a section of a whole-mount stained gastrula (stage 10½). goosecoid mRNA is found in cells lying in the deep layer of the upper lip of the dorsal blastopore. The fate of these cells is to form mostly prechordal (head) mesoderm and notochord in later development (Keller, 1976; Slack, 1991). However, neither the “bottle cells” of Hamburger, located directly at the leading edge of the blastoporal invagination (arrow in Figure 1D), nor the superficial layer of cells located directly above the lip express goosecoid mRNA. The fate of bottle cells is to become pharyngeal endoderm, and the fate of superficial cells is to form the dorsal endoderm of the digestive tract (Keller, 1975). The localization observed is consistent with goosecoid being expressed in cells that give rise to head and notochordal mesoderm.
The in situ hybridizations indicate that the border of the patch of goosecoid expression in the marginal zone is not sharply defined and tends to taper off at the edges (e.g., Figures 1B and 2A), but it is difficult to conclude whether it is graded or not. The Drosophila homeodomain protein bicoid, which mediates the first step in anteroposterior axis formation, forms a gradient in the anterior region of the fly embryo (Driever and Nüsslein-Volhard, 1988). In addition, vertebrate homeobox proteins are known to form gradients of nuclear proteins in fields of differentiating cells such as limbs or feather buds (reviewed by De Robertis et al., 1991). Homeoprotein gradients are best visualized when the distribution of proteins is examined with specific antibodies rather than by in situ hybridization; it will therefore be of interest to examine the distribution of goosecoid protein, particularly during mesoderm invagination, once a specific antiserum becomes available.
A time course of goosecoid expression by in situ hybridization (not shown) reveals that while it is not detectable at mid-blastula, a localized patch of dorsal expression is visible at late blastula at least 1 hr before there are any external signs of gastrulation. Northern blots of staged embryos (data not shown) support this analysis, except that the expression is detected earlier (already by stage 8.5, which is negative by in situ hybridization). This suggests that goosecoid transcription starts with the initial wave of zygotic transcription at the mid-blastula transition (Newport and Kirschner, 1982). goosecoid mRNA accumulates much earlier than that of the other homeobox genes that are also expressed in the dorsal lip, Xlab, Xcad1, and Xcad2, whose transcripts are detectable by Northern analysis only after gastrulation is under way (Blumberg et al., 1991). goosecoid mRNA expression is transient and is no longer detectable by the time neurulation starts.
From these descriptive studies in the undisturbed Xenopus embryo, we conclude that the region of goosecoid expression correlates well with the location expected for the organizer field.
goosecoid Expression Correlates with the Amount of Organizer Tissue Present in the Embryo
To be considered a faithful marker for the organizer in Xenopus, goosecoid should fulfill at least two criteria. First, expression should be increased in embryos treated with the dorsalizing agent LiCI. It has been shown that when Xenopus embryos are immersed in a LiCI solution during cleavage, a great enhancement of dorsoanterior structures (particularly notochord) occurs. Transplantation studies have shown that the entire marginal zone behaves as Spemann’s organizer in embryos treated with LiCI (Kao and Elinson, 1988). The mechanism of action of LiCI is not well understood, but is thought to involve the phosphoinositol intracellular signaling pathway (Busa and Gimlich, 1989). Second, goosecoid expression should be decreased or abolished by irradiation of the vegetal pole of fertilized eggs with ultraviolet (UV) light. This treatment prevents cortical rotation and, in consequence, formation of the Nieuwkoop center in vegetal and dorsal blastomeres (Gerhart et al., 1989). Formation of the organizer is therefore not induced in UV-treated embryos. The molecular mechanism by which the egg cortical rotation leads to formation of the body axis is not known.
Treatment of Xenopus embryos at the 32-cell stage with LiCI does indeed produce expression of goosecoid mRNA in the entire marginal zone (Figure 2B; compare with control embryo in Figure 2A). In some embryos, expression at the early gastrula stage was not completely radial, but in all cases examined it covered more than 180° of arc. The dorsoanterior index (DAI, Kao and Elinson, 1988), a measure of the effectiveness of the treatment, varied between 7 and 10 for our LiCI-treated embryos. In the DAI scale a normal tadpole is given a value of 5, a completely dorsalized one has a value of 10, and a totally ventralized one a value of 0 (Kao and Elinson, 1987). The increase in goosecoid mRNA expression induced by LiCI was confirmed by Northern blot analysis.
UV treatment before first cleavage inhibited goosecoid expression to levels undetectable by the whole-mount in situ procedure in the 15 embryos that were examined (Figure 2C). Northern blot analysis confirmed that goosecoid mRNA was greatly decreased. It should be pointed out that under our particular conditions UV treatment resulted in a majority of intermediate phenotypes. The head was affected or absent in all embryos, but substantial numbers (83%, n = 106) still developed tail structures containing somites and other axial structures (DAI ranging from 4 to 2). This suggests that tail development, but not head development, may be able to proceed in the absence–or in the presence of very small amounts–of goosecoid mRNA.
We conclude from these studies in experimentally perturbed embryos that goosecoid expression correlates well with the known changes in organizer activity induced by dorsalizing (LiCI) or ventralizing (UV) treatments.
Retinoic Acid Inhibits goosecoid Expression
Retinoic acid (RA) has potent teratogenic effects in Xenopus embryos (Durston et al., 1989; Sive et al., 1990), resulting in the truncation of head structures. This effect is mediated at least in part by the mesoderm (Ruiz i Altaba and Jessel, 1991; Cho et al., 1991; Sive and Cheng, 1991). Because goosecoid expression correlates with the formation of dorsoanterior structures, we decided to test the effect of RA on its expression.
When embryos were treated continuously with RA from the 2-cell stage until the onset of gastrulation, severe anterior deficiencies occurred (average DAI of 1.8, n = 83). Whole-mount in situ analysis at the early gastrula stage showed that in about half of these embryos goosecoid expression was substantially decreased but not entirely abolished (Figure 2D; compare with control in 2A), while in the other half goosecoid mRNA was undetectable (not shown). Northern blot analysis confirmed that goosecoid mRNA is decreased, but not abolished, by RA treatment (not shown).
We conclude that exposure of Xenopus embryos to RA, which produces truncations of head structu res, inhibits the expression of goosecoid. This inhibition contrasts with the stimulatory effect of RA on the expression of many other homeobox genes (Simeone et al., 1990; Cho and De Robertis, 1990; Sive and Cheng, 1991).
goosecoid Is a Primary Response Gene Induced by Activin
To examine what signals are responsible for the activation of goosecoid expression, we carried out induction experiments in animal cap fragments isolated at the mid-blastula stage. Animal cap cells cultured in saline solution do not express goosecoid (Figure 3, “untreated” lane). When XTC-MIF was added (a dorsoanterior inducer whose active agent is activin; Green and Smith, 1990), goosecoid mRNA was induced after 2 hr and decreased thereafter (Figure 3). In contrast, the ventroposterior inducer bFGF was unable to induce goosecoid mRNA accumulation (Figure 3). RA inhibited, but did not block completely, the accumulation of goosecoid mRNA induced by XTC-MIF (Figure 3). The same results were obtained when pure recombinant activin A was used instead of XTC-MIF-conditioned medium (data not shown).
Figure 3. goosecoid Expression in Animal Cap Fragments Treated with Peptide Growth Factors and RA.
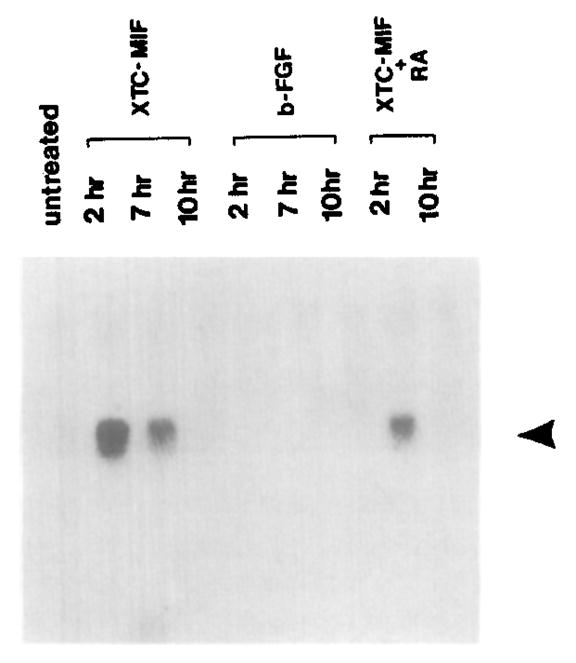
Note that goosecoid mRNA (arrowhead) is induced transiently by XTC-MIF (activin), that it is not induced at all by bFGF, and that XTC-MIF induction is inhibited by RA (compare lanes incubated with growth factor for 2 hr). XTC-MIF was used at 1:3 dilution. FGF was used at 200 ng/ml and RA at 10−6 M. Total RNA extracted from 20 animal caps was loaded in each lane and processed as described (Blumberg et al., 1991).
The induction of goosecoid mRNA by XTC-MIF (or activin) is very rapid. As shown in Figure 4A, transcript accumulation can be detected 30 min after addition of the growth factor. This suggested that goosecoid could be a primary response gene in the induction processes triggered by activin-like growth factors.
Figure 4. Time Course and Protein Synthesis Independence of the Induction of goosecoid mRNA by XTC-MIF in Animal Caps.
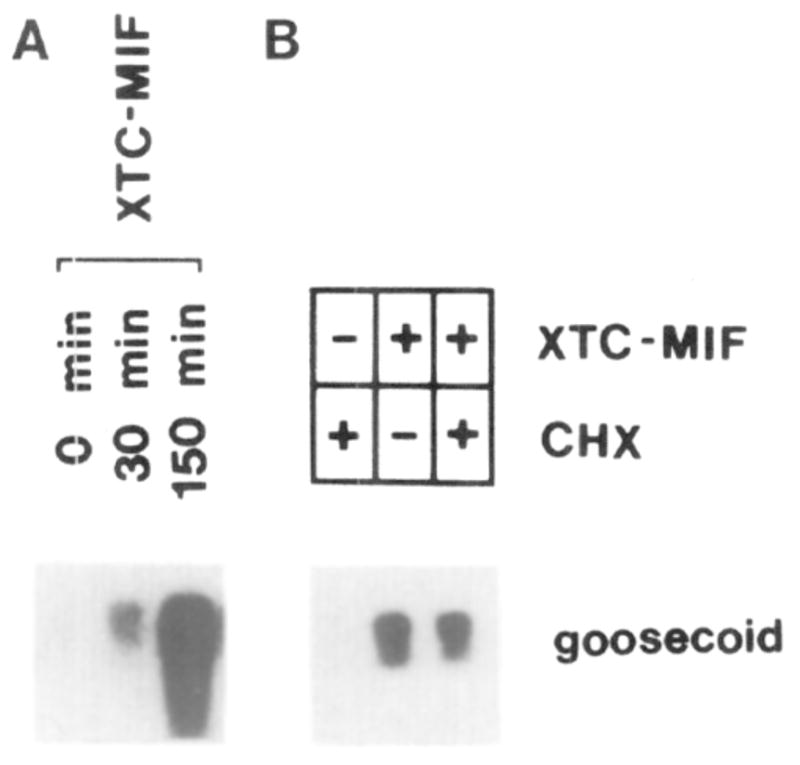
(A) Groups of 20 animal caps isolated from stage 8 blastulae were incubated with XTC-MIF for the indicated times and analyzed by Northern blot. Note that goosecoid induction is detectable after 30 min. (B) Inhibition of protein synthesis by cycloheximide (CHX) does not prevent goosecoid expression. Animal caps were preincubated for 30 min with or without cycloheximide and then induced for 90 min with XTC-MIF, following the protocol of Rosa (1989).
We next tested whether goosecoid induction could take place in the absence of protein synthesis. Animal caps were preincubated in 5 μg/ml cycloheximide for 30 min before adding the growth factor and incubating at 20°C for an additional 90 min (Rosa, 1989). In our hands these conditions prevented the incorporation of [35S]methionine into proteins by over 95%, measured both by scintillation counting and by polyacrylamide gel electrophoresis (not shown). Figure 4B shows that cycloheximide was not able to block the induction of goosecoid transcripts by XTC-MIF.
We conclude that the expression of goosecoid mRNA can be induced by dorsoanterior mesoderm–inducing factors of the activin type, but not by bFGF, an inducer of ventroposterior mesoderm. goosecoid induction is a primary response to activin, not requiring ongoing protein synthesis.
goosecoid mRNA Induces Secondary Axes
The studies on normal localization and experimental manipulation by LiCI, UV, RA, and activin described thus far suggest that goosecoid expression closely follows the properties of the organizer. This raises the question of whether the goosecoid homeodomain protein itself might be able to execute organizer function rather than being a mere marker of position. The most direct approach to this problem in Xenopus embryos is by the microinjection of synthetic mRNA (Krieg and Melton, 1984).
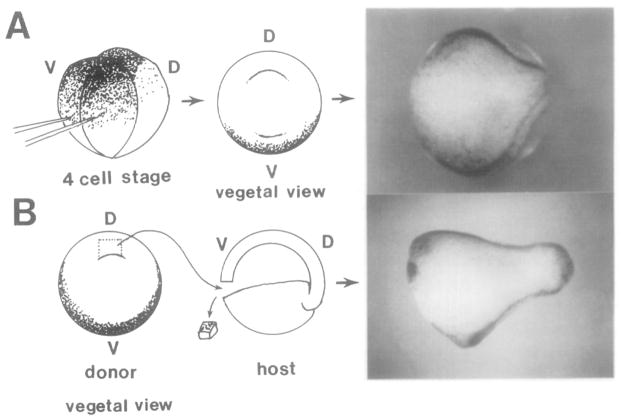
Initial exploratory microinjection experiments using goosecoid full-length mRNA failed to produce conclusive evidence of induction of head and notochordal structures: injection into the 1-cell embryo resulted in very abnormal gastrulation, and “einsteck” transplantations of microinjected animal caps into the blastocoel of host embryos (Cho et al., 1991) induced secondary axes with some anteriorly located structures but lacking clear head markers such as eyes or auditory vesicles. We eventually found a functional assay, however, in which goosecoid had a potent effect, inducing an entire body axis, including massive notochords. To achieve this it was necessary to microinject goosecoid mRNA into the region where it is not normally expressed, i.e., the ventral half of the embryo, as shown in Figure 5A.
Figure 5. Comparable Results Are Obtained by goosecoid mRNA Microinjection and by Dorsal Lip Transplantation.
Experimental diagram and embryos resulting from (A) microinjection of goosecoid mRNA into the two ventral blastomeres (as close as possible to the first cleavage plane) and (B) a traditional Spemann organizer transplantation experiment. Note that the resulting embryos resemble each other and have extensive secondary neural tubes (dark lines) at the late neurula stage. In both embryos the two axes originate independently from each other in the posterior region, i.e., two sites of dorsal invagination were present during gastrulation.
Table 1 shows the results of microinjecting goosecoid mRNA or a control construct lacking the homeobox (called Δ_gsc_; see Experimental Procedures) into the two dorsal or the two ventral blastomeres of 4-cell embryos. Regularly cleaving embryos in which the less pigmented dorsal and the darker ventral side were particularly distinct from each other were selected as described (Klein, 1987; Yuge et al., 1990). While this method of assigning the dorsal and ventral sides is not 100% accurate, it is effective in about 85% of the cases (Niehrs and De Robertis, 1991). Extensive secondary axes were induced by goosecoid mRNA in 75% of the embryos injected into the ventral blastomeres, while microinjection into the dorsal side resulted in a majority of normal tadpoles (Table 1). The secondary axes present in 12% of the embryos resulting from the dorsal injections in Table 1 can be explained by inaccurate assignment of the dorsal side on the basis of embryo pigmentation. Two control mRNAs, Δ_gsc_ and a complete mRNA encoding the unrelated homeodomain protein XIHbox 6 (Cho et al., 1991), did not induce secondary axes. The induction of secondary axes by microinjection of goosecoid mRNA into ventral blastomeres was dose dependent (see Table 1 legend).
Table 1.
Microinjection of goosecoid mRNA into Both Blastomeres of the Ventral Side of the 4-Cell Xenopus Embryos Induces Secondary Axes
| mRNA | Blastomeres | Number of Embryos with Single Axis | Number of Embryos with Two Axes | Percent Secondary Axes |
|---|---|---|---|---|
| Full-Length goosecoid | Ventral | 9 | 27 | 75% |
| Dorsal | 29 | 4 | 12% | |
| Δ_gsc_ | Ventral | 18 | 1? (Weak axis) | 0% or 5%? |
| Dorsal | 20 | 0 | 0 | |
| Complete XIHbox 6 | Ventral | 26 | 0 | 0 |
| Dorsal | 8 | 0 | 0 |
Figure 5A shows that the secondary axes induced on the ventral side by goosecoid mRNA are rather extensive. The two neural tubes can be seen to originate independently from the posterior region and to extend anteriorly as mirror images of each other. Their appearance is very similar to that of embryos obtained by transplanting Spemann’s organizer to the ventral side of a gastrula, as shown in Figure 5B.
The first change seen after goosecoid mRNA microinjection is the formation of an additional dorsal lip–like structure in the gastrula (compare Figures 6A and 6B in the color plate). At the late neurula stage two axes can be discerned (Figure 6C, top two embryos). Note that embryos injected with Δ_gsc_ mRNA (Figure 6C, bottom two embryos) do not form secondary axes. Because we lack a goosecoid antibody, we have been unable to show whether Δ_gsc_ mRNA is translated into a stable protein in vivo, but a similar truncation of the XIHbox 6 protein is known to produce stable products in Xenopus embryos (Cho et al., 1991). Furthermore, in vitro translation of goosecoid and Δ_gsc_ mRNAs produces proteins of the expected size, which are equally stable in the reticulocyte lysate system (not shown).
Figure 6. Phenotypic Effect of Microinjecting goosecoid or Δ_gsc_ mRNA into the Two Ventral Blastomeres at the 4-Cell Stage.

(A) Δ_gsc_ control, only one dorsal lip is present at early gastrula. (B) goosecoid mRNA injection, two dorsal lip–like structures are present (arrows). (C) Top, two embryos that received goosecoid mRNA; secondary neural axes are visible. The two bottom embryos were injected with Δ_gsc_ mRNA, and no secondary axis is present. (D) Twinned embryo produced by goosecoid mRNA injection; note that a complete head structure containing eyes, hatching gland, and cement gland has been induced.
By the third day of development, it can be seen that some of the secondary axes resulting from overexpression of goosecoid mRNA form complete head structures including auditory vesicles, forebrain, eyes, hatching gland, and cement gland (Figure 6D; and histological data not shown). Although only 10% of the secondary axes had all the aforementioned head markers, this observation is important because it shows that ectopic expression of goosecoid is sufficient to trigger formation of even the most anterior elements of the body axis.
goosecoid overexpression seems to compete with the proper formation of tail structures. At the swimming tadpole stage, the body length of embryos with large _goosecoid-_induced axes is shortened considerably, with the additional anterior (head and trunk) structures being formed at the expense of the tail (Figure 6D). This could conceivably be considered a transformation of the homeotic type, although the observations can be equally well described as a competition between the head and tail fields.
When _goosecoid_-injected embryos are examined histologically, the salient feature is the massive notochord usually present in the secondary axis (Figure 7A), which is much larger than that of the primary axis. (The primary axis can be readily identified in serial histological sections because it has more complete eye and forebrain structures.) The secondary axis sometimes has two notochords (Figure 7B), perhaps reflecting the double injection into the ventral side. In one case we found additional neural tubes forming in close proximity to the enlarged notochord. This embryo contained a total of three notochords and four neural tubes (Figure 7B). Histological analysis therefore shows that cells in the ventral side of the injected embryo tend to adopt a notochordal (dorsal) fate. Interestingly, goosecoid mRNA is normally expressed in the Xenopus gastrula in cells destined to become notochord (Figure 1D).
Figure 7. The Additional Axis Induced by goosecoid mRNA Contains Massive Notochord Structures.
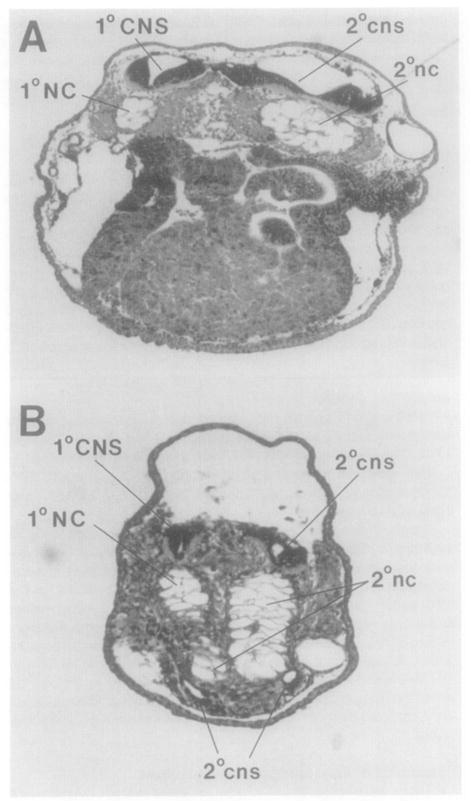
Two transverse sections from the same animal are shown. Note that the notochord is much larger in the secondary axis (2°nc) than in the primary axis (1 °NC). Note that in more posterior regions (B), two small additional neural tubes (cns) have formed in close proximity to the ectopic notochordal tissue in the ventral side of the embryo.
We conclude from these studies that expression of the goosecoid homeodomain protein in the ventral side of the embryo is sufficient to start formation of a new body axis at high frequency. The same protein introduced into the dorsal side, where goosecoid is normally expressed, results in normal embryos. The secondary axes resulting from goosecoid mRNA injection can form complete head structures, compete with tail development, and contain unusually large amounts of notochordal tissue.
Discussion
The Organizer Field
goosecoid expression closely follows the expected distribution of organizer tissue in normal and experimentally treated embryos. At the start of gastrulation, goosecoid transcripts are found in a patch of cells encompassing 60° of arc on the dorsal marginal zone. Treatment with UV light or RA inhibits, while treatment with LiCI greatly enhances, goosecoid mRNA expression. goosecoid mRNA accumulation is induced by activin but not by bFGF. Furthermore, this induction can take place in the absence of protein synthesis, i.e., goosecoid expression is a primary response to activin. The area of goosecoid expression is thus a marker for the region of the embryo where the putative dorsal inductor is most active.
The Head Organizer
Our working hypothesis is that goosecoid is responsible for the development of the head region, which is deleted in RA-treated embryos, while the genes of the Antennapedia-type Hox complexes are involved in the development of trunk and tail regions, which are resistant to RA treatment. An observation that supports this notion is that overexpression of an Antennapedia-type homeodomain protein, XIHbox 6, in uncommitted embryonic cells can cause the induction of tail-like structures in transplantation experiments (Cho et al., 1991). Consistent with this hypothesis, goosecoid expression is absent in embryos resulting from partial UV treatment, which lack heads but have well-formed tails. Spemann distinguished between a “head” organizer, present in the early dorsal lip, and a “tail” organizer, present in the dorsal lip of later gastrulation stages (Spemann, 1931). It might be useful in the future to reexamine this issue in detail, now that molecular markers are available.
Microinjection of goosecoid mRNA Can Induce a Complete Body Axis
Translation of goosecoid mRNA in the ventral half of the embryo, where it is normally not expressed, is sufficient to cause the formation of a new dorsal lip, which in turn generates a secondary axis including complete head structures and massive notochords. The specificity of axis induction by goosecoid is underscored by the fact that when microinjection is performed into the dorsal side, the majority of the embryos are normal. One might find it surprising that these tadpoles do not display, for example, large heads. It should be kept in mind, however, that the organizer field has regulative powers. For instance, when a second dorsal lip is transplanted close to the resident blastopore lip, entirely normal embryos will result, despite having two organizers instead of one (Cooke, 1972).
The dorsal lip is thought to be the source of additional signals that further regionalize the mesoderm (Smith et al., 1985; Cooke, 1989; Slack, 1991) and that spread through the plane of the ectoderm to facilitate the induction of neural tissue (Spemann, 1938; Savage and Phillips, 1989; Dixon and Kintner, 1989). Thus, it can be expected that the organizer itself will be the source of additional growth factors (or other morphogens). Perhaps some of these might be activated, directly or indirectly, by the goosecoid homeodomain protein. goosecoid mRNA is clearly sufficient to trigger dorsal development when injected into the ventral half of a 4-cell embryo. We have not yet shown, however, whether _goosecoid-_expressing cells are able to recruit noninjected neighboring cells into the secondary axis. Grafting experiments using lineage-traced cells should answer this question.
Positional Specification in the Xenopus Gastrula
A biochemical pathway for the formation of the Xenopus anteroposterior axis seems to be emerging. Activin, which acts through a receptor with serine kinase activity (Mathews and Vale, 1991), activates goosecoid, a homeobox gene possibly involved in head development. FGF, which acts through a tyrosine kinase receptor (Amaya et al., 1991), is unable to activate goosecoid, but preferentially activates homeobox genes that are active in the posterior of the embryo such as Xhox 3 (Ruiz i Altaba and Melton, 1989) and XIHbox 6 (Cho and De Robertis, 1990). Interestingly, a dominant-negative mutation of the FGF receptor, which blocks action of this growth factor in vivo, interferes more with tail formation than with that of the head region (Amaya et al., 1991). In addition, RA cooperates with growth factor action, for example, by potentiating the induction of XIHbox 6 by bFGF (Cho and De Robertis, 1990) and by inhibiting goosecoid induction by activin.
An embryological model explaining how the organizer phenomenon is generated in Xenopus is also emerging. As a result of egg cortical rotation (Gerhart et al., 1989), the Nieuwkoop center (vegetal and dorsal) blastomeres would acquire the ability to release a dorsal growth factor (Smith et al., 1989; Thomsen et al., 1990; Slack, 1991). Localized expression or release of growth factors in vegetal and dorsal cells has not yet been demonstrated, but this is an active area of research at the moment. Release of this dorsal growth factor would result in the induction of organizer tissue in the overlying marginal zone cells. Within the organizer region proper, a key role seems to be played by goosecoid, a homeodomain protein. Microinjection experiments suggest that the goosecoid protein is an integral component of the biochemical machinery that executes Spemann’s organizer function. Using a different terminology (Wolpert, 1989), the growth factor released by the Nieuwkoop center would provide the positional information, while the goosecoid homeodomain protein would provide the positional specification that determines the fate of dorsoanterior mesoderm.
Experimental Procedures
Isolation and Characterization of cDNA and Genomic Clones
Because most of the 23 goosecoid cDNA clones isolated previously (Blumberg et al., 1991) were quite short, an unamplified gastrula cDNA library was constructed from poly(A)+ RNA isolated from stage 10½ and 11½ gastrula embryos. Approximately 106 plaques were screened with a random-primed 1.1 kb goosecoid cDNA probe. Hybridization was carried out as described (Cho et al., 1988). Washing was carried out in 0.5 × SSC at 65°C. Fifteen additional positive plaques were repurified and plasmids excised as described (Blumberg et al., 1991). As is the case for many Xenopus genes (e.g., Fritz et al., 1989), two major types of cDNAs were found; these were designated A (18 clones) and B (20 clones). The sequence of a type A clone was published previously (Blumberg et al., 1991; GenBank/EMBL accession number M63872). In all experiments reported here, a full-length goosecoid type B clone (designated p_gsc_) was utilized (GenBank/EMBL number M81481). To clone the genomic counterparts of the goosecoid cDNAs, an unamplified Xenopus genomic library was screened (Cho et al., 1988). A genomic clone corresponding to type A was mapped and subcloned. All intron and exon boundaries were confirmed by sequencing the genomic counterpart. The coding region of the goosecoid gene is a relatively compact 2.5 kb and contains three exons. Unlike most other homeobox genes, in goosecoid the homeobox is interrupted by an intron in the middle of the highly conserved helix 3; this may explain why previous screens of genomic libraries for vertebrate _bicoid_-related genes were unsuccessful.
Preparation of Synthetic mRNAs for Microinjection
The goosecoid cDNA lacking the homeobox region (pΔ_gsc_) was constructed by subcloning a 525 bp Pstl–Pstl fragment of p_gsc_ into the Pstl site of the pBluescript II KS vector (Stratagene, Inc.). Full-length goosecoid sense mRNA was synthesized by linearizing p_gsc_ with Xhol and transcribing with T3 RNA polymerase. The control goosecoid mRNA lacking the homeobox was synthesized by transcribing Xhol linearized pΔ_gsc_ plasmid with T7 RNA polymerase. XIHbox 6 sense mRNA was made by transcribing BamHI linearized pSP64-XIHbox 6 cDNA with SP6 RNA polymerase (Cho et al., 1991). All mRNAs used for microinjection were capped. After two ethanol precipitations and washing in 70% ethanol, they were resuspended in injection buffer (88 mM NaCI, 1 mM KCI, 15 mM Tris–HCI [pH 7.5]). Secondary axes were produced by goosecoid mRNA in five independent experiments, using several independent preparations of synthetic goosecoid mRNA.
Microinjection and Histological Analysis of Embryos
Embryos were fertilized in vitro, and 4-cell stage embryos showing the first cleavage plane bisecting the less pigmented dorsal area and then cleaving perpendicularly to this plane were selected for microinjection (Klein, 1987) and transferred into 1 × modified Barth saline (MBS). RNA was injected into the equatorial region of the two adjacent dorsal (lightly pigmented) or ventral (darkly pigmented) blastomeres, as close as possible to the plane of first cleavage. The concentration of RNA was 40 ng/μl and the injection volume was 4 nl into each blastomere unless otherwise indicated. Thirty minutes after microinjection, embryos were transferred back into 0.1 × MBS and allowed to develop.
Embryos were fixed at the indicated stages in Bouin’s fixative (75 parts saturated picric acid, 25 parts formalin, 5 parts glacial acetic acid) for 2 hr, washed with 70% ethanol, embedded in wax, sectioned, and stained as described previously (Cho et al., 1991).
LICI, UV, and RA Treatments of Embryos
Embryos were treated continuously with 10−6 M RA (All trans RA, Sigma) from the 4-cell stage to the gastrula stage. LiCI (0.12 M) was applied to the 32-cell stage embryos for 40 min, washed in 0.1 × MBS, and allowed to develop to the gastrula stage. For UV treatment, embryos were carefully placed in a narrow plastic box filled with 0.1 × MBS, covered with Saranwrap, and sealed with a rubber band. Embryos were UV irradiated through the Saranwrap for 60 s using a UV GL25 lamp 30 min after fertilization (Sive et al., 1990) and transferred into fresh 0.1 × MBS. Care was taken to minimize rotation of UV-irradiated embryos. RNAs were isolated from these embryos at stage 10½ gastrula.
Animal Cap Assays
Animal caps were isolated from stage 8 blastula embryos and treated with growth factors as described previously (Cho and De Robertis, 1990). The concentrations of bFGF and recombinant purified activin A used were 200 ng/ml and 50 ng/ml, respectively. The XTC-MIF-conditioned medium was heat activated, diluted 1:3 in 1 × MBS, and applied to animal caps.
Cycloheximide block experiments were carried out essentially as described by Rosa (1989). To measure the effectiveness of cycloheximide to block protein synthesis, animal caps that had been preincubated with cycloheximide for 30 min were incubated for 90 min in 1 × MBS buffer containing [35S]methionine (400 μCi/ml), cycloheximide, and growth factors. Animal caps were homogenized in a buffer containing 50 mM Tris (pH 7.6) and 150 mM NaCI and cleared by centrifugation. A 2 μl aliquot of the supernatant was taken to 0.25 ml with 1 N NaOH and aminoacyl-tRNAs hydrolyzed at 37°C for 10 min followed by TCA precipitation and scintillation counting. Other aliquots were analyzed by protein gel electrophoresis followed by autoradiography.
Preparation of Xenopus Egg Blocking Extract
Laid eggs were collected in 1 × MBS, dejellied in 0.2% cysteine HCI (pH 7.8), washed five times in 0.1 × MBS, and washed twice in extract buffer (100 mM Tris–HCI, 150 mM NaCI). Eggs were homogenized in a loose-fitted glass homogenizer by ten strokes together with the equal volume of extract buffer. The homogenate was cleared three times by 25 min centrifugation at 30,000 × g and stored in 1 ml aliquots at −20°C. It was used for blocking nonspecific antibody staining in whole mounts.
Whole-Mount In Situ Hybridization
The localization of goosecoid transcripts in Xenopus embryos was analyzed using in situ hybridization in whole mounts (Hemmati-Brivanlou et al., 1990; Tautz and Pfeifle, 1989). To work with early embryonic stages, some technical modifications had to be introduced: puncturing of the animal cap to prevent accumulation of protein precipitates in the blastocoel; heating of the embryos at 65°C for 1 hr to decrease endogenous background; inclusion of 1 mM levamisol (an inhibitor of endogenous alkaline phosphatase) in all antibody washing solutions; and the addition of soluble Xenopus egg extract into the blocking and antibody-binding solutions.
The pΔ_gsc_ subclone (which contains the amino terminus of pgsc but excludes the homeobox) was linearized with Xhol and Smal, transcribed by T7 and T3 RNA polymerases to synthesize sense and anti-sense RNAs, respectively. Digoxigenin-labeled RNA was synthesized using the Genius kit (Boehringer Mannheim) according to manufacturer’s instructions. The RNA probes were stored as ethanol precipitates at −20°. Albino embryos of X. laevis were obtained by in vitro fertilization and developmental stages determined according to Nieuwkoop and Faber (1967). Embryos were manually dechorionated in 1 × MBS at the indicated stages, fixed in freshly prepared MEMFA (0.1 M MOPS [pH 7.4], 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde; Hemmati-Brivanlou et al., 1990) at room temperature for 90 min on an end-over-end rotator, and stored at −20° in methanol.
Whole-mount in situ hybridization was carried out at room temperature, unless otherwise indicated. The ectodermal roof of the embryos was punctured with a needle in a solution containing 90% methanol and 10% 0.5 M EGTA. The disrupted embryos were rehydrated stepwise, washed three times (10 min each) in PTw (1 × PBS plus 0.1% Tween 20; Hemmati-Brivanlou et al., 1990), treated with proteinase K (10 μg/ml in PTw) for 20 min, and washed twice with rotation, followed by additional washing without agitation (10 min each) in PTw. Embryos were refixed in a phosphate-buffered saline solution containing 4% paraformaldehyde for 20 min, washed four times in PTw, and brought stepwise to 500 μl prehybridization buffer (50% formamide, 5 × SSC, 2% blocking reagent (Boehringer Mannheim), 50 μg/ml heparin, 0.1 % Tween 20, 1 mg/ml Torula tRNA). Embryos were incubated at 65°C for 1 hr to reduce the activity of endogenous alkaline phosphatase and prehybridized for an additional 2 hr at 55°C. The prehybridization solution was replaced with fresh hybridization solution containing the probe (50–100 μg/ml digoxigenin-labeled sense or antisense RNA). Embryos were incubated overnight at 55°C. Washing was done at 37°C once with prehybridization buffer, followed by a series of stepwise washes to 2 × SSC. The nonhybridized excess RNA was removed by treatment with 20 μg/ml RNAase A in 2 × SSC for 15 min at 37°C followed by two washes at 60°C in 0.2 × SSC. The samples were brought stepwise to TNT (100 mM Tris–HCI [pH 7.5], 150 mM NaCI, 0.1% Tween 20), transferred to 0.65 ml microfuge tubes, and incubated for 2 hr at 4°C in 0.5 ml of blocking solution (150 mM NaCI, 100 mM Tris–HCI [pH 7.5], 0.1% Tween, 2 mg/ml blocking reagent (Boehringer Mannheim), 15% heat-inactivated goat serum; 5% Xenopus egg extract prepared as described above. To prevent nonspecific antibody binding, the anti-digoxigenin alkaline phosphatase conjugate was diluted 1:600 in this blocking solution and preincubated for 2 hr at 4°C before use. The embryos were gently rocked overnight at 4°C in 500 μl of the antibody solution, washed three times for 90 min in TNT containing 1 mM levamisol, followed by a 30 min wash in a solution containing 100 mM Tris–HCI (pH 9.5), 100 mM NaCI, 50 mM MgCl2, and 1 mM levamisol. The color reaction for alkaline phosphatase (Hemmati-Brivanlou et al., 1990) was carried out in 24-well tissue culture plates (coated in 1% agar) for 1–2.5 hr at 20°C in the dark. The reaction was stopped by transferring the embryos into 10 mM Tris–HCI (pH 8.0), 1 mM EDTA, followed by dehydration in methanol. Embryos can be cleared to detect staining in the deep layers (Dent et al., 1989), but this treatment has a tendency to dissolve less intense staining. If cleared, embryos are always returned to methanol for storage.
Acknowledgments
We thank F. Rosa and I. Dawid for a gift of XTC-MIF and Genentech for pure recombinant activin A. The manuscript was greatly improved by the comments of Dennis Bittner, Michael Carey, Beatrice Jegalian, Judith Lengyel, Christof Niehrs, and Larry Zipursky. B. B. was supported by postdoctoral fellowships of the NIH (HD 07273) and the Lucille Markey Trust. H. S. was supported by an Alexander von Humboldt fellowship. This work was funded by grant HD 21502-06 of the NIH.
References
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Wright CVE, De Robertis EM, Cho KWY. Organizer-specific homeobox genes in Xenopus laevis embryos. Science. 1991;253:194–196. doi: 10.1126/science.1677215. [DOI] [PubMed] [Google Scholar]
- Busa WB, Gimlich RL. Lithium-induced teratogenesis in frog embryos prevented by a polyphosphoinositide cycle intermediate or a diacylglycerol analog. Dev Biol. 1989;132:315–324. doi: 10.1016/0012-1606(89)90228-5. [DOI] [PubMed] [Google Scholar]
- Cho KWY, De Robertis EM. Differential activation of Xenopus homeobox genes by mesoderm inducing growth factors and retinoic acid. Genes Dev. 1990;4:1910–1916. doi: 10.1101/gad.4.11.1910. [DOI] [PubMed] [Google Scholar]
- Cho KWY, Goetz J, Wright CVE, Fritz A, Hardwicke J, De Robertis EM. Differential utilization of the same reading frame in a Xenopus homeobox gene encodes two proteins sharing the same DNA-binding specificity. EMBO J. 1988;7:2139–2149. doi: 10.1002/j.1460-2075.1988.tb03053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KWY, Morita EA, Wright CVE, De Robertis EM. Overexpression of a homeodomain protein confers axis-forming activity to uncommitted Xenopus embryonic cells. Cell. 1991;65:55–64. doi: 10.1016/0092-8674(91)90407-p. [DOI] [PubMed] [Google Scholar]
- Cooke J. Properties of the primary organization field in the embryo of Xenopus laevis. II. Positional information for axial organization in embryos with two head organizers. J Embryol Exp Morph. 1972;28:27–46. [PubMed] [Google Scholar]
- Cooke J. Mesoderm-inducing factors and Spemann’s organizer phenomenon in amphibian development. Development. 1989;107:229–241. doi: 10.1242/dev.107.2.229. [DOI] [PubMed] [Google Scholar]
- Cooke J, Smith JC, Smith EJ, Yaqoob M. The organization of mesodermal pattern in Xenopus laevis: experiments using a Xenopus mesoderm-inducing factor. Development. 1987;101:893–908. doi: 10.1242/dev.101.4.893. [DOI] [PubMed] [Google Scholar]
- Dent JA, Polson AG, Klimkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Oliver G, Wright CVE. Homeobox genes and the vertebrate body plan. Scientific American. 1990;263:46–52. doi: 10.1038/scientificamerican0790-46. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Morita EA, Cho KWY. Gradient fields and homeobox genes. Development. 1991;112:669–678. doi: 10.1242/dev.112.3.669. [DOI] [PubMed] [Google Scholar]
- Dixon JE, Kintner CR. Cellular contacts required for neural induction in Xenopus embryos: evidence for two signals. Development. 1989;106:749–758. doi: 10.1242/dev.106.4.749. [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Durston AJ, Timmermans JP, Hage WJ, Hendriks HF, de Vries NJ, Heideveld M, Nieuwkoop PD. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–144. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- Elinson RP, Kao KR. The location of dorsal information in frog early development. Dev Growth Diff. 1989;31:423–430. doi: 10.1111/j.1440-169X.1989.00423.x. [DOI] [PubMed] [Google Scholar]
- Fritz AF, Cho KWY, Wright CVE, Jegalian BG, De Robertis EM. Duplicated homeobox genes in Xenopus. Dev Biol. 1989;131:584–588. doi: 10.1016/s0012-1606(89)80029-6. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. Homeo boxes in the study of development. Science. 1987;236:1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Danilchik M, Doniach T, Roberts S, Rowning B, Stewart R. Cortical rotation of the Xenopus egg: consequences for the anteroposterior pattern of embryonic dorsal development. Development (Suppl) 1989;107:37–51. doi: 10.1242/dev.107.Supplement.37. [DOI] [PubMed] [Google Scholar]
- Gimlich RL. Acquisition of developmental autonomy in the equatorial region of the Xenopus embryo. Dev Biol. 1986;115:340–352. doi: 10.1016/0012-1606(86)90254-x. [DOI] [PubMed] [Google Scholar]
- Gimlich RL, Gerhart JC. Early cellular interactions promote embryonic axis formation in Xenopus laevis. Dev Biol. 1984;104:117–130. doi: 10.1016/0012-1606(84)90042-3. [DOI] [PubMed] [Google Scholar]
- Green JBA, Smith JC. Graded changes in dose of a Xenopus activin A homologue elicit stepwise transitions in embryonic cell fate. Nature. 1990;347:391–394. doi: 10.1038/347391a0. [DOI] [PubMed] [Google Scholar]
- Hamburger V. The Heritage of Experimental Embryology. Oxford, England: Oxford University Press; 1988. [Google Scholar]
- Hemmati-Brivanlou A, Frank D, Bolce ME, Brown BD, Sive HL, Harland RM. Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development. 1990;110:325–330. doi: 10.1242/dev.110.2.325. [DOI] [PubMed] [Google Scholar]
- Kao KR, Elinson RP. The entire mesodermal mantle behaves as Spemann’s organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev Biol. 1988;127:64–77. doi: 10.1016/0012-1606(88)90189-3. [DOI] [PubMed] [Google Scholar]
- Keller RE. Vital dye mapping of the gastrula and neurula of Xenopus laevis. I. Prospective areas and morphogenic movements of the superficial layer. Dev Biol. 1975;42:222–241. doi: 10.1016/0012-1606(75)90331-0. [DOI] [PubMed] [Google Scholar]
- Keller RE. Vital dye mapping of the gastrula and neurula of Xenopus laevis. II. Prospective areas and morphogenic movements of the deep layer. Dev Biol. 1976;51:118–137. doi: 10.1016/0012-1606(76)90127-5. [DOI] [PubMed] [Google Scholar]
- Kessel M, Gruss P. Murine developmental control genes. Science. 1990;249:374–379. doi: 10.1126/science.1974085. [DOI] [PubMed] [Google Scholar]
- Klein SL. The first cleavage furrow demarcates the dorsal-ventral axis in Xenopus embryos. Dev Biol. 1987;120:299–304. doi: 10.1016/0012-1606(87)90127-8. [DOI] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNA. Nucl Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews LS, Vale WW. Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell. 1991;65:973–982. doi: 10.1016/0092-8674(91)90549-e. [DOI] [PubMed] [Google Scholar]
- Melton DA. Pattern formation during animal development. Science. 1991;252:234–241. doi: 10.1126/science.1672778. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Niehrs C, De Robertis EM. Ectopic expression of a homeobox gene changes cell fate in Xenopus embryos in a position-specific manner. EMBO J. 1991 doi: 10.1002/j.1460-2075.1991.tb04928.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD. The “organization center” of the amphibian embryo: its spatial organization and morphogenic action. Adv Morphogen. 1973;10:1–39. doi: 10.1016/b978-0-12-028610-2.50005-8. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. A Normal Table of Xenopus laevis (Daudin) Amsterdam: North Holland Publishing Co; 1967. [Google Scholar]
- Nüsslein-Volhard C. Determination of the embryonic axes of Drosophila. Development (Suppl) 1991;1:1–10. [PubMed] [Google Scholar]
- Rosa FM. Mix. 1, a homeobox mRNA inducible by mesoderm inducers, is expressed mostly in the presumptive endodermal cells of Xenopus embryos. Cell. 1989;57:965–974. doi: 10.1016/0092-8674(89)90335-8. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Jessell T. Retinoic acid modifies mesodermal patterning in early Xenopus embryos. Genes Dev. 1991;5:175–187. doi: 10.1101/gad.5.2.175. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Melton DA. Interaction between peptide growth factors and homeobox genes in the establishment of anteroposterior polarity in frog embryos. Nature. 1989;341:33–38. doi: 10.1038/341033a0. [DOI] [PubMed] [Google Scholar]
- Savage R, Phillips CR. Signals from the dorsal blastopore lip region during gastrulation bias the ectoderm toward a non-epidermal pathway of differentiation in Xenopus laevis. Dev Biol. 1989;133:157–168. doi: 10.1016/0012-1606(89)90307-2. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora P, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. Sequential activation of Hox 2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346:763–766. doi: 10.1038/346763a0. [DOI] [PubMed] [Google Scholar]
- Sive HL, Cheng PF. Retinoic acid perturbs the expression of Xhox.lab genes and alters mesodermal determination in Xenopus laevis. Genes Dev. 1991;5:1321–1332. doi: 10.1101/gad.5.8.1321. [DOI] [PubMed] [Google Scholar]
- Sive HL, Draper BW, Harland R, Weintraub H. Identification of retinoic acid–sensitive period during primary axis formation in Xenopus laevis. Genes Dev. 1990;4:932–942. doi: 10.1101/gad.4.6.932. [DOI] [PubMed] [Google Scholar]
- Slack JMW. From Egg to Embryo. Cambridge, England: Cambridge University Press; 1991. [Google Scholar]
- Slack JMW, Darlington BG, Gillespie LL, Godsave SF, Isaacs HV, Paterno GD. The role of fibroblast growth factor in early Xenopus development. Development (Suppl) 1989;107:141– 148. doi: 10.1242/dev.107.Supplement.141. [DOI] [PubMed] [Google Scholar]
- Smith JC, Dale L, Slack JMW. Cell lineage labels and region-specific markers in the analysis of inductive interactions. J Embryo Exp Morph (Suppl) 1985;89:317–331. [PubMed] [Google Scholar]
- Smith JC, Cooke J, Green JBA, Howes G, Symes K. Inducing factors and the control of mesodermal pattern in Xenopus laevis. Development. 1989;107:149–159. doi: 10.1242/dev.107.Supplement.149. [DOI] [PubMed] [Google Scholar]
- Spemann H. Über den Anteil von Implantat und Wirtskeim an der Orientierung und Beschaffenheit der induzierten Embryonalage. Roux’ Arch f Entw Mech. 1931;123:389–517. doi: 10.1007/BF01380646. [DOI] [PubMed] [Google Scholar]
- Spemann H. Embryonic Development and Induction. New Haven, Connecticut: Yale University Press; 1938. [Google Scholar]
- Spemann H, Mangold H. Über Induktion von Embryon-alanlagen durch Implantation artfremder Organisatoren. Roux’ Arch f Entw Mech. 1924;100:599–638. [Google Scholar]
- Stewart RM, Gerhart JC. The anterior extent of dorsal development of the Xenopus embryonic axis depends on the quantity of organizer in the late blastula. Development. 1990;109:363–372. doi: 10.1242/dev.109.2.363. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Thomsen G, Woolf T, Whitman M, Sokol S, Vaughan J, Vale W, Melton DA. Activins are expressed early in Xenopus embryogenesis and can induce axial mesoderm and anterior structures. Cell. 1990;63:485–493. doi: 10.1016/0092-8674(90)90445-k. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Positional information revisited. Development (Suppl) 1989;107:3–12. doi: 10.1242/dev.107.Supplement.3. [DOI] [PubMed] [Google Scholar]
- Yuge M, Kobayakawa Y, Fujisue M, Yamana K. A cytoplasmic determinant for dorsal axis formation in an early embryo of Xenopus laevis. Development. 1990;110:1051–1056. doi: 10.1242/dev.110.4.1051. [DOI] [PubMed] [Google Scholar]