Control of the nuclear localization of Extradenticle by competing nuclear import and export signals (original) (raw)
Abstract
The Drosophila PBC protein Extradenticle (Exd) is regulated at the level of its subcellular distribution: It is cytoplasmic in the absence of Homothorax (Hth), a Meis family member, and nuclear in the presence of Hth. Here we present evidence that, in the absence of Hth, Exd is exported from nuclei due to the activity of a nuclear export signal (NES). The activity of this NES is inhibited by the antibiotic Leptomycin B, suggesting that Exd is exported by a CRM1/exportin1-related export pathway. By analyzing the subcellular localization of Exd deletion mutants in imaginal discs and cultured cells, we identified three elements in Exd, a putative NES, a nuclear localization sequence (NLS), and a region required for Hth-mediated nuclear localization. This latter region coincides with a domain in Exd that binds Hth protein in vitro. When Exd is uncomplexed with Hth, the NES dominates over the NLS. When Exd is expressed together with Hth, or when the NES is deleted, Exd is nuclear. Thus, Hth is required to overcome the influence of the NES, possibly by inducing a conformational change in Exd. Finally, we provide evidence that Hth and Exd normally interact in the cytoplasm, and that Hth also has an NLS. We propose that in Exd there exists a balance between the activities of an NES and an NLS, and that Hth alters this balance in favor of the NLS.
Keywords: Extradenticle, Homothorax, nuclear localization, Drosophila
Regulated nuclear localization has been described for many transcription factors, including rel domain proteins such as NFκB and Dorsal (Blank et al. 1992; Gillespie and Wasserman 1994), the MAD proteins that relay TGFβ and BMP signals (Cho and Blitz 1998), and the STAT family of cytokine signal mediators (Hoey and Schindler 1998). The regulation of nuclear localization serves many different functions, including limiting transcriptional activity to the presence (e.g., NF-AT; Shibasaki et al. 1996; Timmerman et al. 1996) or absence (e.g., Smad7; Itoh et al. 1998) of a signal, a specific stage of the cell cycle (e.g., cyclin B1; Li et al. 1997), or state of differentiation (e.g., E2F4; Puri et al. 1998). Just as the regulation of nuclear localization serves multiple functions, the means of regulation are also varied. Phosphorylation can block the interaction of a nuclear localization sequence (NLS) with a cytoplasmic retention factor (e.g., NFκB; Blank et al. 1992), mask (Jans et al. 1991) or unmask (Rihs et al. 1991) an NLS, or enhance the interaction of the protein with importin α, a subunit of the NLS receptor (Briggs et al. 1998). Finally, nuclear localization can be regulated by nuclear export signals (NESs), which are short peptides that mediate the nuclear to cytoplasmic translocation of proteins (for review, see Nakielny and Dreyfuss 1997). Canonical NESs bind to an export receptor, CRM1/exportin1, which is related in sequence to importin β, another subunit of the NLS receptor (Fornerod et al. 1997; Fukuda et al. 1997; Ossareh-Nazari et al. 1997; Stade et al. 1997).
Extradenticle (Exd), a Drosophila member of the PBC family of Hox cofactors (Mann and Affolter 1998), was the first homeodomain protein shown to exhibit regulated nuclear import (Mann and Abu-Shaar 1996; Aspland and White 1997). In the wild type, Exd is nuclear only in the presence of Homothorax (Hth), another homeodomain protein (Rieckhof et al. 1997; Kurant et al. 1998; Pai et al. 1998). In Drosophila, both hth and exd are necessary for proximal leg development but are not required for distal leg development (Gonzalez-Crespo and Morata 1996; Casares and Mann 1998; Gonzalez-Crespo et al. 1998). Accordingly, Exd is normally cytoplasmic in cells that will give rise to distal leg fates (Gonzalez-Crespo and Morata 1996; Rieckhof et al. 1997; Casares and Mann 1998; Gonzalez-Crespo et al. 1998). However, when Exd is expressed at high levels, the protein is able to enter distal leg nuclei (Gonzalez-Crespo and Morata 1996; Gonzalez-Crespo et al. 1998), even in the absence of Hth (M. Abu-Shaar et al., unpubl.). This observation suggests that there exists a saturable mechanism that normally functions to keep Exd out of the nucleus. Further, these data are consistent with the presence of a consensus classical nuclear localization sequence (Dingwall and Laskey 1991) in the Exd ORF, located within the homeodomain (Fig. 1).
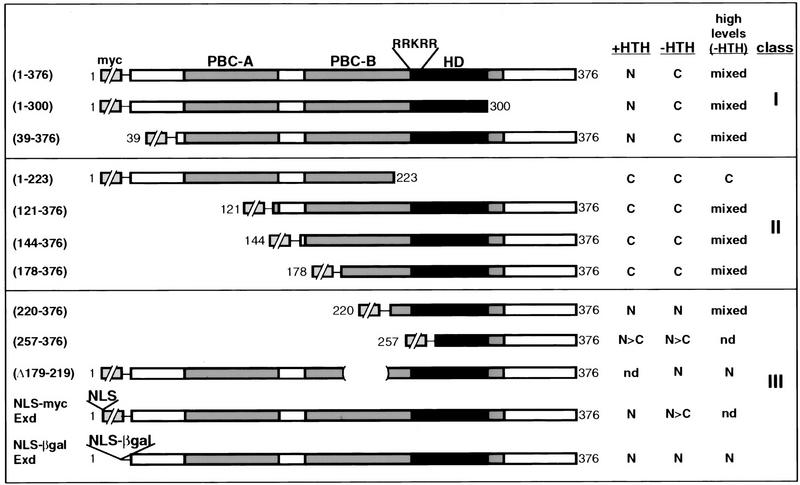
Figure 1.
Schematic diagram of Exd protein variants and summary of their subcellular localization. Full-length Exd(1–376) contains three major domains of conservation with vertebrate Pbx1 and C. elegans Ceh-20; PBC-A, PBC-B, and the homeodomain (HD) as indicated. In addition, these proteins are similar for an additional 13 residues carboxy-terminal to the homeodomain (dark gray shading). A classical NLS (RRKRR) is present in the amino-terminal arm of the homeodomain. NLS–myc–Exd has the NLS sequence from the SV40 large T antigen inserted amino-terminal to the myc tag and NLS–β gal–Exd has the same NLS and the nearly full-length β-galactosidase ORF inserted in place of the myc tag. All proteins except for NLS–β gal–Exd were myc-tagged at their amino termini (small broken boxes). Protein domains are shown to scale with the exception of the myc epitope, which is 86 amino acids. The columns at right summarize the subcellular localization of these proteins in the presence (+HTH) or absence (−HTH) of Homothorax, and when expressed at high levels in the absence of Hth. (N) Nuclear; (C) cytoplasmic; (mixed) nuclear and cytoplasmic; (N > C) more nuclear than cytoplasmic; (nd) not determined. Class I proteins are nuclear in the presence of Hth but generally cytoplasmic in its absence; class II proteins are not regulated by Hth and generally cytoplasmic; class III proteins are not regulated by Hth and generally nuclear.
To investigate the mechanism controlling Exd’s subcellular regulation, we generated a series of epitope-tagged mutant Exd proteins. We analyzed the subcellular distribution of these deletion mutants in both cultured cells and imaginal discs. The data indicate that, in addition to an NLS, there are two separable regulatory domains in Exd, a nuclear export signal and a Hth interaction domain. The interplay between these sequences in Exd, and the ability of Hth to alter their relative activities, suggests a novel mechanism for controlling the nuclear localization of these two homeodomain proteins.
Results
Analysis of Exd deletion mutants in imaginal discs
Exd shares extensive homology with its vertebrate (Pbx1) and Caenorhabditis elegans (Ceh-20) homologs throughout much of its coding sequence (Burglin and Ruvkun 1992; Rauskolb et al. 1993). In addition to the homeodomain, these homology domains have been referred to as PBC-A and PBC-B (Fig. 1). To address the role of these domains, we constructed a series of myc-tagged Exd deletion mutants (Fig. 1). Transgenic flies carrying these mutants under the control of the yeast transcription factor Gal4 (Brand and Perrimon 1993) were crossed to flies expressing Gal4 from the patched (ptc)–Gal4 driver line, which is expressed as a stripe across all leg imaginal discs. This stripe of _ptc_–Gal4 expression includes both Hth-expressing and nonexpressing cells (Fig. 2). Therefore, the subcellular localization of the Exd mutants could be analyzed in the presence and absence of Hth by staining individual leg discs. The resultant third instar larvae were dissected and the leg discs stained for Hth and myc-Exd proteins. In addition, the discs were stained for Dachshund (Dac), which served as a nuclear marker for leg disc cells that do not express Hth and therefore normally have Exd in the cytoplasm (Mardon et al. 1994; Abu-Shaar and Mann 1998; Wu and Cohen 1999).
Figure 2.
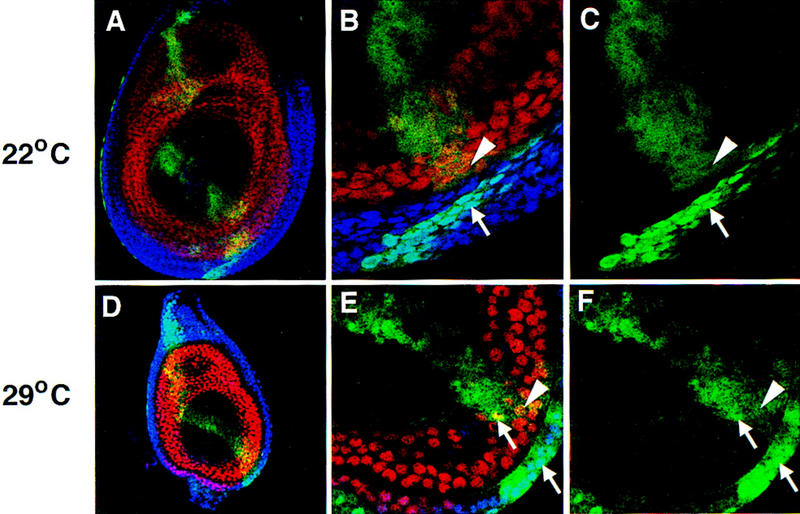
Subcellular localization of Exd(1–376) in leg imaginal discs. Shown are leg imaginal discs in which Exd(1–376) expression was induced by _ptc_–Gal4 and monitored by staining with an anti-myc antibody (green). These discs were costained for Hth (blue) and Dac (red), which, in these focal planes, serves as a nuclear marker for non-Hth-expressing cells. (A–C) A leg disc grown at 22°C; (D–F) a leg disc grown at 29°C, which results in higher levels of Exd(1–376). (A,D) Low magnification views of the entire disc; the _ptc_–Gal4 induced expression of Exd(1–376) can be seen as a green stripe extending across the entire disc. (B,C,E,F) High magnification views showing regions of these discs with Hth+, Dac− cells next to Hth−, Dac+ cells; (C,F) show only the signal from the myc staining. At 22°C, nearly all of the Exd(1–376) is in the cytoplasm in the absence of Hth (A–C) whereas at 29°C (D–F), Exd(1–376) is partially localized to nuclei in the absence of Hth (arrows in F). In this and subsequent figures, arrows point to examples of nuclear localization and arrowheads point to examples of cytoplasmic localization.
To confirm that the myc tag does not interfere with the regulation of Exd’s nuclear localization, we tested the subcellular distribution of a myc-tagged full-length Exd protein, Exd(1–376) (Fig. 2). This protein behaved like wild-type Exd: It was nuclear in the presence of Hth, cytoplasmic in the absence of Hth (Fig. 2A–C), but was capable of entering nuclei in the absence of Hth when expressed at high levels (Fig. 2D–F).
The Exd deletion mutants can be divided into three main classes, depending on their localization in the presence or absence of Hth (Fig. 1). The first class of mutants are those whose localization resembles that of full-length Exd. The very amino or carboxyl termini of the protein do not contribute to the regulation of Exd’s localization, because proteins with the first 38 (Fig. 3A–C) or the last 76 (Fig. 3D–F) residues deleted behaved like wild type. In addition, like full-length Exd, higher levels of these proteins resulted in their partial nuclear localization, even in the absence of Hth (Fig. 1; data not shown).
Figure 3.
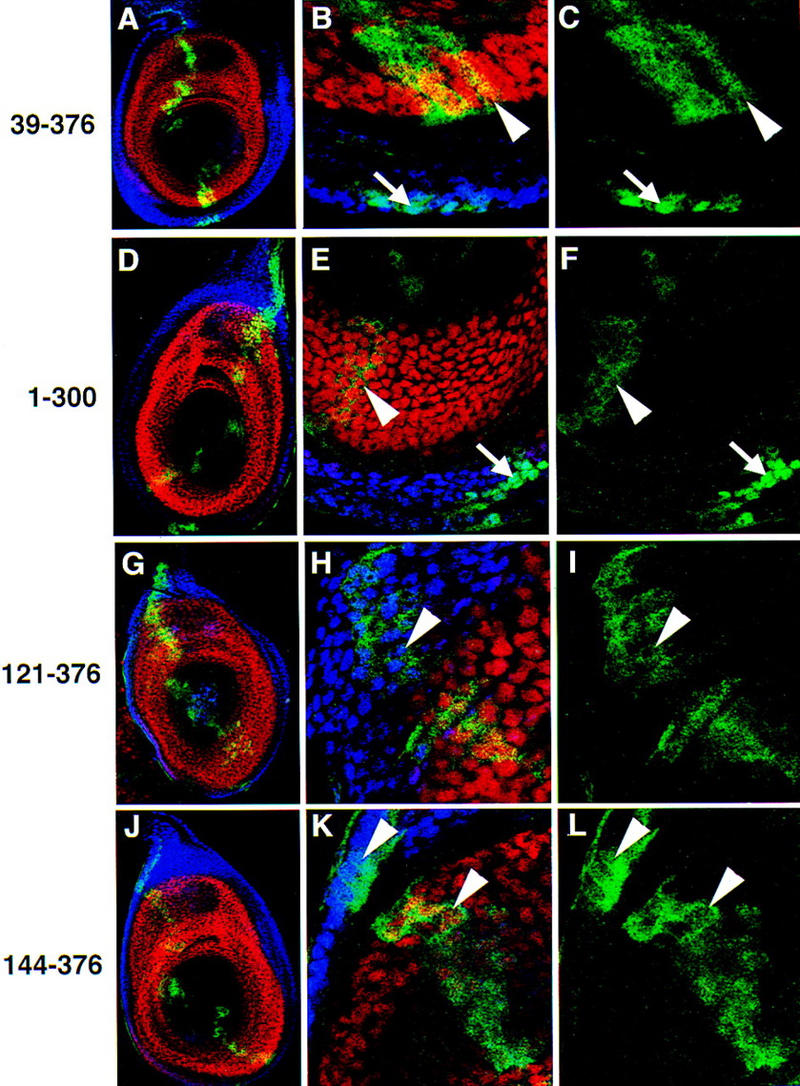
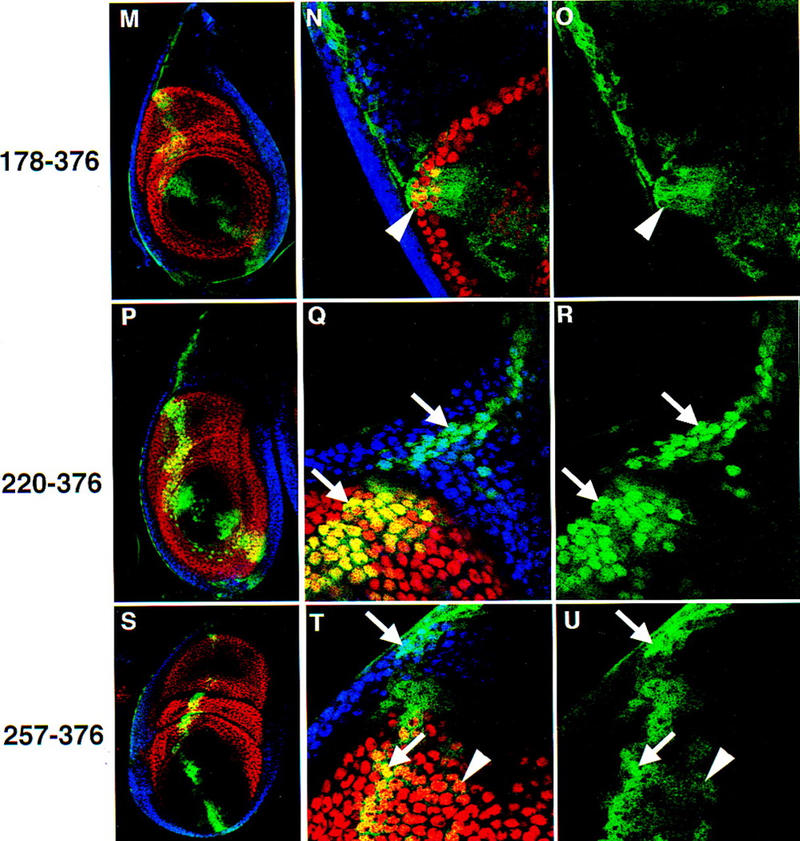
Subcellular localization of Exd truncations in leg imaginal discs. Antibody stains and views of leg discs are as described in Fig. 2. Exd(39–376) (A–C); Exd(1–300) (D–F); Exd(121–376) (G–I); Exd(144–376) (J–L); Exd(178–376) (M–O); Exd(220–376) (P–R); Exd(257–376) (S–U). All discs were grown at 22°C. See Fig. 1 for schematic illustrations of these mutants. (Arrows) Examples of nuclear localization; (arrowheads) examples of cytoplasmic localization.
The second class of mutants results from further deletions of the amino terminus of Exd, and consist of proteins that are cytoplasmic in the presence or absence of Hth (Fig. 1). These mutants are still able to enter nuclei when expressed at very high levels. Deletions in this class remove the first 120 (Fig. 3G–I), 143 (Fig. 3J–L), or 177 (Fig. 3M–O) amino acids of Exd. These deletions all remove at least part of the PBC-A domain, which has been mapped as the site of protein–protein interaction between Pbx1 and Meis1, a vertebrate Hth homolog (Chang et al. 1997; Knoepfler et al. 1997). We have confirmed that this domain in Exd interacts with Hth (see below).
The third class of deletions is represented by Exd(220–376), which is a predominantly nuclear protein in the presence or absence of Hth (Fig. 3P–R). This distribution indicates that Exd(220–376) is missing an element that normally prevents Exd’s stable nuclear localization. The nuclear localization of this mutant is consistent with the presence of a consensus NLS (RRKRR) at position 239–243, which is still present in Exd(220–376) (Fig. 1). As the previous deletion mutant, Exd(178–376), is a constitutively cytoplasmic protein, the domain that inhibits nuclear localization maps to residues 178–220. On the basis of additional experiments (see below), this sequence appears to function as an NES.
The final deletion, Exd(256–376), resulted in a protein that is present in both the nucleus and the cytoplasm (Fig. 3S–U). Because this protein has a preference for the nucleus and is not regulated by Hth, we consider it a class III protein (Fig. 1). The mixed distribution of this protein may in part be due to its small size (predicted molecular mass = ∼23 kD), which in principle has the ability to diffuse between the cytoplasm and the nucleus in an unregulated fashion. However, the partial nuclear localization of this protein suggests that there may be another weak NLS in this part of Exd. Consistent with this possibility, the sequence KRIRYKK is present at position 292–298, and may be functioning as a weak NLS.
The results from the analysis of this deletion series suggest that Exd has three separable elements controlling its subcellular localization, at least one NLS (residues 239–243), a sequence that inhibits nuclear localization (located between amino acids 178 and 220), and a Hth interaction sequence (located in part between amino acids 39 and 120).
Exd is excluded from the nucleus by nuclear export
The element located between amino acids 178 and 220 that inhibits the stable nuclear localization of Exd could be an NES, or it could bind to a protein that contains an NES. Alternatively, this sequence could function as a cytoplasmic retention element, perhaps by binding a cytoplasmic anchoring protein. To determine whether Exd is normally cytoplasmic because of the activity of an NES, we tested its sensitivity to the antibiotic leptomycin B (LMB) which blocks the export of many NES-containing proteins. LMB has been shown to block NES function in Schizosaccharomyces pombe by directly binding to the NES receptor, CRM1/exportin1, and competitively inhibiting its interaction with this class of NES (Fukuda et al. 1997; Kudo et al. 1998).
To test for an LMB-sensitive NES in Exd, we transfected into Schneider line 2 (S2) cells plasmids capable of expressing myc-tagged Exd(1–376) or myc-tagged Exd(178–376) in the presence or absence of LMB. In the absence of LMB, these proteins behave in S2 cells as they do in embryos: Exd(1–376) is cytoplasmic at low levels (Fig. 4A) but nuclear in the presence of Hth (Fig. 4B) or when expressed at high levels (data not shown). Also consistent with our studies in imaginal discs, Exd(178–376), which is missing the Hth-interaction domain, is cytoplasmic in the presence or absence of Hth (Fig. 4F–G) but is partially nuclear when expressed at high levels (data not shown). Strikingly, even when expressed at low levels, both of these proteins were fully nuclear in the presence of LMB (Fig. 4C,H). The ability of LMB to induce Exd’s nuclear translocation indicates that Exd is normally cytoplasmic because of the activity of an NES that depends on a CRM1-related export system.
Figure 4.
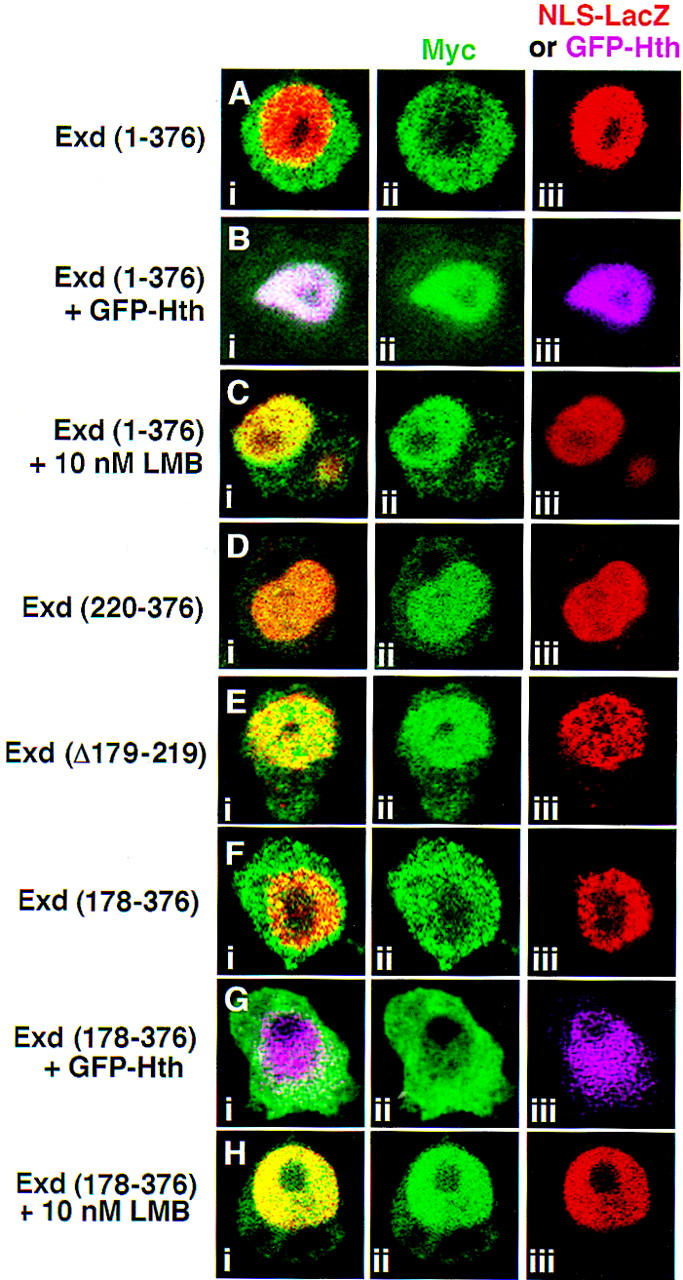
Exd is excluded from nuclei because of an LMB-sensitive NES. Shown are S2 cells immunostained for proteins expressed from transiently transfected plasmids; all Exd derivatives are myc tagged and shown in green. Expression of NLS–βgal served as a nuclear marker except for B and G, in which GFP–Hth was a nuclear marker. (A) Exd(1–376) + NLS–βgal (red); (B) Exd(1–376) + GFP–Hth (fuchsia); (C) Exd(1–376) + NLS–βgal (red) + 10 nm LMB; (D) Exd(220–376) + NLS–βgal (red); (E) Exd(Δ179–219) + NLS–βgal (red); (F) Exd(178–376) + NLS–βgal (red); (G) Exd(178–376) + GFP–Hth (fuchsia); (H) Exd(178–376) + NLS–βgal (red) + 10 nm LMB. (i,ii,iii) The merged, green, and red or fuchsia channels, respectively.
The in vivo mapping experiments suggest that sequences located between residues 178 and 220 include an element that inhibits Exd’s nuclear localization, perhaps an NES. One prediction from these results is that an Exd protein that does not contain these sequences should be constitutively nuclear in S2 cells. To test this hypothesis, we analyzed the distribution of Exd(220–376) (Fig. 4D) and Exd(Δ179–219) (Fig. 4E), which harbors an internal deletion of this sequence (Fig. 1). Both of these proteins are nuclear in S2 cells, thus confirming the localization of this element to between amino acids 178 and 220. Although there are additional possibilities (see Discussion), for simplicity we refer to this sequence as an NES.
Curiously, unlike Exd synthesized from a transfected plasmid, the endogenously expressed Exd in S2 cells, which is normally cytoplasmic (Rieckhof et al. 1997), is only partially shifted to the nucleus in the presence of LMB (Fig. 5A–C). We found that at all concentrations tested (10, 25, 100, and 400 nm), a 4-hr incubation with LMB resulted in a shift of most, but not all, of the Exd protein into the nucleus (Fig. 5B,C; data not shown). In a small percentage of the LMB-treated cells, the endogenous Exd protein was entirely nuclear (not shown). Incubations of up to 8 hr in the presence of 25 nm LMB did not increase the ratio of nuclear/cytoplasmic Exd. In contrast, 100% of the endogenous Exd in S2 cells became nuclear when Hth was present (Fig. 5D). Thus, in addition to nuclear export, there may be a second mechanism inhibiting the nuclear localization of the endogenously expressed Exd in S2 cells.
Figure 5.
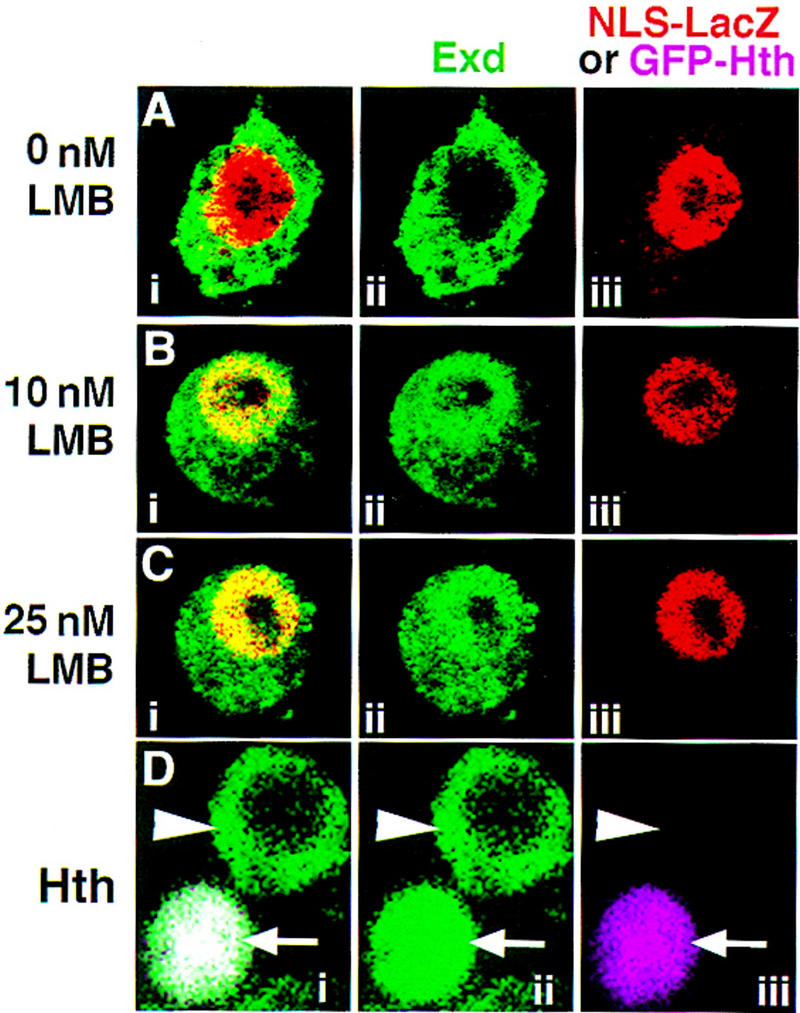
The endogenously expressed Exd protein in S2 cells is only partially sensitive to LMB. Shown are S2 cells, transfected with NLS–β gal or GFP–Hth, stained for Exd (green) and β gal (red) or GFP–Hth (fuchsia) proteins. Prior to fixation, the cells were incubated with either no LMB (A,D), 10 nm LMB (B), or 25 nm LMB (C). Without LMB (A) Exd was in the cytoplasm. In the presence of LMB (B,C), although most of the Exd was in nuclei, some remained in the cytoplasm. Similar results were obtained with concentrations of LMB up to 400 nm (not shown). When GFP–Hth was expressed in S2 cells, all of the endogenous Exd protein became nuclear (D, arrow), whereas in untransfected cells, Exd was cytoplasmic (D, arrowhead). (i,ii,iii) The merged, green, and red/fuchsia channels, respectively.
Mechanism of Hth action
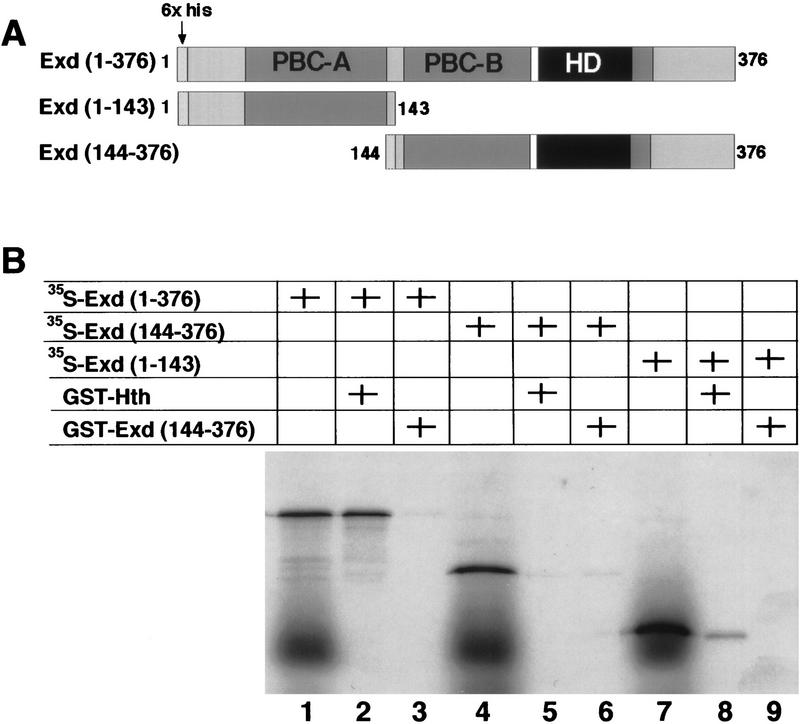
Meis1, a vertebrate Hth homolog, has been shown to bind Exd in vitro with high affinity (Rieckhof et al. 1997). On the basis of the behavior of amino-terminal deletions of Exd described above and from studies on Pbx1–Meis1 interactions (Chang et al. 1997; Knoepfler et al. 1997), we anticipated that Hth would interact with a domain located in the amino terminus of Exd, perhaps within PBC-A. To test this hypothesis, we carried out GST pull-down experiments with 35S-labeled Exd proteins (Fig. 6). GST–Hth bound Exd(1–376), but not Exd(144–376), which is missing the PBC-A domain (Fig. 6B, lanes 2,5). Interestingly, GST–Hth was also able to bind Exd(1–143), although this interaction appeared to be weaker than the interaction between Hth and full-length Exd (Fig. 6B, cf. lanes 2 and 8). As a negative control, GST–Exd(144–376) was unable to bind any of the 35S-labeled Exd proteins in this assay (Fig. 6B, lanes 3,6,9). These data suggest that residues 1–143 of Exd are required to bind Hth, but that other regions of Exd may contribute to the interaction.
Figure 6.
The PBC-A domain of Exd binds Hth. (A) Schematic illustration of the his-tagged 35S-labeled-Exd proteins used in this experiment. (B) GST pull-down experiment. Crude Escherichia coli extracts containing GST–Hth (lanes 2,5,8) or GST–Exd(144–376) (lanes 3,6,9) were incubated with 35S-labeled-Exd(1–376) (lanes 2,3), 35S-labeled-Exd(144–376) (lanes 5,6), or 35S-labeled-Exd(1–143) (lanes 8,9). Bound proteins were analyzed by SDS-PAGE and autoradiography. For each of the three 35S-labeled Exd proteins, 25% of the amount used in the binding reactions was directly loaded in lanes 1, 4, and 7, respectively.
The behavior of Exd(1–223), which is lacking the homeodomain and carboxy-terminal sequences (Fig. 1), is also informative. From the mapping experiments, Exd(1–223) lacks an NLS but retains the Hth-interaction domain and the NES. As with the previous Exd mutants, this protein was tagged at its amino terminus with the myc epitope and expressed in the ptc domain of leg discs by the Gal4–UAS system (Fig. 7A–C). Immunostaining for the myc epitope revealed a fully cytoplasmic protein in the presence or absence of Hth. Unlike other forms of Exd, this protein remained predominantly cytoplasmic, even when expressed at high levels (Fig. 1). However, despite the fact that Exd(1–223) contains the Hth interaction domain, we failed to observe an effect on the localization of the endogenous Hth or Exd proteins in imaginal discs (Fig. 7B; data not shown). Consistent with these findings, expression of Exd(1–223) in vivo driven by _ptc_–Gal4 did not lead to any obvious mutant phenotypes in embryos or adults (data not shown).
Figure 7.
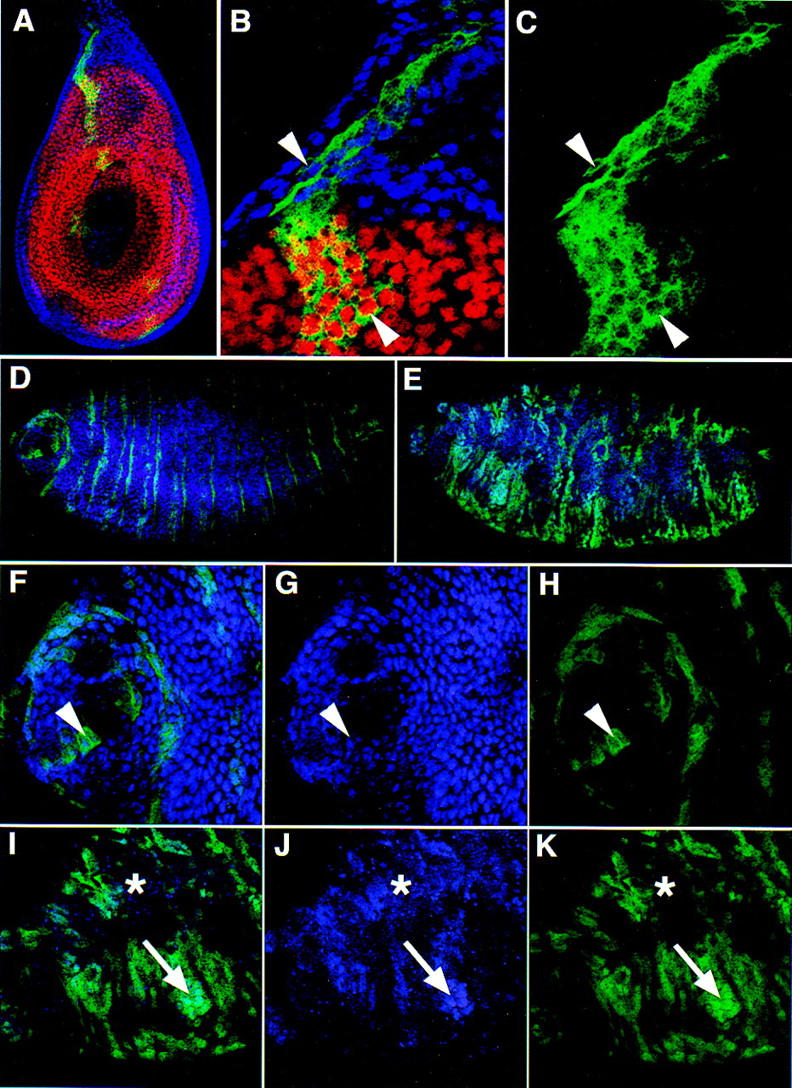
The subcellular localization of Exd(1–223) in the presence and absence of endogenous Exd. (A–C) _ptc_–Gal4 driven expression of Exd(1–223) in otherwise wild-type (exd+) leg discs; views and stains are as in Fig. 2 [Hth, blue; Dac, red; and Exd(1–223), green]. Exd(1–223), which includes the Hth-interaction domain and the NES, but excludes the NLS, is cytoplasmic in Hth-expressing and nonexpressing cells (arrowheads). (D–K) Embryos expressing Exd(1–223) in stripes with the _ptc_–Gal4 driver line. These embryos were stained for Hth (blue) and the myc epitope of Exd(1–223) (green). (D,F–H) An embryo that had no maternal exd expression but had wild-type zygotic (paternal) exd expression; (E,I–K) a sibling embryo that had no maternal or zygotic exd expression. In these _exd_− embryos, the stripes of _ptc_–Gal4-induced Exd(1–223) expression are disorganized because of a breakdown in engrailed expression (Peifer and Wieschaus 1990). (D,E) Low magnification views of the entire embryos (anterior is left); (F–K) higher magnification views of the anterior regions of these embryos. In the presence of zygotic Exd (F–H), Hth protein is detected in many nuclei (G) and Exd(1–223) is predominantly cytoplasmic (H, arrowhead). In the absence of maternal and zygotic Exd (I–K), Hth was not detected in most cells (asterisk) but could be detected in cells that also express Exd(1–223) (J,K; arrow). In these cells, both proteins are predominantly nuclear.
The inability of Exd(1–223) to alter the localization of the endogenous Hth protein in imaginal discs could be explained in two ways. One possibility is that Exd and Hth normally interact in the nucleus but, because Exd(1–223) has no NLS and is always found in the cytoplasm, it never has the opportunity to interact with Hth. A second possibility is that Exd(1–223) has the potential to interact with Hth, but it does so much less efficiently than full-length Exd and therefore cannot effectively compete for this interaction with the endogenous Exd protein. To distinguish between these possibilities, we expressed Exd(1–223) in embryos that are devoid of any endogenously expressed (maternal or zygotic) Exd protein. In the complete absence of Exd, Hth protein is difficult to detect because it is unstable (Fig. 7J; Abu-Shaar and Mann 1998; Kurant et al. 1998). However, when Exd(1–223) is expressed in _exd_− embryos, Hth is partially stabilized, and both proteins can be detected in nuclei (Fig. 7E,I–K). In contrast, in the presence of endogenous Exd protein, Exd(1–223) is predominantly cytoplasmic (Fig. 7D,E–H). These results suggests that Exd(1–223) can interact with Hth in vivo, but that this interaction is less efficient than the interaction between Hth and full-length Exd. Further, these results suggest that Hth normally interacts with Exd in the cytoplasm and that Hth has a functional NLS that is capable of importing the Hth–Exd(1–223) complex into nuclei.
Because Exd has both an NES and an NLS, we suggest that a balance between these two control elements dictates Exd’s subcellular localization. This balance may be sensitive to the overall structure of the protein, and the relative positions of the Hth-interaction domain, the NES, and the NLS. It may be possible to perturb this balance by the addition of another NLS to Exd. To test this idea, we fused additional NLS sequences to Exd, and determined the subcellular distribution of these fusion proteins. In one case (NLS–myc–Exd), we added the NLS from the SV40 large T antigen together with the myc tag (∼100 amino acids, total) to the amino terminus of full-length Exd (Fig. 1). In the second case (NLS–β gal–Exd), we added the same NLS fused to the nearly full-length β-galactosidase protein (∼1000 amino acids) to the amino terminus of full-length Exd (Fig. 1). When expressed in leg discs using _ptc_–Gal4, NLS–myc–Exd was weakly nuclear in the absence of Hth (Fig. 8A–C), whereas NLS–β gal–Exd was fully nuclear in the absence of Hth (Fig. 8D–F). Consistent with the localization of these proteins, expression of NLS–β gal–Exd with _ptc_–Gal4 (at 22°C) generated severe defects in leg development, whereas NLS–myc–Exd or Exd(1–376) did not (data not shown). These results suggest that the context of the native NLS in Exd is important for its regulated activity because the addition of a second NLS, at least in some cases, can induce the Hth-independent nuclear localization of Exd.
Figure 8.
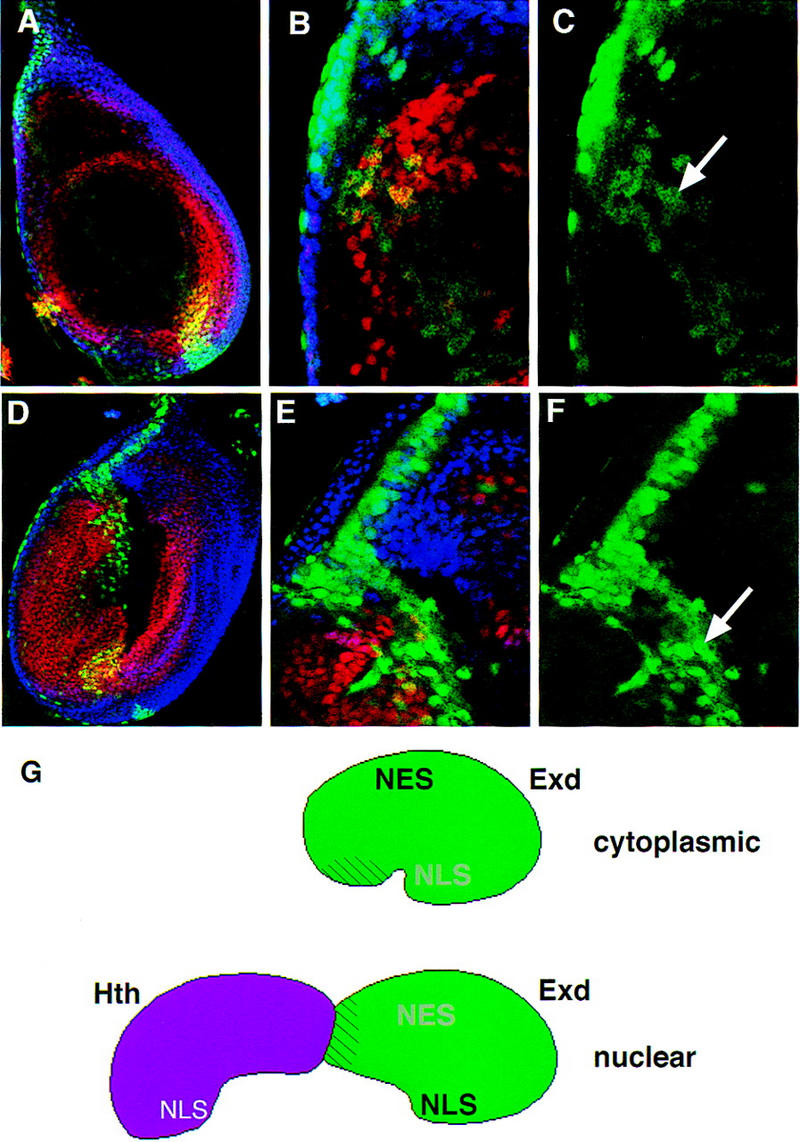
The regulated nuclear import of Exd depends on a balance between nuclear export and localization signals. (A–C) The subcellular localization of NLS–myc–Exd in leg discs; (D–F) the subcellular localization of NLS–β gal–Exd in leg discs. The stains and views of these discs are as described in Fig. 2 (Hth, blue; Dac, red; and NLS–myc–Exd or NLS–β gal–Exd, green). In the absence of Hth, NLS–myc–Exd is weakly nuclear (C, arrow), whereas NLS–β gal–Exd is predominantly nuclear (F, arrow). The nuclear localization of NLS–β gal–Exd is incompatible with Dac expression (Abu-Shaar and Mann 1998); therefore, these nuclei appear green (E). (G) A model summarizing these findings. We propose that Exd contains both an NES and an NLS and that, due to its conformation, the activity of the NES dominates when Hth is not present. In the presence of Hth, we suggest that Hth binds to a region within the PBC-A domain of Exd (striped region) and induces a conformational change, resulting in the NLS dominating.
Discussion
A model for the control of Exd’s subcellular localization
By progressively deleting sequences from the amino terminus, we have been able to uncover three qualitatively different control elements in Exd. The first element, located at least in part between residues 39 and 120, is required for Hth-mediated nuclear localization. Removing the same region of Exd also resulted in the loss of Exd’s ability to physically interact with Hth in vitro. Thus, this region of Exd also contains a Hth-interaction domain. The second element, located at least in part between residues 178 and 220, is required for Exd’s cytoplasmic localization, which is its normal localization in the absence of Hth. Our data further suggest that this element might be an NES because its activity is inhibited by the antibiotic LMB, which specifically binds to the NES receptor, CRM1. The third element, located between residues 220 and 256, behaves as an NLS. Inspection of this sequence suggests that there is a consensus NLS located within the amino terminus of the Exd homeodomain.
On the basis of these findings, we suggest a model for how Exd’s subcellular localization is controlled (Fig. 8G). In this model, we suggest that, although Exd contains both an NES and an NLS, in the native protein (uncomplexed with Hth), the NES is more active than the NLS, resulting in the net nuclear export and cytoplasmic localization of the protein. The balance between the NES and NLS is perhaps determined by the conformation of the native protein; one possibility is that the NLS is physically blocked by another region of the protein. The model further suggests that this balance can be altered by the binding of Hth to its binding site in Exd. We suggest that this shift in balance could be due to a Hth-induced change in Exd’s conformation, such that the NLS becomes more accessible to the transport machinery than the NES.
An alternative model is that, by binding Exd, Hth simply provides a second NLS, and that this NLS is sufficient to alter the balance in favor of nuclear localization. One argument against this model is that a truncated form of Hth containing the highly conserved HM domain (Rieckhof et al. 1997) is able to induce Exd’s nuclear localization in imaginal discs (H.D. Ryoo and R.S. Mann, in prep.). From its sequence, the HM domain does not appear to contain a monopartite or bipartite NLS (Dingwall and Laskey 1991). Thus, it is more likely that the nuclear localization of Exd induced by Hth or its isolated HM domain is primarily due to their ability to alter the balance between the NES and NLS within Exd. However, because we suggest that Hth also has an NLS (see below) it may be that this NLS contributes to the nuclear localization of the Hth–Exd complex.
A novel NES in Exd
The experiments with the antibiotic LMB suggest that Exd is cytoplasmic because of the activity of an NES that uses the CRM1 export pathway (for review, see Nakielny and Dreyfuss 1997). The deletion studies indicate that Exd requires a sequence located between residues 178 and 220 to be exported. Although this sequence could itself be an NES, at this time we cannot exclude the possibility that it mediates an interaction with another NES-containing protein. A third possibility is that residues 178–220 are required for the activity of an NES that is located elsewhere in Exd. Examination of the primary Exd sequence in this region does not reveal a LxxLxxLxL motif, which is the consensus sequence for the LMB-sensitive, Leucine-rich class of NES (Fischer et al. 1995; Wen et al. 1995). However, nonconsensus NES sequences that are inhibited by LMB have been described, suggesting that there is significant flexibility in the NES sequences bound by CRM1 (e.g., Otero et al. 1998). Like the consensus NES, these atypical NESs also have clusters of hydrophobic amino acids. Although finer point mutagenesis will be required to fully resolve this question, we note that this region of Exd contains clusters of hydrophobic amino acids, and that these residues are conserved among the PBC family members.
Additional controls of Exd’s subcellular distribution
When S2 cells were treated with LMB, the endogenous Exd protein shifted from the cytoplasm to the nucleus. However, even in the presence of 400 nm LMB, the endogenously synthesized protein did not become fully nuclear. This result is in marked contrast to the response of Exd protein synthesized from a transiently transfected plasmid, which is completely localized to the nucleus after LMB treatment. Moreover, the endogenous Exd protein is completely shifted to the nucleus after expression of Hth or Meis1 (Fig. 5; Rieckhof et al. 1997). Thus, a fraction of the endogenous Exd in S2 cells is resistant to LMB, but none is resistant to Hth. This suggests that, in addition to nuclear export, there may be a second mechanism inhibiting Exd’s nuclear localization. For example, Exd might be tethered to a cytoplasmic anchoring protein. The different behaviors of the endogenously and transiently expressed proteins may be explained in two ways: In the first, it could be due to a difference in the amount of Exd because the endogenous Exd in S2 cells is present at lower levels than Exd expressed transiently (M. Abu-Shaar and R.S. Mann, unpubl.). Higher levels of Exd may not be limited by the second mechanism, which may already be saturated by the endogenous Exd. A second possibility is that the second mechanism might depend on a post-translational modification of Exd, which may not immediately occur on newly synthesized protein. This modification could, for example, stabilize an interaction with a cytoplasmic anchoring protein. Although this interaction would be resistant to LMB, it would be disrupted by Hth.
The role of Hth in Exd’s nuclear localization
In the absence of Hth, Exd is cytoplasmic (Rieckhof et al. 1997; Kurant et al. 1998; Pai et al. 1998) and in the absence of Exd, Hth is unstable (Abu-Shaar and Mann 1998; Kurant et al. 1998). Therefore, these two homeodomain proteins appear to require each other for their stable nuclear localization. The model proposed here suggests that Exd is constantly shuttling in and out of the nucleus but, because of its conformation in the absence of Hth, the majority of Exd is cytoplasmic. The model further proposes that Hth, by directly interacting with Exd, alters this equilibrium in favor of nuclear localization.
If, as we suggest, Exd is shuttling in and out of the nucleus, Hth could in principle interact with Exd in either the cytoplasm or the nucleus. If the initial interaction occurs in the cytoplasm, then the heterodimer would enter the nucleus as a complex. If the initial interaction occurs in the nucleus, Hth would have to enter the nucleus on its own and then bind Exd, effectively trapping it in the nucleus. In either case, the interaction between Exd and Hth would block Hth’s degradation. Interestingly, this aspect of the model has a striking parallel in yeast in which the homeoproteins MATα2 and MATa1 are both protected from degradation by their physical association with each other (Johnson et al. 1998).
The behavior of Exd(1–223) suggests that Exd and Hth normally associate with each other in the cytoplasm. This protein has the Hth-interaction domain and the putative NES, but lacks the NLS. In imaginal discs, this protein is cytoplasmic even when expressed at high levels and appears to have no effect on the endogenous Exd or Hth proteins. However, when expressed in embryos in the absence of any full-length Exd, this protein is found in nuclei. Further, expression of Exd(1–223) in _exd_− embryos can at least partially stabilize Hth protein, and this Hth protein is also found in nuclei. We suggest that, in the absence of endogenous Exd, Exd(1–223) interacts with Hth in the cytoplasm, and that the Hth–Exd(1–223) complex is imported into nuclei due to an NLS in Hth. In the presence of endogenous Exd, Exd(1–223) does not associate with Hth because the Hth–Exd(1–223) interaction is less strong than the Hth–Exd (full-length) interaction. Consistent with this idea is our finding that although the PBC-A domain can, on its own, bind Hth, full-length Exd binds more tightly (Fig. 6).
There is additional evidence in favor of the idea that the initial Hth–Exd interaction occurs in the cytoplasm. One piece of suggestive data is that in S2 cells only some of the endogenous Exd is translocated to the nucleus in the presence of LMB, but all of it becomes nuclear in the presence of Hth. Hth’s ability to translocate the LMB-resistant fraction, which may not be shuttling between the nucleus and cytoplasm, suggests that it has the ability to interact with Exd in the cytoplasm. Second, as mentioned above, the HM domain of Hth is unlikely to contain an NLS, yet it is sufficient to induce Exd’s nuclear translocation in imaginal discs (H.D. Ryoo and R.S. Mann, in prep.). We suggest that, like full-length Hth, the HM domain binds to Exd and alters the balance between the NES and NLS in Exd, resulting in nuclear import.
Implications for leukemogenesis
Because the sequences of Exd and Pbx proteins are >80% identical within the PBC-A, PBC-B, and homeodomains, it is likely that the elements characterized here in Exd are probably conserved within the Pbx proteins. When the amino terminus of Pbx1 is replaced by the activation domain from the helix-loop-helix protein E2A, the hybrid protein is leukemogenic (Kamps et al. 1980; Nourse et al. 1990). Perhaps significantly, the breakpoint in Pbx1 occurs within the PBC-A domain which, from the results presented here, contains a Meis–Hth interaction domain. In addition, our model suggests that in the native conformation of Exd or Pbx (in the absence of Meis–Hth) the PBC-A domain may also interfere with the action of the NLS. In the E2A–Pbx1 hybrid, the breakpoint in Pbx1 may affect PBC-A’s ability to interact with Meis and, more importantly, may alter the balance between the NES and NLS. Therefore, unlike Pbx1, which may require a Meis-like cofactor for nuclear localization, the E2A–Pbx1 hybrid may be constitutively nuclear. We suggest that this effect on subcellular localization may be an important contributing factor to leukemogenesis.
Materials and methods
Construction of Exd deletion mutants
p131 is a Drosophila transformation vector in which the 6Xmyc epitope from MT6 (Roth et al. 1991) was inserted between the _Kpn_I and _Xba_I sites of pUAST (Brand and Perrimon 1993). In addition, the ATG start codon at the start of the myc epitope was optimized for expression in Drosophila and the _Stu_I site of pUAST was converted to an _Nhe_I site. This vector can be used to make amino-terminal 6Xmyc–epitope fusions of any ORF under the control of yeast GAL4.
Exd(1–376)
The Exd coding region (in pBluescript) was engineered to contain an _Nde_I site at the 5′ATG (CCATGG). This plasmid was digested with _Nde_I and the self-annealed oligonucleotide TATGTGAATTCACA ligated into the vector, creating an _Eco_RI site at the 5′ end of the coding region. The full coding region was then cloned into p131 as an _Eco_RI fragment.
Exd(1–300)
The Exd coding region was PCRd with MA18 (a top strand oligonucleotide spanning the first 18 bases of the Exd coding region with an _Nde_I site at its 5′ end) and E2 from S.K. Chan (Columbia University, NY) (a bottom strand oligonucleotide which ends outside the Exd homeobox and carries a _Bgl_II site at its 5′ end). The PCR product was digested with _Nde_I and _Bgl_II and cloned into the _Nde_I and _Bam_HI sites of SKSN3, a pBluescript vector in which the _Sph_I site had been changed into an _Nde_I site. The _Nde_I site in this construct was changed into an _Eco_RI site as for Exd(1–376) and the insert cloned into p131 as an _Eco_RI fragment.
Exd(1–223)
Exd(1–223) was constructed by ligating the _Asp_718–_Sal_I fragment of p131–Exd(1–376) into p131 that was digested with _Asp_718 plus _Pae_R71.
Exd(39–376)
The coding region of Exd in pBluescript was partially digested with _Fsp_I, the overhangs were phosphatased, and a phosphorylated _Xho_I linker (CCACGTGG) was ligated into the site. Next, the _Hin_dIII site in the polylinker, 3′ to the Exd coding region, was changed to an _Nhe_I site by cutting with the enzyme, filling in the overhangs, and reclosing the DNA. Subsequently, the _Xho_I–_Nhe_I fragment was cloned into p131.
Exd(121–376)
The Exd coding region (in pBluescript) was partially digested with _Ava_I, filled in, phosphatased, and reclosed with an 8-bp-phosphorylated _Xho_I linker (CCACGTGG). The remaining steps were identical to the construction of Exd(39–376).
Exd(144–376)
The Exd coding region (in pBluescript) was digested with _Cla_I and _Eco_RV (in the 3′ polylinker) and the overhangs filled in. The piece was ligated into the MT6 vector after cutting with _Xho_I, filling in the overhangs, and phosphatasing the vector. This piece of the Exd coding sequence was released from the MT6 vector as an _Eco_RI–_Xba_I fragment and ligated into p131.
Exd(178–376)
The Exd coding region (in pBluescript) was digested with _Sph_I, the overhangs removed with T4 polymerase, the vector phosphatased, and a phosphorylated 12-bp-_Eco_RI linker (CCGGATTCCGG) was ligated into the blunt site. This portion of Exd was then released as an _Eco_RI fragment and cloned into p131.
Exd(220–376)
The Exd coding region (in pBluescript) was digested with _Pvu_II and _Eco_RV, and this piece was placed into the _Stu_I site of p131.
Exd(257–376)
The Exd coding region (in pBluescript) was partially digested with _Sca_I, phosphatased, and a phosphorylated _Xho_I linker (CCACGTGG) was ligated into the vector. The subsequent steps were as for Exd(39–376).
Exd(Δ179–219)
The Exd coding region in pBluescript was amplified by PCR with the oligos MA95 (TTAGCTGCCTGAAGCAGTCGACCTGC) and T7. The product was digested with _Sph_I and _Hin_dIII and the fragment used to replace the corresponding fragment in the Exd coding region of p131–Exd(1–376).
NLS–β gal–Exd has been described previously (Abu-Shaar and Mann 1998). NLS–myc–Exd was constructed by inserting an oligo containing an optimized ATG for Drosophila and the NLS from SV40 large T antigen (HPPKKKRKVED) (Riddihough and Ish-Horowicz 1991) amino-terminal to the myc tag in MT6. The NLS–myc fragment was cloned into pUAST as described for the construction of p131.
Generation of cell culture constructs
Exd(1–376), Exd(178–376), and Exd(220–376) were placed under metallothionin (MT) control in the same manner: The cell culture vector with the MT promoter, pRMHA3, was digested with _Asp_718 and _Xba_I and the appropriate myc-tagged Exd region, together with the SV40 3′ UTR, was cloned into as an _Asp_718–_Nhe_I fragment. Exd(Δ179–219) was cloned into pRMHA3 by replacing the ORF of MT–Exd(1–376): MT–Exd(1–376) was digested with _Stu_I, and then partially digested with _Eco_RI and ligated with the _Eco_RI–_Sma_I fragment from Exd(Δ179–219). The GFP–Hth and NLS–β gal ORFs were also cloned in pRMHA3 to generate MT–GFP–Hth and MT–NLS–β gal.
Cell culture experiments
S2 transfection assays were done by standard procedures (Rieckhof et al. 1997). Exd expression plasmids were cotransfected with expression plasmids encoding GFP–Hth or NLS–β gal, which served as nuclear markers. For LMB treatment, initially a range of LMB concentrations (10–400 nm) and times (2–8 hr) were tested on S2 cells; these ranges were based on previous reports, (e.g., Taagepera et al. 1998). Two to three hours post-induction by CuSO4, LMB (generously provided by Minoru Yoshida, University of Tokyo, Japan) was added at the appropriate concentration and the cells incubated for 3 hr before fixation. The presence of LMB did not alter S2 cell morphology or the nuclear localization of marker proteins, suggesting that this treatment was not harmful to the integrity of the nucleus.
Immunostains
These were conducted by standard procedures. The primary antibodies used in this study were as follows: rabbit anti-Exd (Mann and Abu-Shaar 1996); chicken anti-Hth (Casares and Mann 1998); rabbit anti-Myc and mouse monoclonal anti-Myc (Serotec); mouse monoclonal anti-β gal (Promega); and rabbit anti-β gal (Cappel). Mouse anti-Dac was a gift from G. Mardon (Baylor College of Medicine, Houston, TX). The Hth stains in embryos were carried out with a guinea pig anti-Hth antibody, which was generated against full-length, his-tagged Hth protein (Cocalico Biologicals, Reamstown, PA). Unless otherwise stated, imaginal discs were grown at 22°C before dissection and staining. The stained leg discs or cultured cells were analyzed by three channel confocal microscopy (Bio-Rad).
Different levels of a particular Exd variant in imaginal discs were initially characterized by staining different insertion lines, which expressed different amounts of protein. When differences were observed, they were confirmed by testing a single insertion line at two different temperatures (22°C and 29°C), which results in different levels of Gal4 activity (Gonzalez-Crespo et al. 1998).
GST pull-downs
The Exd ORFs, Exd(1–376), Exd(144–376), and Exd(1–143), were cloned in the his-tagged pET14b vector (Novagen) and labeled in vitro with 35S by the Single Tube Protein System 3 (Novagen). From 8 ml of bacterial culture expressing either GST–Hth (full length) or GST–Exd(144–376), crude extracts were generated and mixed with 30 μl of glutathione agarose beads. After 2 hr of incubation at 4°C, the beads were washed three times in binding buffer (PBS + 0.1% NP-40, 10% glycerol, 0.1% BSA, 1 mm DTT, PMSF, aprotinin, and leupeptin), then mixed with 700 μl of binding buffer plus 20 μl of 35S-labeled protein, and incubated for an additional 2 hr at 4°C. The beads were washed twice with binding buffer and washed an additional two times with binding buffer minus BSA. A total of 20 μl of SDS loading buffer was added to the beads, which were boiled, spun, and the supernatant loaded onto a 12% SDS–polyacrylamide gel. After electrophoresis, the gel was dried and exposed overnight onto X-ray film.
Acknowledgments
We thank R. Cisse for technical assistance, M. Featherstone for communicating unpublished results, S. Greenwood for plasmids, G. Mardon for the Dac antibody, G. Struhl for flies, C. Tomkins for injections, Z. Wang for embryos, M. Yoshida for LMB, and members of the Mann lab for comments on this work. This work was supported by grants from the National Institutes of Health and Human Frontiers Science Program to R.S.M., who is a Scholar of the Leukemia Society of America.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rsm10@columbia.edu; FAX (212) 305-7924.
References
- Abu-Shaar M, Mann RS. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development. 1998;125:3821–3830. doi: 10.1242/dev.125.19.3821. [DOI] [PubMed] [Google Scholar]
- Aspland SE, White RA. Nucleocytoplasmic localisation of extradenticle protein is spatially regulated throughout development in Drosophila. Development. 1997;124:741–747. doi: 10.1242/dev.124.3.741. [DOI] [PubMed] [Google Scholar]
- Blank V, Kourilsky P, Israel A. NF-kappa B and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci. 1992;17:135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Briggs LJ, Stein D, Goltz J, Corrigan VC, Efthymiadis A, Hubner S, Jans DA. The cAMP-dependent protein kinase site (Ser312) enhances dorsal nuclear import through facilitating nuclear localization sequence/importin interaction. J Biol Chem. 1998;273:22745–22752. doi: 10.1074/jbc.273.35.22745. [DOI] [PubMed] [Google Scholar]
- Burglin TR, Ruvkun G. New motif in PBX genes. Nat Genet. 1992;1:319–320. doi: 10.1038/ng0892-319. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KW, Blitz IL. BMPs, Smads and metalloproteases: Extracellular and intracellular modes of negative regulation. Curr Opin Genet Dev. 1998;8:443–449. doi: 10.1016/s0959-437x(98)80116-0. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Gillespie SK, Wasserman SA. Dorsal, a Drosophila Rel-like protein, is phosphorylated upon activation of the transmembrane protein Toll. Mol Cell Biol. 1994;14:3559–3568. doi: 10.1128/mcb.14.6.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Crespo S, Morata G. Genetic evidence for the subdivision of the arthropod limb into coxopodite and telopodite. Development. 1996;122:3921–3928. doi: 10.1242/dev.122.12.3921. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Crespo S, Abu-Shaar M, Torres M, Martínez-Arias C, Mann RS, Morata G. Antagonism between extradenticle function and Hedgehog signalling in the development limb. Nature. 1998;394:196–200. doi: 10.1038/28197. [DOI] [PubMed] [Google Scholar]
- Hoey T, Schindler U. STAT structure and function in signaling. Curr Opin Genet Dev. 1998;8:582–587. doi: 10.1016/s0959-437x(98)80015-4. [DOI] [PubMed] [Google Scholar]
- Itoh S, Landstrom M, Hermansson A, Itoh F, Heldin CH, Heldin NE, ten Dijke P. Transforming growth factor beta1 induces nuclear export of inhibitory Smad7. J Biol Chem. 1998;273:29195–29201. doi: 10.1074/jbc.273.44.29195. [DOI] [PubMed] [Google Scholar]
- Jans DA, Ackermann MJ, Bischoff JR, Beach DH, Peters R. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV- 40 T antigen proteins. J Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- Kamps MP, Murre C, Sun X-h, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1:19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- Knoepfler P, Calvo K, Chen H, Antonarakis S, Kamps M. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci. 1997;94:14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- Kurant E, Pai C-Y, Sharf R, Halachmi N, Sun Y, Salzberg A. dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- Li J, Meyer AN, Donoghue DJ. Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc Natl Acad Sci. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R, Abu-Shaar M. Nuclear import of the homeodomain protein Extradenticle in response to Decapentaplegic and Wingless signalling. Nature. 1996;383:630–633. doi: 10.1038/383630a0. [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- Nourse J, Mellentin J, Galili N, Wilkinson J, Stanbridge E, Smith S, Cleary M. Chromosomal translocation t(1:19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Otero GC, Harris ME, Donello JE, Hope TJ. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C-Y, Kuo T, Jaw T, Kurant E, Chen C, Bessarab D, Salzberg A, Sun Y. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes & Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes & Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- Puri PL, Cimino L, Fulco M, Zimmerman C, La Thangue NB, Giordano A, Graessmann A, Levrero M. Regulation of E2F4 mitogenic activity during terminal differentiation by its heterodimerization partners for nuclear translocation. Cancer Res. 1998;58:1325–1331. [PubMed] [Google Scholar]
- Rauskolb C, Peifer M, Wieschaus E. extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx1. Cell. 1993;74:1–20. doi: 10.1016/0092-8674(93)90731-5. [DOI] [PubMed] [Google Scholar]
- Riddihough G, Ish-Horowicz D. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes & Dev. 1991;5:840–854. doi: 10.1101/gad.5.5.840. [DOI] [PubMed] [Google Scholar]
- Rieckhof G, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of Extradenticle requires homothorax, which encodes an Extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Rihs HP, Jans DA, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 1991;10:633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki F, Price ER, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, Hope TJ. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Clipstone NA, Ho SN, Northrop JP, Crabtree GR. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Proximodistal axis formation in the Drosophila leg: Subdivision into proximal and distal domains by Homothorax and Distal-less. Development. 1999;126:109–117. doi: 10.1242/dev.126.1.109. [DOI] [PubMed] [Google Scholar]