Genotype Probabilities at Intermediate Generations in the Construction of Recombinant Inbred Lines (original) (raw)
Abstract
The mouse Collaborative Cross (CC) is a panel of eight-way recombinant inbred lines: eight diverse parental strains are intermated, followed by repeated sibling mating, many times in parallel, to create a new set of inbred lines whose genomes are random mosaics of the genomes of the original eight strains. Many generations are required to reach inbreeding, and so a number of investigators have sought to make use of phenotype and genotype data on mice from intermediate generations during the formation of the CC lines (so-called pre-CC mice). The development of a hidden Markov model for genotype reconstruction in such pre-CC mice, on the basis of incompletely informative genetic markers (such as single-nucleotide polymorphisms), formally requires the two-locus genotype probabilities at an arbitrary generation along the path to inbreeding. In this article, I describe my efforts to calculate such probabilities. While closed-form solutions for the two-locus genotype probabilities could not be derived, I provide a prescription for calculating such probabilities numerically. In addition, I present a number of useful quantities, including single-locus genotype probabilities, two-locus haplotype probabilities, and the fixation probability and map expansion at each generation along the course to inbreeding.
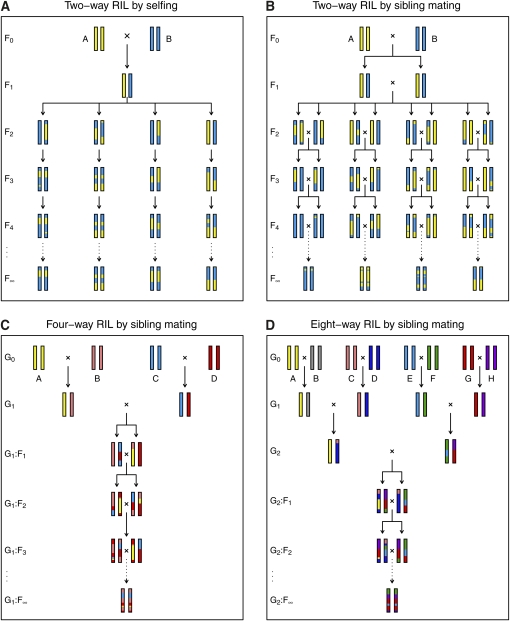
THE mouse Collaborative Cross (CC) is a panel of eight-way recombinant inbred lines (RIL): eight diverse parental strains are intermated, followed by repeated sibling mating, many times in parallel (see Figure 1D), to create a new set of inbred lines whose genomes are random mosaics of the genomes of the original eight strains (Complex Trait Consortium 2004; Collaborative Cross Consortium 2012). There are similar efforts for Drosophila (Macdonald and Long 2007) and Arabidopsis (Kover et al. 2009); the panels will serve as important reference populations for the systemic genetic analysis of complex traits.
Figure 1 .
(A–D) The generation of two-way RIL by selfing (A), two-way RIL by sibling mating (B), four-way RIL by sibling mating (C), and eight-way RIL by sibling mating (D). A single autosome is shown. In A and B, the generation of multiple RIL in parallel is shown, while C and D illustrate the generation of a single RIL.
Many generations are required for inbreeding of such RIL, and so a number of investigators have sought to make use of phenotype and genotype data on mice from intermediate generations during the formation of the CC lines: the pre-CC mice (e.g., see Aylor et al. 2011). The mapping of quantitative trait loci (QTL) with data on pre-CC mice, whether by interval mapping (Lander and Botstein 1989) or Haley–Knott regression (Haley and Knott 1992), requires the calculation of conditional genotype probabilities given incompletely informative marker data (e.g., at single-nucleotide polymorphisms). Such probabilities are generally derived using a hidden Markov model (HMM). The construction of an HMM for pre-CC mice formally requires the calculation of two-locus diplotype probabilities at arbitrary generations along the course to inbreeding. Thus, I sought to calculate single-locus genotype probabilities and two-locus diplotype probabilities at generation G2 : F_k_ (see Figure 1D), with the latter being a function of the recombination fraction between the two loci.
Previous work on genotype probabilities in RIL has focused largely on the final lines (Haldane and Waddington 1931; Broman 2005; Teuscher and Broman 2007), although Haldane and Waddington (1931) did calculate a portion of the probabilities for intermediate generations in two-way RIL by selfing. More recently, Johannes and Colomé-Tatché (2011) fully derived the two-locus genotype probabilities for two-way RIL by selfing and described numerical calculations for the autosome in two-way RIL by sibling mating.
Here I extend these results to the case of four- and eight-way RIL by selfing and sibling mating, including consideration of the X chromosome. The basic problem is to calculate the k_-step probabilities of a Markov chain with many states. While I was not able to obtain closed-form solutions for the two-locus diplotype probabilities at F_k in RIL by sibling mating, I do provide recipes for calculating the probabilities numerically. And I was able to obtain closed-form solutions for single-locus genotype probabilities and two-locus haplotype probabilities. Further, I derived the fixation probability and map expansion at F_k_. These latter results have important applications: the single-locus genotype probabilities could be useful in efforts to identify regions under selection, through the comparison of observed to expected genotype frequencies; the fixation probability can be interpreted as the expected proportion of the genome that is fixed; and the map expansion results indicate the accumulation of recombination events over generations.
Methods
The generation of two-way RIL by selfing and of two-, four-, and eight-way RIL by sibling mating is shown in Figure 1. The notation for generation numbers for RIL can be confusing. The numbering indicated in Figure 1 is used throughout, with F1 being the first generation in which all parental alleles are present in a single individual. In the following, I abbreviate G1 : F_k_ in four-way RIL and G2 : F_k_ in eight-way RIL as simply F_k_. In particular, G1 in four-way RIL and G2 in eight-way RIL is called F0.
Consider a particular crossing strategy, and let Xk denote the parental type at generation F_k_. For RIL by selfing, this is the diplotype of the individual; for RIL by sibling mating, this is the pair of diplotypes for the two siblings. For example, in considering two loci in four-way RIL by sibling mating, one possible state is the starting state at F0, AA | BB × CC | DD. (In this notation, the pairs of letters on each side of the vertical bar denote the two haplotypes for an individual; the first and second letters in each haplotype correspond to the alleles at the first and second loci, respectively.) The sequence _X_0, _X_1, _X_2, … , forms a Markov chain. That is, Xk+1 is conditionally independent of _X_0, _X_1, … , _Xk_−1, given Xk.
Let P denote the transition matrix of the Markov chain, defined by Pij = Pr(Xk+1 = j | Xk = i). Our goal is to calculate the k_-step probabilities, π_k = π0_Pk_, where π0 is the starting distribution (at F0), which contains 1 at the fixed starting state and 0 for all other states.
First note that, for RIL by selfing, it is sufficient to consider two-way RIL, and for RIL by sibling mating, it is sufficient to consider four-way RIL. This is due to the bottleneck with two chromosomes in RIL by selfing at generation F1 and with four chromosomes in RIL by sibling mating at generation F0. The results may be extended from two-way RIL by selfing to four-way RIL by selfing or from four-way RIL by sibling mating to eight-way RIL by sibling mating, by considering an additional generation of recombination. One may obtain the results for two-way RIL by sibling mating from the results for four-way RIL by sibling mating by collapsing states: let A ≡ B and C ≡ D.
The major technique for deriving the k_-step probabilities, π_k, is to derive the eigen decomposition of the transition matrix: P = _V_Λ_V_−1, where Λ is the diagonal matrix of eigenvalues and V is a matrix whose columns are the corresponding eigenvectors. Then Pk = V_Λ_kV_−1, and Λ_k is obtained from Λ by taking the _k_th powers of the eigenvalues.
Such an eigen decomposition is straightforward in theory but is unwieldy in practice, due to the extremely large number of possible states. And so the second major technique is to take account of various symmetries to collapse the states into a smaller number. For two-way RIL by selfing with two loci, the simplest formulation would give 24 = 16 possible states (two possible alleles at each locus on each of the two chromosomes). But considering that the order of the two haplotypes is immaterial, these may be reduced to 10 possible diplotypes. As shown in Haldane and Waddington (1931), these may be further reduced to just five states, by taking account of two additional symmetries: the order of the two loci may be ignored, and the symbols A and B may be switched.
Let us formalize this idea. (For a more rigorous approach, see Burke and Rosenblatt 1958.) Let the possible states of the chain be S = {_s_1, … , sn}. Partition S into m subsets of equivalent states, Si ⊂ S, so that, for any pair i and j, Pr(Xk+1 ∈ Sj | Xk = s) = qij for all s ∈ Si. The qij form an m × m transition matrix, Q, for the collapsed states. Let Z denote the n × m incidence matrix defined by zij = 1 if si ∈ Sj and 0 otherwise. Then
and so PkZ = ZQk. As a result, π_kZ_ = π0_PkZ_ = π0_ZQk_. Thus, one may work with the m × m transition matrix Q in place of the n × n transition matrix P.
For this collapse of states to be useful, the multiple states within each equivalence class, Si, need to have equal probabilities at each generation, so that the probabilities of the individual states may be derived from the probabilities of the collapsed states. This will depend on the starting distribution. For example, consider one locus in two-way RIL by sibling mating. If the starting state is AA × BB, then at any future generation, the chance of being in state AA × AB is the same as that of being in state AB × BB. However, if the starting state is AA × AB, then there will be a lack of symmetry between A and B. (For the asymmetric case of two-way RIL initiated from a backcross, see Johannes and Colomé-Tatché 2011.)
Kimura (1963) described a further technique that has been critical in this work. In many instances, we do not need the full distribution π_k_, but only various linear combinations, say π_kz_ = π0_Pkz_, where z is an n × 1 vector. Kimura (1963) demonstrated how to expand z to an n × m matrix Z in such a way that there exists a matrix Q satisfying Equation 1. Then we again have π_kZ_ = π0_PkZ_ = π0_ZQk_ and may work with the m × m matrix Q in place of the n × n matrix P. Here, the matrix Q is not a transition matrix but simply defines a recursion. The first element of π_kZ_ is the target quantity, π_kz_.
Consider, for example, the probability of a random two-locus haplotype drawn from generation F_k_ in the formation of RIL by sibling mating. Let Ck(AA) denote that chance that AA is drawn. This could either be an intact haplotype, transmitted without recombination from generation F_k_−1, or be the result of recombination between the two haplotypes in a random F_k_–1 individual. Consider drawing a single random allele at the first locus from generation k and then taking the allele at the second locus but on the opposite chromosome in that individual. Let Sk(AA) denote the probability that these two alleles are both A. Then Ck(AA) = (1 − r)_Ck_−1(AA) + _rSk_−1(AA), where r is the recombination fraction between the two loci. Further, Sk(AA) = Tk_−1(AA), where Tk(AA) is the chance that, if one draws a random allele at the first locus from generation F_k and then a random allele from the opposite individual at the second locus, both alleles are A. We may further write Tk(AA) as a function of _Ck_−1, _Sk_−1, and Tk_−1, forming the recursion matrix, Q, which is shown in Supporting Information, Table S1. Moreover, this same recursion applies for all of the other haplotypes; one just needs to use different starting distributions, π0_Z. For the three distinct cases for four-way RIL by sibling mating, these are shown in Table S2.
A particularly useful aspect of Kimura's technique is that the recursion matrix can be constructed by probabilistic arguments, without the need to form the full transition matrix, P, or even the n × m matrix Z. For two autosomal loci in four-way RIL by sibling mating, there are 48 = 65, 536 diplotype pairs without accounting for any symmetries. This may be reduced to 9316 after accounting for the obvious symmetries (exchange the two haplotypes in each individual and exchange the two individuals) and then to 700 diplotype states after accounting for the less obvious symmetries (exchange the two loci, exchange alleles A and B, exchange alleles C and D, and exchange both A for C and B for D). By the technique of Kimura (1963), one may work with a 3 × 3 matrix in place of the 700 × 700 transition matrix, if only haplotype probabilities are desired.
Throughout this work, Maxima (http://maxima.sourceforge.net) was used for symbolic algebra, and R (R Development Core Team 2010) and Perl (Wall et al. 2000) were used for additional verifications of the results.
Results
Two-way RIL by selfing
As noted in the previous section, it is sufficient to consider two-way RIL by selfing, due to the bottleneck with two chromosomes at F1. Let us jump directly to two-locus diplotype probabilities, as the results are fairly simply obtained. As noted in Haldane and Waddington (1931) and in the previous section, if one takes account of the various symmetries, one may collapse the two-locus diplotype states to a Markov chain with five states. The transition matrix of this chain is shown in Table S3, with r denoting the recombination fraction between the two loci.
I obtained the eigen decomposition of this transition matrix (not shown) and, noting that the starting state (at generation F1) is AA | BB, derived the two-locus diplotype probabilities at generation F_k_ (that is, after k − 1 steps), π_k_−1 = π0_Pk_−1 = π0_V_Λ_k_−1_V_−1, presented in Table 1. For each group of states, a single prototype and the number of states in that group are provided. For example, in the third row, with prototype AA | AB, the cited probability is for that prototype as well as each of the other three states in that group: AA | BA, BB | AB, and BB | BA.
Table 1 .
Two-locus diplotype probabilities at generation F_k_ (for k ≥ 1) in the formation of two-way RIL by selfing
| Prototype | No. states | Probability of each |
|---|---|---|
| AA | AA | 2 | 12(1+2r)−(12)k+1[2−(1−2r+2r2)k−1+(1−2r)k1+2r] |
| AB | AB | 2 | r1+2r−(12)k+1[2−(1−2r+2r2)k−1− (1−2r)k1+2r] |
| AA | AB | 4 | (12)k[1−(1−2r+2r2)k−1] |
| AA | BB | 1 | (12)k[(1−2r+2r2)k−1+(1−2r)k−1] |
| AB | BA | 1 | (12)k[(1−2r+2r2)k−1−(1−2r)k−1] |
Haldane and Waddington (1931) also derived these results for intermediate generations in two-way RIL by selfing. They displayed just the two cases AA | AA and AB | AB (Haldane and Waddington 1931, equation 1.4), but the results match those in Table 1, though note that their results are a factor of 2 larger, as they concern the combined states. Also note that Haldane and Waddington (1931) allowed a sex difference in the recombination fraction, whereas I assume no sex difference in recombination.
Four-way RIL by sibling mating, one locus
Autosome:
For a single autosomal locus in four-way RIL by sibling mating, there are 55 genotype pairs, after accounting for the obvious symmetries. These may be reduced to 13 states, after accounting for the less obvious symmetries. The transition matrix for this reduced Markov chain is shown in Table S4. The starting state at generation F0 is AB × CD. I calculated the eigen decomposition of the transition matrix, and from that π_k_ = π0_Pk_ = π0_V_Λ_kV_−1. The results are shown in Table S5, which also indicates the number of genotype pairs corresponding to each of the 13 states. (The sum of the second column in Table S5 is 55.)
The probabilities of single-locus genotype pairs at generation F_k_, shown in Table S5, are complex and not of particular interest in themselves (hence their inclusion in the Supporting Information). However, the single-locus probabilities for a random F_k_ individual follow immediately from these results; they are shown in Table 2. These are of considerably greater interest, as they constitute the “initiation” probabilities for an HMM. The single-locus genotype probabilities for the first several generations are plotted in Figure S1A.
Table 2 .
Single-locus genotype probabilities at generation F_k_ in the formation of four-way RIL by sibling mating
| Chromosome | Individual | Prototype | No. states | Probability of each |
|---|---|---|---|---|
| A | Random | AA | 4 | 14−(5+3540)(1+54)k−(5−3540)(1−54)k |
| AB | 2 | (5−520)(1+54)k+(5+520)(1−54)k | ||
| AC | 4 | 510[(1+54)k− (1−54)k] | ||
| X | Female | AA | 2 | 13+16(−12)k−(5+520)(1+54)k−(5−520)(1−54)k |
| AB | 1 | (5−510)(1+54)k+ (5+510)(1−54)k | ||
| AC | 2 | 55[(1+54)k−(1−54)k] | ||
| CC | 1 | 13+16(−12)k−1− (5+520)(1+54)k−1− (5−520)(1−54)k−1 | ||
| X | Male | A | 2 | 13[1−(−12)k] |
| C | 1 | 13[1+2(−12)k] |
X chromosome:
In considering the X chromosome, it is important to consider the order of the initial crosses. In four-way RIL by sibling mating, I assume that a female A was crossed to a male B, and a female C was crossed to a male D, and then a female from the A × B F1 was crossed to a male from the C × D F1 (see Figure S2B). In the F1, there are three X chromosomes, A, B, and C; the D allele is lost.
After accounting for the obvious symmetries, there are 18 possible single-locus genotype pairs. These may be reduced to 10 states, after accounting for the less obvious symmetries. The transition matrix for this reduced Markov chain is shown in Table S6. The starting state at generation F0 is AB × C. Through the eigen decomposition of the transition matrix, the results in Table S7 are obtained.
The marginal probabilities for the female and male are displayed in Table 2 and are plotted in Figure S1, B and C. The oscillations in the male X chromosome probabilities are interesting, but not particularly surprising.
Fixation probabilities:
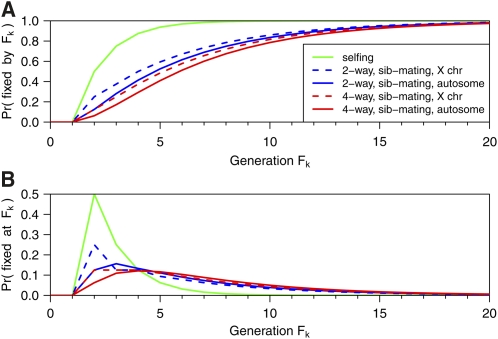
The detailed single-locus results in Table S5 (for autosomes) and Table S7 (for the X chromosome) immediately provide the fixation probability in four-way RIL by sibling mating. For an arbitrary autosomal locus, the chance that a four-way RIL has been fixed at or before generation F_k_ is 4 · Pr(AA × AA). For an arbitrary X chromosome locus, the chance of fixation by generation F_k_ is 2 · Pr(AA × A) + Pr(CC × C). These are shown in Table 3 and are further plotted in Figure 2A. Note that the probability that an arbitrary locus in four-way RIL has become fixed at exactly generation F_k_ may be derived as the difference between the results for k and k − 1. These are shown in Figure 2B. Note that the fixation probabilities for eight-way RIL are identical to those for four-way RIL.
Table 3 .
Fixation probability at generation F_k_ in the formation of RIL
| Cross | Chromosome | Probability of fixation at or before F_k_ |
|---|---|---|
| Two-way selfing | 1−(12)k−1 | |
| Four-way sibling mating | A | 1+(12)k−(15)(14)k−(9+4510)(1+54)k−(9−4510)(1−54)k |
| Two-way sibling mating | A | 1+(25)(14)k−(7+3510)(1+54)k−(7−3510)(1−54)k |
| Four-way sibling mating | X | 1+(12)k+1−(15+7520)(1+54)k−(15−7520)(1−54)k |
| Two-way sibling mating | X | 1−(5+3510)(1+54)k−(5−3510)(1−54)k |
Figure 2 .
Fixation probability at generation F_k_ for an arbitrary locus as a function of k. (A) The cumulative probability. (B) The probability of fixation precisely at F_k_. The results for RIL by selfing are in green. The results for two-way and four-way RIL by sibling mating are in blue and red, respectively, with the solid curves corresponding to an autosomal locus and the dashed curves corresponding to an X chromosome locus.
The fixation probabilities for two-way RIL by sibling mating are also simply derived: collapse alleles A ≡ B and C ≡ D. Thus, for an autosomal locus, one adds up the rows in Table S5 that contain only A or B.
The fixation probability for an X chromosome locus is slightly larger than that for an autosomal locus, and that for two-way RIL is slightly larger than that for four-way RIL. The fixation probability for two-way RIL by sibling mating at generation k is quite similar to that for four-way RIL at generation k + 1. Fixation in RIL by selfing occurs much more rapidly.
For a large genome, the fixation probabilities for an arbitrary autosomal locus, displayed in Figure 2A, may be interpreted as the approximate proportion of the autosomal genome that will be fixed at generation F_k_. Nevertheless, as shown in Broman (2005) via computer simulation, there will be considerable variation across lines.
Four-way RIL by sibling mating, two-locus haplotypes
Autosome:
The technique of Kimura (1963), described above, may be used to derive probabilities of random two-locus haplotypes drawn from generation F_k_ in the formation of four-way RIL by sibling mating. There are three distinct cases to consider (AA, AB, and AC), which share a common recursion matrix (shown in Table S1) but require consideration of different starting states. For each case, the starting probabilities, π0_Z_, form a 1 × 4 row vector with a single nonzero entry (see Table S2).
I obtained the eigen decomposition of the recursion matrix, Q, which is shown in Table S1, and through the equation π_kZ_ = π0_ZQk_ = π0_ZV_Λ_kV_−1 obtained the two-locus haplotype probabilities in Table 4. The equations for autosomal haplotypes in Table 4 are valid only for r < 12, but the results for r = 12 are obvious by symmetry: Pr(AA) = 14 at F0, Pr(AA) = Pr(AB) = 18 at F1, and Pr(AA) = Pr(AB) = Pr(AC) = 116 at F_k_ for k ≥ 2.
Table 4 .
Two-locus haplotype probabilities at generation F_k_ in the formation of four-way RIL by sibling mating
| Chr. | Individual | Prototype | No. states | Probability of each |
|---|---|---|---|---|
| A | Random | AA | 4 | 14(1+6r)−[6r2−7r−3rs4(1+6r)s](1−2r+s4)k+[6r2−7r+3rs4(1+6r)s](1−2r−s4)k |
| AB | 4 | r2(1+6r)+[10r2−r−rs4(1+6r)s](1−2r+s4)k−[10r2−r+rs4(1+6r)s](1−2r−s4)k | ||
| AC | 8 | r2(1+6r)−[2r2+3r+rs4(1+6r)s](1−2r+s4)k+[2r2+3r−rs4(1+6r)s](1−2r−s4)k | ||
| X | Female | AA | 2 | 13(1+4r)+16(1+r)(−12)k−[4r3−(4r2+3r)t+3r2−5r4(4r2+5r+1)t](1−r+t4)k+[4r3+(4r2+3r)t+3r2−5r4(4r2+5r+1)t](1−r−t4)k |
| AB | 2 | 2r3(1+4r)+r3(1+r)(−12)k+[2r3+6r2−(2r2+r)t2(4r2+5r+1)t](1−r+t4)k−[2r3+6r2+(2r2+r)t2(4r2+5r+1)t](1−r−t4)k | ||
| AC | 4 | 2r3(1+4r)−r6(1+r)(−12)k−[9r2+5r+rt4(4r2+5r+1)t](1−r+t4)k+[9r2+5r−rt4(4r2+5r+1)t](1−r−t4)k | ||
| CC | 1 | 13(1+4r)−13(1+r)(−12)k+[9r2+5r+rt2(4r2+5r+1)t](1−r+t4)k−[9r2+5r−rt2(4r2+5r+1)t](1−r−t4)k | ||
| X | Male | AA | 2 | 13(1+4r)−13(1+r)(−12)k+[r3−(8r3+r2−3r)t−10r2+5r2(4r4−35r3−29r2+15r+5)](1−r+t4)k+[r3+(8r3+r2−3r)t−10r2+5r2(4r4−35r3−29r2+15r+5)](1−r−t4)k |
| AB | 2 | 2r3(1+4r)−2r3(1+r)(−12)k+[r4+(5r3−r)t−10r3+5r24r4−35r3−29r2+15r+5](1−r+t4)k+[r4−(5r3−r)t−10r3+5r24r4−35r3−29r2+15r+5](1−r−t4)k | ||
| AC | 4 | 2r3(1+4r)+r3(1+r)(−12)k−[2r4+(2r3−r2+r)t−19r3+5r2(4r4−35r3−29r2+15r+5)](1−r+t4)k−[2r4−(2r3−r2+r)t−19r3+5r2(4r4−35r3−29r2+15r+5)](1−r−t4)k | ||
| CC | 1 | 13(1+4r)+23(1+r)(−12)k+[2r4+(2r3−r2+r)t−19r3+5r4r4−35r3−29r2+15r+5](1−r+t4)k+[2r4−(2r3−r2+r)t−19r3+5r4r4−35r3−29r2+15r+5](1−r−t4)k |
The autosomal haplotype probabilities as a function of the recombination fraction, r, are displayed in the left panels in Figure S3.
X chromosome:
To calculate the probability of a random two-locus X chromosome haplotype drawn from the female at F_k_ in four-way RIL by sibling mating and of the single X chromosome haplotype in the corresponding male, there are four cases to consider (AA, AB, AC, and CC). Application of the technique of Kimura (1963) for the X chromosome requires consideration of a set of four states, with the recursion matrix shown in Table S8. Each of the four cases uses the same recursion matrix but different starting probabilities (shown in Table S9). Calculation of the probabilities of a random haplotype drawn from the female and of the single haplotype in the male uses the same set of equations, but for a random female haplotype one takes the first element of π_kZ_, while for the male haplotype one takes the third element. Again, following the eigen decomposition of the matrix in Table S8, the haplotype probabilities in Table 4 were obtained.
The female and male X chromosome haplotype probabilities, as a function of the recombination fraction, r, are displayed in the center and right panels, respectively, in Figure S3. The probabilities of haplotypes AA and CC in females, and all of the haplotype probabilities in males, show pronounced oscillations across generations. For example, the CC haplotype is common in males for even k and common in females for odd k and vice versa for haplotype AA.
Map expansion:
The multiple generations of recombination in the formation of RIL lead to genetic map expansion. The map expansion as a function of generation is easily obtained from the haplotype probabilities. Let R denote the probability of a recombinant haplotype. [For the autosome, take 1 − 4 · Pr(AA).] The map expansion relative to a single meiosis is (dR / dr)|r=0 (see Teuscher and Broman 2007). For the X chromosome, I calculated the map expansion separately in females and males and then obtained a combined map expansion by averaging the two values, giving the female value weight 23.
The map expansion for the two-way RIL by selfing case may be obtained from the values in Table 1. To obtain the map expansion for two-way RIL by sibling mating, equate alleles A ≡ B and C ≡ D; the recombinant haplotypes correspond to the AC case in Table 4. To obtain the map expansion for eight-way RIL by sibling mating, note that the chance of each nonrecombinant haplotype would be (1 − r)/2 times the probability for haplotype AA shown in Table 4. A similar calculation applies for four-way and eight-way RIL by selfing.
The map expansions for two-, four-, eight-, and 2_n_-way RIL by selfing and for the autosome by sibling mating are shown in Table 5 and are further illustrated in Figure 3. The results for the X chromosome in RIL by sibling mating are simply two-thirds those for the autosome and so are not shown. (I have verified, via Maxima, that this is true for 2_n_-way RIL up to n = 98. A general proof continues to elude me.)
Table 5 .
Map expansion at generation F_k_ in the formation of RIL
| Selfing | Sibling mating (autosome) | |
|---|---|---|
| Two-way | 2−(12)k−2 | 4−(10+655)(1+54)k−(10−655)(1−54)k |
| Four-way | 3−(12)k−2 | 6−(15+755)(1+54)k−(15−755)(1−54)k |
| Eight-way | 4−(12)k−2 | 7−(15+755)(1+54)k−(15−755)(1−54)k |
| 2_n_-way | n+1−(12)k−2 | n+4−(15+755)(1+54)k−(15−755)(1−54)k |
Figure 3 .
Map expansion at generation F_k_ as a function of k, for two-way (blue), four-way (red), and eight-way (black) RIL by selfing (dashed curves) and by sibling mating (solid curves). The displayed results for RIL by sibling mating are for the autosomes; values for the X chromosome are exactly two-thirds those for the autosomes.
Teuscher et al. (2005) also sought to calculate the map expansion by generation for two-way RIL by sibling mating. My results match those of Teuscher et al. (2005), were more easily obtained, and provide a closed-form solution.
Four-way RIL by sibling mating, two-locus diplotypes
Autosome:
I now turn to the calculation of the distribution of the two-locus diplotype on an autosome, for a random individual drawn from generation F_k_ in the formation of four-way RIL by sibling mating. There are 18 cases falling into three groups: diplotypes of the form AA | AA, with both loci being homozygous; AA | AB, with one locus being homozygous; and AA | BB, with both loci being heterozygous.
Let us start with the AA | AA case. Following the approach of Kimura (1963), I obtained a 13 × 13 recursion matrix, Q, whose transpose is shown in Table S10. Because of the size and sparsity of the matrix, only the nonzero elements are indicated. The starting states for the three related diplotypes are shown in Table S11. (For each diplotype pattern, the starting distribution π0_Z_ has a single nonzero entry.)
The next step is to derive the eigen decomposition of Q. However, while 7 of the 13 eigenvalues can be obtained, the other 6 eigenvalues are the roots of a sixth degree polynomial (whose coefficients are polynomials of r of degree up to 5). This prevented the calculation of the diplotype probabilities at F_k_.
Nevertheless, the reduction of states represented in Table S10 and Table S11 is considerable and is useful for the numeric calculation of these probabilities. Direct calculation would require consideration of the full transition matrix for pairs of diplotypes, which, even after accounting for all possible symmetries, is a 700 × 700 matrix.
Turning to the AA | AB case, with one locus being homozygous and the other heterozygous, the recursion matrix is 17 × 17; the nonzero elements of its transpose are shown in Table S12. The starting states for the four related diplotype patterns are shown in Table S13.
Finally, for the AA | BB case, with both loci being heterozygous, the recursion matrix is 14 × 14; its transpose is shown in Table S14. The starting states for the 11 related diplotype patterns are shown in Table S15.
X chromosome:
It should not be surprising that closed-form solutions for the two-locus diplotype probabilities for the female on the X chromosome at generation F_k_ in the formation of four-way RIL by sibling mating could not be derived. (But note that the two-locus haplotype probabilities for the male X chromosome could be calculated; see Table 5.)
Nevertheless, the recursion matrices and starting states, derived by the approach of Kimura (1963), may be useful for numerical computations. After consideration of the various symmetries, there are 17 two-locus diplotype patterns falling into the same three groups as for the autosome.
For the AA | AA case, the recursion matrix is 13 × 13; its transpose is shown in Table S16. The starting states for the four related diplotype patterns are shown in Table S17. Six of the 13 eigenvalues may be derived; the other 7 are roots of a pair of polynomials, one of degree 3 and the other of degree 4.
For the AA | AB case, the recursion matrix is 18 × 18; its transpose is shown in Table S18. The starting states for the five related diplotype patterns are shown in Table S19. For the AA | BB case, the recursion matrix is 12 × 12; its transpose is shown in Table S20. The starting states for the eight related diplotype patterns are shown in Table S21.
Eight-way RIL
The calculation of genotype or diplotype probabilities for eight-way RIL from those for four-way RIL is straightforward, but also tedious and potentially confusing. For clarity, lowercase letters are used for the alleles in eight-way RIL, while uppercase letters denote the alleles in four-way RIL.
First, consider the genotype at an autosomal locus for a random individual drawn from generation F_k_ in the construction of eight-way RIL by sibling mating. Due to the bottleneck at generation G2, the genotypes ab, cd, ef, and gh are not possible. (At any one locus, only one allele from each of these pairs will be transmitted from G1 to G2.) The probability of genotype aa is the chance that the G2 individual receives the allele a (which is 12) times the probability for the genotype AA in the construction of four-way RIL by sibling mating. The probabilities of genotypes ac and ae are 14 the probabilities of genotypes AB and AC, respectively, in the construction of four-way RIL by sibling mating.
Two-locus haplotype probabilities are obtained in a similar way, noting that the haplotype in the position of the A haplotype at G2 in the construction of four-way RIL is aa or bb with probability (1 − r)/2 each and is ab or ba with probability r/2 each. Thus, for example, the chance of obtaining ab as a two-locus autosomal haplotype, drawn at random from generation F_k_ in the construction of eight-way RIL by sibling mating, is r/2 times the probability of drawing AA from generation F_k_ in the construction of four-way RIL by sibling mating. The chance of drawing the haplotype cg is 14 times the probability of drawing BD from the corresponding generation in the construction of four-way RIL.
Table S22 contains a complete prescription for the calculation of two-locus autosomal diplotype probabilities for intermediate generations in the construction of eight-way RIL, on the basis of the corresponding probabilities for four-way RIL. The first column contains the possible diplotype patterns. The second column contains the numbers of diplotype states corresponding to each pattern. To calculate the probability of the pattern in the first column, for an eight-way cross, take the corresponding probability for the pattern in the third column, for a four-way cross, multiplied by the value in the fourth column.
Table S23 contains a similar prescription for calculating probabilities of the two-locus X chromosome diplotype in the female at intermediate generations in the construction of eight-way RIL. A key feature to note is that, at a single X chromosome locus at generation G2, the female is either ac or bc, while the male is hemizygous e or f (see Figure S2C).
Discussion
I sought to calculate the two-locus diplotype probabilities for a random individual drawn from generation G2 : F_k_ in the formation of eight-way RIL by sibling mating, as these could form the basis for an HMM for reconstructing the genotype probabilities in pre-CC individuals given incompletely informative marker data. While I was not able to obtain closed-form solutions for these probabilities, the results in Table S10, Table S11, Table S12, Table S13, Table S14, Table S15, Table S16, Table S17, Table S18, Table S19, Table S20, and Table S21 provide a recipe for numerical calculations.
A more careful reading of Haldane and Waddington (1931) would have indicated that closed-form solutions for these probabilities would not be possible. As they state (Haldane and Waddington 1931, p. 367), regarding the calculation of related probabilities for two-way RIL by sibling mating, “These equations can, in part at least, be reduced to quartics, but at least one quartic is irreducible. Hence only numerical calculation is practicable.”
Moreover, Liu et al. (2010) described a general HMM for the treatment of complex pedigrees with inbreeding, appropriate for pre-CC individuals, that does not require the explicit derivation of these two-locus probabilities. Further, with the high-density genotype data available on the pre-CC mice (Aylor et al. 2011), a relatively simple HMM, such as that in HAPPY (Mott et al. 2000), which does not take formal account of the varying recombination patterns as a function of cross direction or generation, is likely sufficient for genotype reconstruction.
Nevertheless, an HMM making use of these calculations, as well as functions for simulating partially inbred lines, will be implemented in a future version of R/qtl (Broman et al. 2003). With this implementation, we will be able to assess the relative advantages of such a specially tailored HMM over both the more general approach of Liu et al. (2010) and the simpler approach of Mott et al. (2000).
In practice, the genetic analysis of RIL generally proceeds prior to full inbreeding, with calculations based on the haplotype frequencies at fixation, and with remaining heterozygous genotypes often omitted and treated as missing. The calculations herein might be used to deal with residual heterozygosity, but it is unlikely that it will give much improvement in the analysis. After only a few generations, the haplotype probabilities are quite close to the values at fixation. Moreover, the consideration of heterozygotes can lead to problems in the QTL analysis if the frequency of heterozygotes is low, although this can be alleviated by an assumption of additive allele effects at a QTL. Finally, the remaining regions of heterozygosity in RIL may have survived due to selection, whereas these calculations rely on assumptions of no selection or mutation and so are not appropriate to capture such phenomena.
I derived closed-form solutions for a number of quantities that are of considerable interest. The single-locus genotype probabilities (Table 2) could be useful in efforts to identify regions under selection, through the comparison of observed to expected genotype frequencies. The fixation probability (Table 3 and Figure 2) can be interpreted as the expected proportion of the genome that is fixed. The map expansion results (Table 5 and Figure 3) indicate the accumulation of recombination events over generations, which can be valuable for study design: If an investigator intervenes at an intermediate generation to speed up the process toward inbreeding, what proportion of the final recombination breakpoints might be lost, and so to what extent might mapping precision be eroded?
The single-locus genotype probabilities (Table 2) were derived by brute force, using the full transition matrix for the pair of genotypes from generation to generation. The approach of Kimura (1963) could also be used in these cases, to provide considerable simplification. For example, in the single-locus autosome case, one may use a 3 × 3 recursion matrix in place of the 13 × 13 matrix in Table S4.
There is also a simpler way to calculate the fixation probabilities for four-way RIL by sibling mating. One may consider a single locus in a two-way RIL, but starting at a different state (e.g., start at AB × BB, to calculate the chance that a four-way RIL is fixed at allele A). This requires the consideration of a 6 × 6 transition matrix.
There is an interesting connection between this work and the Fibonacci sequence {0, 1, 1, 2, 3, 5, 8, …}, defined by the recursive formula xk = _xk_−1 + _xk_−2, with starting values _x_0 = 0 and _x_1 = 1 (see Graham et al. 1994, Section 6.6). To obtain a closed-form solution for xk, one may write the recursion in matrix form and apply the same techniques used herein to obtain xk=[φk−(1−φ)k]/5, where φ is the “golden ratio”, (1+5)/2. Note that φ/2=(1+5)/4 appears numerous times in the results. (When I first derived the single-locus probabilities in Table 2, I was confused about why the results involve 5, when the numbers are all clearly rational.)
The great effort expended here to derive symbolic results raises the question of the relative merits of computer simulations, numeric calculations, and symbolic calculations. Simulations are most flexible and are generally simpler to obtain, but lack precision. Numeric calculations can be precise, but can be computationally intensive. Symbolic results are more general than numeric calculations, can enable quicker calculations in software, and have the potential to provide more clear insight. Ultimately, the effort toward symbolic results largely serves to satisfy a personal compulsion.
Acknowledgments
I thank James Crow, Bret Larget, and Timo Seppäläinen for valuable discussions and Maria Colomé-Tatché, Frank Johannes, Lauren McIntyre, Tracey DePellegrin Connelly, and two anonymous reviewers for comments on the manuscript. This work was supported in part by National Institutes of Health grant GM074244.
Footnotes
Edited by Lauren M. McIntyre, Dirk-Jan de Koning, and 4 dedicated Associate Editors
Literature Cited
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., et al. , 2011. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21: 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Burke C. J., Rosenblatt M., 1958. A Markovian function of a Markov chain. Ann. Math. Stat. 29: 1112–1122 [Google Scholar]
- Collaborative Cross Consortium, 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Complex Trait Consortium, 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Graham R.L., Knuth D.E., Patashnik O., 1994 Concrete Mathematics, Ed. 2. Addison-Wesley, Reading, MA/Menlo Park, CA
- Haldane J. B. S., Waddington C. H., 1931. Inbreeding and linkage. Genetics 16: 357–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Johannes F., Colomé-Tatché M., 2011. Quantitative epigenetics through epigenomic pertubation of isogenic lines. Genetics 188: 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1963. A probability method for treating inbreeding systems, especially with linked genes. Biometrics 19: 1–17 [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Zhang Q., McMillan L., de Villena F. P., Wang W., 2010. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics 26: i199–i207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald S. J., Long A. D., 2007. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176: 1261–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2010 R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna
- Teuscher F., Broman K. W., 2007. Haplotype probabilities for multiple-strain recombinant inbred lines. Genetics 175: 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher F., Guiard V., Rudolph P. E., Brockmann G. A., 2005. The map expansion obtained with recombinant inbred strains and intermated recombinant inbred populations for finite generation designs. Genetics 170: 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall L., Christiansen T., Orwant J., 2000 Programming Perl, Ed. 3. O'Reilly Media, Sebastopol, CA