Metagenomic Analysis of the Viral Flora of Pine Marten and European Badger Feces (original) (raw)
Abstract
A thorough understanding of the diversity of viruses in wildlife provides epidemiological baseline information about potential pathogens. Metagenomic analysis of the enteric viral flora revealed a new anellovirus and bocavirus species in pine martens and a new circovirus-like virus and geminivirus-related DNA virus in European badgers. In addition, sequences with homology to viruses from the families _Paramyxo_- and Picornaviridae were detected.
TEXT
Emerging and (re)emerging viruses pose major threats not only to public health but also to the food supply, economy, and environment (19, 38, 40, 41). Animals, and particularly wild animals, are thought to be the source of the majority of all emerging infections (19, 38, 40, 41). Virus surveillance in wild animals is generally confined to pathogens with known impact, explaining how viruses such as severe acute respiratory syndrome (SARS) coronavirus and the pandemic H1N1 influenza A virus in 2009 escaped detection prior to causing epidemics in humans (12, 14, 15). A thorough understanding of virus diversity in wild animals provides epidemiological baseline information about potential pathogens and may lead to the identification of newly emerging human pathogens in the future. In this study, we used next-generation sequencing to gain insight in the fecal viral populations from wild pine martens (Martes martes) and European badgers (Meles meles) in The Netherlands (Table 1), as was previously performed for other wildlife species such as California sea lions and bats (11, 22, 23).
Table 1.
Viral sequences identified in pine marten and European badger rectal swabs
| Host and viral sequence sample | Animal | No. of reads | Virus [no. of reads_b_] | Length/identity_c_ | |
|---|---|---|---|---|---|
| Sex_a_ | Wt (g) | ||||
| Martes martes | |||||
| VS4700001 | M | 1,650 | 5,246 | _Myo_-, _Podo_-, Siphoviridae | |
| VS4700002 | M | 1,650 | 4,592 | Myoviridae | |
| Microviridae | |||||
| Parvoviridae (porcine bocavirus [2]) | 212/75 | ||||
| Picornaviridae (porcine kobuvirus [2]) | 527/95 | ||||
| VS4700003 | M | 1,805 | 8,290 | Microviridae (Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 [20]) | 1,182/53 |
| VS4700004 | M | 1,590 | 13,463 | _Myo_-, Siphoviridae | |
| Anelloviridae (torque teno virus [68]) | 4,401/48 | ||||
| Meles meles | |||||
| VS4700005 | F | 800 | 8,000 | Myoviridae | |
| Anelloviridae (torque teno virus [2]) | 544/41 | ||||
| VS4700006 | M | 4,820 | 3,810 | Myoviridae | |
| Microviridae | |||||
| Reoviridae (Bombyx mori cypovirus [30]) | 2,668/49 | ||||
| Circoviridae (columbid circovirus [4]) | 781/49 | ||||
| VS4700007 | F | 6,650 | 8652 | Paramyxoviridae (canine distemper virus [7]) | 1,039/97 |
| (Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 [2]) | 230/47 |
Large-scale molecular virus screening, based on host nucleic acid depletion, viral nucleic acid isolation, sequence-independent amplification, and next-generation sequencing with a 454 GS Junior instrument (Roche) was performed as described previously and by the manufacturer (1, 2, 35, 36, 39) on four rectal swabs from pine martens and three rectal swabs from European badgers. Rectal swabs were centrifuged at 10,000 × g for 3 min, and the supernatant was filtered through a 0.45-μm filter (Millipore). The viral-particle-containing filtrates were digested with a mixture of DNases and RNases (39). Viral RNA and DNA were extracted using the Nucleospin RNA XS kit (Machery-Nagel) and High Pure Viral Nucleic Acid kit (Roche). First- and second-strand syntheses and random PCR amplification were performed as described previously (39). Random PCR products from the RNA and DNA fractions were pooled and purified using the MinElute PCR purification kit (Qiagen). The resulting purified product was prepared for sequencing by use of a GS FLX Titanium library preparation kit (454 Life Science, Roche), and the library of DNA fragments was sequenced on a 454 GS Junior instrument (454 Life Science, Roche). The pyrosequencing reads were sorted into their rectal samples of origin according to their unique sequence tag added by using the GS FLX Titanium Rapid Library MID Adaptors kit (454 Life Science, Roche). Adaptor and primer sequences were trimmed from each read, and more than 52,000 trimmed reads were assembled using de novo assembly in CLC Genomics Workbench 4.5.1 (CLC Bio [24]) and analyzed according to nucleotide (contigs and singletons) and translated nucleotide BLAST searches (contigs) (3). Sequences were classified into eukaryotic viruses, phages, bacteria, and eukaryotes based on the taxonomic origin of the best-hit sequence using MEGAN 4.40 (16, 17). An E value of 0.001 was used as the cutoff value of significant virus hits.
Virome overview.
Most of the identified sequences were of eukaryotic or bacterial origin. All seven samples showed evidence for the presence of bacteriophages from the order Caudovirales and/or family Microviridae (Table 1). In pine marten rectal swabs, eukaryotic viruses with homology to kobuvirus from the Picornaviridae family, bocavirus from the Parvoviridae family, torque teno virus from the Anelloviridae family, and Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 (SSHADV-1) from the _Geminiviridae-_like family were detected (Table 1). In European badgers, eukaryotic viruses with homology to Bombyx mori cypovirus from the Reoviridae family, columbid circovirus from the Circoviridae family, canine distemper virus from the Paramyxoviridae family, SSHADV-1 from the _Geminiviridae_-like family, and torque teno virus from the Anelloviridae family were observed (Table 1). Some of the eukaryotic viruses showed a high identity to known viruses. For example, the obtained canine distemper virus sequences from the L, H, and F genome segments were ∼97% identical to known canine distemper viruses on the amino acid level (Table 1), and this virus is known to cause disease with respiratory, enteric, and neurological manifestations with a high fatality rate in terrestrial carnivores (5, 10). The obtained kobuvirus reads were >95% identical to known porcine kobuviruses on the amino acid level (Table 1), and it is also known that picornaviruses are found in the enteric tract. Samples VS4700002, VS4700004, and VS4700006 were interesting, as sequences with low homology on the protein level to known viruses, bocavirus, torque teno virus (TTV), and circovirus, were identified, and these viruses are known to infect mammalian host species.
Anellovirus.
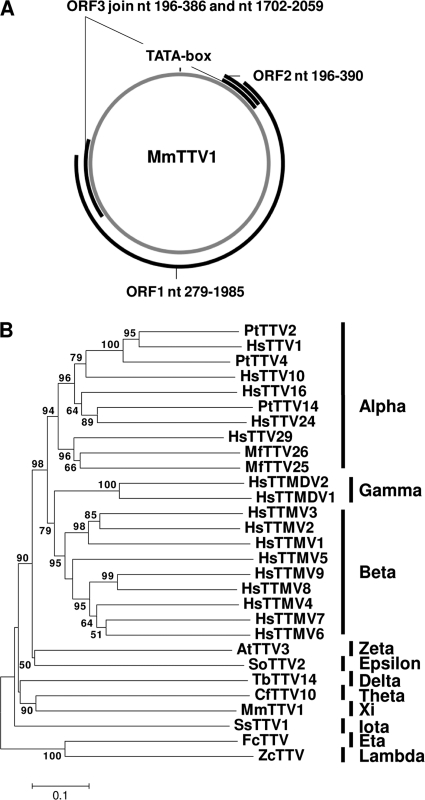
In addition to next-generation sequencing, rolling circle amplification was employed, using Illustra Templiphi 100 amplification kit (GE Healthcare), according to the instructions of the manufacturer, to acquire complete circular viral genome sequences from pine marten rectal swab VS4700004 and European badger rectal swab VS4700006 (7, 26). The anellovirus genome sequence from sample VS4700004 was designated MmTTV1 (for Martes martes torque teno virus 1) and was determined by Sanger sequencing of full-length genomes obtained by rolling circle amplification, which confirmed the sequences obtained via next-generation sequencing (GenBank accession no. JN704611). Open reading frame 1 (ORF1), ORF2, and ORF3 sequences with homology to the corresponding ORFs in other anelloviruses were identified, as well as a TATA box (Fig. 1A) (4, 18, 30, 31). MmTTV1 ORF1 nucleotide or protein sequences were aligned to the complete ORF1 sequences of other anelloviruses in GenBank using Clustal X2 (20). A taxonomic proposal submitted to the International Committee on the Taxonomy of Viruses (ICTV) by the Anelloviridae-Circoviridae Study Group proposes that genera in the family Anelloviridae are defined as having >56% divergence in the nucleotide sequence of ORF1 (25). Divergence analysis using _p_-distance calculated with MEGA4 (37) demonstrated that the anellovirus MmTTV1 was in general >56% divergent on the nucleotide level from anelloviruses identified in wildlife, among dogs, California sea lions, douroucoulis, and tupaias (Table 2 and data not shown), with the torque teno tamarin virus SoTTV2 (Saguinus oedipus torque teno virus 2) being the only exception with 55.3%, which is at the border of defining a new genus. Neighbor-joining phylogenetic trees were generated using the ORF1 amino acid alignments, which underlined the nucleotide divergence analysis (Fig. 1B). Our data therefore suggest that the torque teno virus species identified in pine martens belongs to a new genus.
Fig 1.
Phylogenetic analysis of pine marten torque teno virus. (A) Genome organization of the pine marten torque teno virus. The black boxes represent ORF1 to ORF3. The location of the TATA box is indicated. nt, nucleotide. (B) A phylogenetic tree of the amino acid sequences of anellovirus ORF1 was generated by using MEGA4, the neighbor-joining method with _p_-distance, and 1,000 bootstrap replicates. Significant bootstrap values are shown. The different anellovirus genera are indicated to the right of the phylogenetic tree by the black lines. HsTTVx, human (Homo sapiens) torque teno virus genotype x; PtTTVx, Pan troglodytes torque teno virus genotype x; MfTTVx; Macaca fuscata torque teno virus genotype x; HsTTMDVx, human torque teno midi virus genotype x; HsTTMVx, human torque teno mini virus genotype x; SoTTVx, Saguinus oedipus torque teno virus genotype x; AtTTVx, Aotes trivirgatus torque teno virus genotype x; CfTTVx, Canis familiaris torque teno virus genotype x; TbTTVx, Tupaia belangeri chinensis torque teno virus genotype x; FcTTVx, Felis catus torque teno virus genotype x; SsTTVx, Sus sucrofa torque teno virus genotype x; ZcTTV, Zalophus californianus torque teno virus, MmTTVx, Martes martes torque teno virus genotype x.
Table 2.
Pairwise sequence distance between ORF1 nucleotide sequences for the indicated anelloviruses
| Virus | Pairwise sequence distance (%) | ||||
|---|---|---|---|---|---|
| SoTTV2 | AtTTV3 | CfTTV10 | TbTTV14 | MmTTV1 | |
| SoTTV2 | 0 | 49.7 | 57.3 | 59.7 | 55.3 |
| AtTTV3 | 0 | 60 | 62.8 | 60 | |
| CfTTV10 | 0 | 62.3 | 58.1 | ||
| TbTTV14 | 0 | 60.9 | |||
| MmTTV1 | 0 |
To obtain insight regarding the prevalence of anelloviruses in pine martens, a degenerate universal anellovirus PCR targeting the untranslated region of the anellovirus genome was performed on pine marten rectal swabs by the method of Ninomiya and coworkers (27). All four pine martens were positive for anelloviruses, indicating that anelloviruses are prevalent among pine martens. PCR fragments from pine marten swabs VS4700001 to VS4700003 were cloned, and eight clones were sequenced per sample. Two different anellovirus variants were observed in rectal swab VS4700001, and the PCR fragments of the nontranslated region of pine marten anelloviruses from rectal swabs VS4700001 to VS4700004 showed ∼48 to 100% similarity to each other, suggesting that pine martens are infected with different anellovirus variants.
Geminivirus-like and circovirus-like virus.
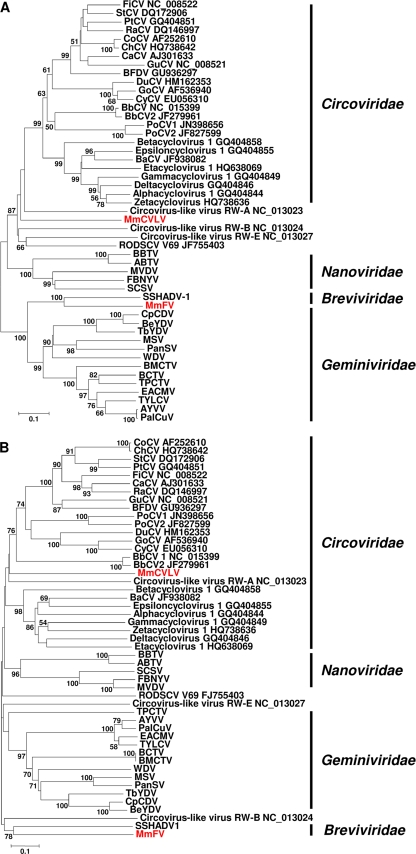
Two viral genomes were obtained by rolling circle amplification from sample VS4700006 and sequenced using Sanger sequencing on multiple clones in both directions. One virus (2,199 bp) showed homology to a recently described geminivirus-related DNA mycovirus SSHADV-1 (42). The fact that the identified European badger (Meles meles) fecal virus (MmFV) was amplified by rolling circle amplification suggests that it contains a circular single-stranded DNA (ssDNA) genome, as was previously proposed for SSHADV-1 (42). MmFV contains two large ORFs, one encoding a putative capsid protein on the sense strand and the other on the complementary-sense strand coding for a putative replication initiation protein (REP). It lacks a movement protein, which is important for cell-to-cell movement of plant geminiviruses. Two intergenic regions separate the two ORFs. One of the intergenic regions of MmFV, corresponding to the large intergenic region (LIR) of SSHADV-1, has an unusual nonanucleotide TAACTTT↓GT at the apex of a potential stem-loop structure like SSHADV-1, which is recognized at the arrow by the REP protein during the initiation of virion DNA replication (data not shown) (19, 39). Phylogenetic analyses, using the neighbor-joining method with _p_-distance in the MEGA4 program (37), of the REP and capsid proteins of MmFV, SSHADV-1, and viruses in the _Gemini_-, _Circo_-, and Nanoviridae families showed that MmFV is most closely related to SSHADV-1 (Fig. 2). The REP proteins of SSHADV-1 and MmFV are most closely related to the REP proteins of geminiviruses, whereas the capsid proteins are distinct from the capsid proteins of all viruses in this analysis. MmFV and SSHADV-1 REP and capsid proteins show >70% or >90% pairwise distance to the corresponding proteins of geminiviruses (data not shown), respectively. Thus, SSHADV-1 and MmFV most likely belong to a new virus family that we provisionally named Breviviridae, after the Latin word brevis for short, which refers to the small genome of these viruses. They may even constitute different genera, based on the amino acid diversity in the capsid protein between SSHADV-1 and MMFV (∼80%), although the diversity between REP proteins (∼52%) is much less.
Fig 2.
Phylogenetic trees of the complete amino acid sequences of the REP (A) and capsid (B) of the two circular identified viruses from European badger, MmFV (GenBank accession no. JN704610) and MmCVLV (GenBank accession no. JQ085285), and selected circular ssDNA viruses in the _Gemini_-, _Nano_-, and Circoviridae families. Phylograms were generated using MEGA4, the neighbor-joining method with _p_-distance, and 1,000 bootstrap replicates. Significant bootstrap values are shown. The accession numbers and full names for individual viruses from the _Nano_- and Geminiviridae and SSHADV-1 are in reference 42. The different families are indicated to the right of the phylogenetic tree by the black lines. SSHADV-1 and MmFV seem to belong to a new family of viruses that was provisionally named Breviviridae, after the Latin word brevis for short referring to the small genome of these viruses.
The second characterized virus from European badgers showed some homology to viruses in the family Circoviridae. Circoviruses are characterized by a small circular ssDNA genome that contains two major ORFs, encoding the REP and capsid proteins, in an ambisense organization (13). The Meles meles circovirus-like virus (MmCVLV) (2,218 bp), however, contains two overlapping ORFs in the same orientation, encoding a putative REP and capsid protein. The MmCVLV genome does contain the conserved nonanucleotide sequence (CAGTATTAC) that is thought to play a role in circovirus replication and has the conserved circovirus DRYP and WWDGY motifs in the REP protein. The genome organization of MmCVLV resembles the type III circovirus-like genomes characterized from the environment and rodents (33, 34). Phylogenetic analyses, using the neighbor-joining method with _p_-distance in the program MEGA4 (37), of the REP and capsid proteins of MmCVLV and viruses in the _Gemini_-, _Circo_-, and Nanoviridae families showed that MmCVLV is most closely related to circoviruses (Fig. 2). However, on the basis of low similarities to the REP and capsid proteins of circoviruses and a genome organization that has been observed only in linear ssDNA viruses, MmCVLV most likely represents a member of a novel virus family as was described recently for the circovirus-like viruses from the environment and rodents (33, 34).
Bocavirus.
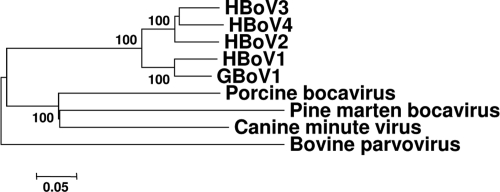
To obtain more pine marten bocavirus sequences, an additional ∼42,000 trimmed reads obtained via next-generation sequencing with a 454 GS Junior instrument (Roche) were analyzed from sample VS4700002, and a few bocavirus reads were identified. Specific primers VS656 (5′-TTCCAGGAGGATGTTTCATTGG-3′) and VS657 (5′-TTCCAGGAGGATGTTTCATTGG-3′) designed on the obtained 454 sequencing reads were used to obtain a 1,048-bp PCR amplicon of the genome region encoding VP2, using AmpliTaq Gold DNA polymerase (Roche), according to the instructions of the manufacturer. The obtained pine marten bocavirus VP2 protein sequence was aligned to the corresponding VP2 protein sequences of other bocaviruses in GenBank using Clustal X2 (20). Phylogenetic analyses, using the neighbor-joining method with _p_-distance in the MEGA4 program (37), showed that the pine marten bocavirus is most closely related to porcine bocavirus and canine minute virus (Fig. 3). The ICTV criteria for classification of bocaviruses establishes that members of each species are probably antigenically distinct, that natural infection is confined to a single host species, and that species are defined as <95% homologous in nonstructural (NS) gene DNA sequence. Although the antigenic properties of the pine marten bocavirus were not studied, the identification in a new natural host in combination with a genetic diversity of pine marten bocavirus VP2 compared to other bocavirus VP2 proteins of ∼48 to 69% on the amino acid level suggests that the pine marten bocavirus is a new bocavirus species.
Fig 3.
Phylogenetic trees of the partial amino acid sequences of the VP2 protein of the pine marten bocavirus and other selected bocaviruses. Phylograms were generated using MEGA4, with the neighbor-joining method with _p_-distance and 1,000 bootstrap replicates. Significant bootstrap values are shown. The viruses and GenBank accession numbers shown in the phylogenetic tree follow: HBoV1, human bocavirus 1 (AB480186); HBoV2, human bocavirus 2 (FJ973558); HBoV3, human bocavirus 3 (GQ867667); HBoV4, human bocavirus 4 (NC_012729); GBoV1, gorilla bocavirus 1 (NC_014358); porcine bocavirus (HM053693); canine minute virus (FJ899734); bovine parvovirus (NC_001540), pine marten bocavirus (JQ085286).
In conclusion, the majority of bacteriophage sequences in pine martens and European badgers belonged to the order Caudovirales and to single-stranded DNA viruses in the family Microviridae, as was observed before in viral metagenomics studies of the feces of horses, humans, and California sea lions (8, 9, 22). The presence of insect viruses (cypovirus) and mycoviruses (MmFV) may be attributable to the host diet. The identified MmFV and the recently described fungal virus SSHADV-1 (42) seem to differ from geminiviruses in natural host range and biological properties of the genome. In addition, the sequence identity of these two viruses to geminiviruses is low. Thus, SSHADV-1 and MmFV most likely belong to a new family of viruses (42) that we named Breviviridae.
Mammalian viruses from the _Anello_-, _Picorna_-, _Paramyxo_-, and Parvoviridae families were identified, as was a circovirus-like virus that potentially constitutes a new virus family. Pine martens and European badgers do not seem to harbor as many different mammalian viruses as California sea lions and bats do (11, 22, 23). The newly identified pine marten anellovirus (MmTTV1) belongs to a new genus that we provisionally name Xitorquevirus in analogy to the classification of torque teno viruses in nine genera named Alpha-, Beta-, Gamma-, Delta-, Epsilon-, Eta-, Iota-, Theta-, and Zetatorquevirus, and the proposed four genera Kappa-, Lambda-, Mu- and Nutorquevirus (6, 25). The results of degenerate universal anellovirus PCR (27) on rectal swabs suggested that anelloviruses are prevalent among pine martens. A high prevalence of TTV has also been described in apparently healthy humans, tupaias, tamarins, douroucoulis, swine, dogs, and cats (25, 28, 29, 32). The circovirus-like virus from a European badger, MmCVLV, most likely represents a member of a novel virus family (33, 34), and the pine marten bocavirus represents a new bocavirus species.
The discovery of a new anellovirus and bocavirus from pine marten rectal swabs and a circovirus-like virus from European badgers is an example of the needed expansion of our knowledge of the virus diversity present in the animal reservoir. In addition, a new potential mycovirus was identified from a European badger rectal swab. Sequence-independent amplification of viral nucleic acid in combination with a next-generation sequencing platform, which we used to discover these viruses, provides a relatively simple, unselective technology to identify new viral species, as was observed previously with similar techniques (8, 11, 21–23, 35, 36, 39).
Nucleotide sequence accession numbers.
GenBank accession numbers for the genomes of Martes martes torque teno virus 1 (MmTTV1), European badger (Meles meles) fecal virus (MmFV), Meles meles circovirus-like virus (MmCVLV), and the partial genome of pine marten bocavirus are JN704611, JN704610, JQ085285, and JQ085286, respectively. The GenBank accession numbers for the anelloviruses in Fig. 1 are NC_014071, NC_012126, NC_014087, EF538877, NC_002076, NC_014076, NC_014091, AB060597, AB038621, NC_014083, AB041958, NC_014480, AB041957, NC_014077, NC_014085, AB057358, AY823990, NC_009225, NC_014093, NC_014097, NC_014086, NC_014088, NC_014090, NC_014089, NC_014095, NC_014082, NC_014068, and NC_002195.
ACKNOWLEDGMENTS
The research leading to these results received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under the project “European Management Platform for Emerging and Re-emerging Infectious disease Entities” (EMPERIE) EC grant agreement number 223498.
We thank Sim Broekhuizen, Centre for Ecosystem Studies of Alterra, Wageningen University and Research Centre, and members of the Study Group on Pine Martens (WBN) of the Dutch Mammal Society for gathering and supplying rectal swab samples.
A. D. M. E. Osterhaus and S. L. Smits are the part-time chief scientific officer and senior scientist, respectively, of Viroclinics Biosciences B.V.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1.Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. U. S. A. 98:11609–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander T, et al. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendinelli M, et al. 2001. Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clin. Microbiol. Rev. 14:98–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benetka V, Leschnik M, Affenzeller N, Mostl K. 2011. Phylogenetic analysis of Austrian canine distemper virus strains from clinical samples from dogs and wild carnivores. Vet. Rec. 168:377. [DOI] [PubMed] [Google Scholar]
- 6.Biagini P. 2009. Classification of TTV and related viruses (anelloviruses). Curr. Top. Microbiol. Immunol. 331:21–33 [DOI] [PubMed] [Google Scholar]
- 7.Biagini P, et al. 2007. Circular genomes related to anelloviruses identified in human and animal samples by using a combined rolling-circle amplification/sequence-independent single primer amplification approach. J. Gen. Virol. 88:2696–2701 [DOI] [PubMed] [Google Scholar]
- 8.Breitbart M, et al. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cann AJ, Fandrich SE, Heaphy S. 2005. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30:151–156 [DOI] [PubMed] [Google Scholar]
- 10.Deem SL, Spelman LH, Yates RA, Montali RJ. 2000. Canine distemper in terrestrial carnivores: a review. J. Zoo. Wildl. Med. 31:441–451 [DOI] [PubMed] [Google Scholar]
- 11.Donaldson EF, et al. 2010. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 84:13004–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong J, Olano JP, McBride JW, Walker DH. 2008. Emerging pathogens: challenges and successes of molecular diagnostics. J. Mol. Diagn. 10:185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faurez F, Dory D, Grasland B, Jestin A. 2009. Replication of porcine circoviruses. Virol. J. 6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garten RJ, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haagmans BL, Andeweg AC, Osterhaus AD. 2009. The application of genomics to emerging zoonotic viral diseases. PLoS Pathog. 5:e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21:1552–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamahora T, Hino S, Miyata H. 2000. Three spliced mRNAs of TT virus transcribed from a plasmid containing the entire genome in COS1 cells. J. Virol. 74:9980–9986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiken T, et al. 2005. Public health. Pathogen surveillance in animals. Science 309:1680–1681 [DOI] [PubMed] [Google Scholar]
- 20.Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 21.Li L, et al. 2011. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 92:2534–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, et al. 2011. The fecal viral flora of California sea lions. J. Virol. 85:9909–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, et al. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84:6955–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losada L, et al. 2011. Genome sequencing and analysis of Yersinia pestis KIM D27, an avirulent strain exempt from select agent regulation. PLoS One 6:e19054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maggi F, Bendinelli M. 2010. Human anelloviruses and the central nervous system. Rev. Med. Virol. 20:392–407 [DOI] [PubMed] [Google Scholar]
- 26.Niel C, Diniz-Mendes L, Devalle S. 2005. Rolling-circle amplification of Torque teno virus (TTV) complete genomes from human and swine sera and identification of a novel swine TTV genogroup. J. Gen. Virol. 86:1343–1347 [DOI] [PubMed] [Google Scholar]
- 27.Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. 2008. Development of PCR assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. J. Clin. Microbiol. 46:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto H, et al. 2000. Species-specific TT viruses and cross-species infection in nonhuman primates. J. Virol. 74:1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto H, et al. 2001. Genomic and evolutionary characterization of TT virus (TTV) in tupaias and comparison with species-specific TTVs in humans and non-human primates. J. Gen. Virol. 82:2041–2050 [DOI] [PubMed] [Google Scholar]
- 30.Okamoto H, et al. 2000. TT virus mRNAs detected in the bone marrow cells from an infected individual. Biochem. Biophys. Res. Commun. 279:700–707 [DOI] [PubMed] [Google Scholar]
- 31.Okamoto H, et al. 1999. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology 259:437–448 [DOI] [PubMed] [Google Scholar]
- 32.Okamoto H, et al. 2002. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J. Gen. Virol. 83:1291–1297 [DOI] [PubMed] [Google Scholar]
- 33.Phan TG, et al. 2011. The fecal viral flora of wild rodents. PLoS Pathog. 7:e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosario K, Duffy S, Breitbart M. 2009. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 90:2418–2424 [DOI] [PubMed] [Google Scholar]
- 35.Smits SL, et al. 2010. Human astrovirus infection in a patient with new-onset celiac disease. J. Clin. Microbiol. 48:3416–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smits SL, et al. 2010. Identification and characterization of deer astroviruses. J. Gen. Virol. 91:2719–2722 [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 38.Taylor LH, Latham SM, Woolhouse ME. 2001. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Leeuwen M, et al. 2010. Human picobirnaviruses identified by molecular screening of diarrhea samples. J. Clin. Microbiol. 48:1787–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woolhouse ME, Gowtage-Sequeria S. 2005. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11:1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woolhouse ME, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20:238–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X, et al. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U. S. A. 107:8387–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]