Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi (original) (raw)
Abstract
Six DNA regions were evaluated as potential DNA barcodes for Fungi, the second largest kingdom of eukaryotic life, by a multinational, multilaboratory consortium. The region of the mitochondrial cytochrome c oxidase subunit 1 used as the animal barcode was excluded as a potential marker, because it is difficult to amplify in fungi, often includes large introns, and can be insufficiently variable. Three subunits from the nuclear ribosomal RNA cistron were compared together with regions of three representative protein-coding genes (largest subunit of RNA polymerase II, second largest subunit of RNA polymerase II, and minichromosome maintenance protein). Although the protein-coding gene regions often had a higher percent of correct identification compared with ribosomal markers, low PCR amplification and sequencing success eliminated them as candidates for a universal fungal barcode. Among the regions of the ribosomal cistron, the internal transcribed spacer (ITS) region has the highest probability of successful identification for the broadest range of fungi, with the most clearly defined barcode gap between inter- and intraspecific variation. The nuclear ribosomal large subunit, a popular phylogenetic marker in certain groups, had superior species resolution in some taxonomic groups, such as the early diverging lineages and the ascomycete yeasts, but was otherwise slightly inferior to the ITS. The nuclear ribosomal small subunit has poor species-level resolution in fungi. ITS will be formally proposed for adoption as the primary fungal barcode marker to the Consortium for the Barcode of Life, with the possibility that supplementary barcodes may be developed for particular narrowly circumscribed taxonomic groups.
Keywords: DNA barcoding, fungal biodiversity
The absence of a universally accepted DNA barcode for Fungi, the second most speciose eukaryotic kingdom (1, 2), is a serious limitation for multitaxon ecological and biodiversity studies. DNA barcoding uses standardized 500- to 800-bp sequences to identify species of all eukaryotic kingdoms using primers that are applicable for the broadest possible taxonomic group. Reference barcodes must be derived from expertly identified vouchers deposited in biological collections with online metadata and validated by available online sequence chromatograms. Interspecific variation should exceed intraspecific variation (the barcode gap), and barcoding is optimal when a sequence is constant and unique to one species (3, 4). Ideally, the barcode locus would be the same for all kingdoms. A region of the mitochondrial gene encoding the cytochrome c oxidase subunit 1 (CO1) is the barcode for animals (3, 4) and the default marker adopted by the Consortium for the Barcode of Life for all groups of organisms, including fungi (5). In Oomycota, part of the kingdom Stramenopila historically studied by mycologists, the de facto barcode internal transcribed spacer (ITS) region is suitable for identification, but the default CO1 marker is more reliable in a few clades of closely related species (6). In plants, CO1 has limited value for differentiating species, and a two-marker system of chloroplast genes was adopted (7, 8) based on portions of the ribulose 1-5-biphosphate carboxylase/oxygenase large subunit gene and a maturase-encoding gene from the intron of the _trn_K gene. This system sets a precedent for reconsidering CO1 as the default fungal barcode.
CO1 functions reasonably well as a barcode in some fungal genera, such as Penicillium, with reliable primers and adequate species resolution (67% in this young lineage) (9); however, results in the few other groups examined experimentally are inconsistent, and cloning is often required (10). The degenerate primers applicable to many Ascomycota (11) are difficult to assess, because amplification failures may not reflect priming mismatches. Extreme length variation occurs because of multiple introns (9, 12–14), which are not consistently present in a species. Multiple copies of different lengths and variable sequences occur, with identical sequences sometimes shared by several species (11). Some fungal clades, such as Neocallimastigomycota (an early diverging lineage of obligately anaerobic, zoosporic gut fungi), lack mitochondria (15). Finally, because most fungi are microscopic and inconspicuous and many are unculturable, robust, universal primers must be available to detect a truly representative profile. This availability seems impossible with CO1.
The nuclear rRNA cistron has been used for fungal diagnostics and phylogenetics for more than 20 y (16), and its components are most frequently discussed as alternatives to CO1 (13, 17). The eukaryotic rRNA cistron consists of the 18S, 5.8S, and 28S rRNA genes transcribed as a unit by RNA polymerase I. Posttranscriptional processes split the cistron, removing two internal transcribed spacers. These two spacers, including the 5.8S gene, are usually referred to as the ITS region. The 18S nuclear ribosomal small subunit rRNA gene (SSU) is commonly used in phylogenetics, and although its homolog (16S) is often used as a species diagnostic for bacteria (18), it has fewer hypervariable domains in fungi. The 28S nuclear ribosomal large subunit rRNA gene (LSU) sometimes discriminates species on its own or combined with ITS. For yeasts, the D1/D2 region of LSU was adopted for characterizing species long before the concept of DNA barcoding was promoted (19–21).
Currently, ∼172,000 full-length fungal ITS sequences are deposited in GenBank, and 56% are associated with a Latin binominal, representing ∼15,500 species and 2,500 genera, derived from ∼11,500 scientific studies in ∼500 journals. An important fraction of the sequences lacking binominals is from environmental samples (22, 23). In a smaller number of environmental studies, ITS has been used combined with LSU (24, 25). ITS is also used in some fungi for providing an indication of delimitation by a measure of the genetic distances (26). However, phylogenetic approaches are also being used to identify taxonomic units in environmental sampling of fungi (27) and are often more effective in comparison (28).
Protein-coding genes are widely used in mycology for phylogenetic analyses or species identification. For Ascomycota (including mold genera such as Aspergillus), they are generally superior to rRNA genes for resolving relationships at various taxonomic levels (29). Specialized identification databases use several markers [e.g., translation elongation factor 1-α for _Fusarium_ (30) and β-tubulin for Penicillium (31)], but there is little standardization. Available primers for such markers usually amplify a narrow taxonomic range. Among protein-coding genes, the largest subunit of RNA polymerase II (RPB1) may have potential as a fungal barcode; it is ubiquitous and single copy, and it has a slow rate of sequence divergence (32). Its phylogenetic use was shown in studies of Basidiomycota, zygomycota, Microsporidia (33–36), and some protists (37). RPB1 primers were developed for the Assembling the Fungal Tree of Life (AFToL) project, and the locus is included in the subsequent AFToL2 (38). However, its use as a barcode remains untested.
This paper stems from a multilaboratory, multinational initiative to formalize a standard DNA barcode for kingdom Fungi (excluding nonfungal organisms traditionally treated as fungi). We compared barcoding performance of three nuclear ribosomal regions (ITS, LSU, and SSU) and one region from a representative protein-coding gene, RPB1, based on probability of correct identification (PCI) and barcode gap analysis using newly generated sequences for representatives of the 17 major fungal lineages (Fig. 1). Contributors used standard primers and protocols developed by AFToL and submitted sequences to a customized database for analysis. Some also contributed sequences from regions of two additional optional genes, namely the second largest subunit of RNA polymerase II (RPB2; also an AFToL marker) (39) and a gene encoding a minichromosome maintenance protein (_MCM_7), which were chosen based on their usefulness in phylogenetic studies and ease of amplification across Ascomycota (40–42).
Fig. 1.
Dendrogram of 17 fungal lineages sampled in this study showing consensus relationships and sampling. Relationships with high levels of uncertainty are indicated by stippled lines. Lineages are labeled and listed together with the approximate number of currently described species. The currently accepted node for delineating Fungi is indicated by F. The phyla Ascomycota and Basidiomycota are indicated by A and B, respectively. Gray bars to the left indicate numbers of strains in the barcode database, with the longest bar equal to 1,176 strains. Black bars indicate the proportions selected for a PCI analysis. The four datasets analyzed for PCI are numbered 1–4: 1, Pezizomycotina; 2, Saccharomycotina; 3, Basidiomycota; 4, early diverging lineages. Pie charts indicate the proportion of success from attempts to amplify the four-marker regions in the following order: ITS, LSU, SSU, and RPB1. Black, successful PCRs and sequences; gray, uncertain cases where no report was given; white, unsuccessful PCR.
Results
We compared the barcoding performance of four markers using newly generated sequences from 742 strains or specimens, with two additional protein-coding markers analyzed for a smaller subset of about 200 fungi. Our taxon sampling was comprehensive and covered the main fungal lineages, with heavier sampling in the most speciose clades. Comparisons of PCI for all combinations of ITS, LSU, SSU, and RPB1 for all Fungi are shown in Figs. S1 and S2. We attempted to include Glomeromycota in the four-marker comparison, but RPB1 could only be amplified for some species of Glomeraceae. A simplified analysis of ITS vs. LSU in Glomeromycota (Fig. S3) indicated high levels of intraspecific variation in this group. We were unable to include Neocallimastigomycota because of the absence of sufficient sequence data spanning the full length of the ribosomal cistron. We omitted the Cryptomycota and Microsporidia clades; arguments for and against their inclusion within Fungi continue (43, 44), although they presently are classified within the kingdom. For practical reasons, we had to assume that species concepts used by the taxonomists in the consortium were accurate and consistent, relying on the current circumscription of each species as assessed by the participants’ expertise. Genealogical concordance phylogenetic species recognition is commonly applied in mycology (45).
PCR Success.
The survey (Fig. S4) showed that PCR amplifications of ribosomal RNA genes were more reliable across the Fungi than the protein-coding markers (Fig. 1). As expected, the success varied by taxonomic group [e.g., ITS PCR amplification success ranged from 100% (Saccharomycotina) to 65% (early diverging lineages)]. Ranges for the other ribosomal markers were similar. In comparison, success for RPB1 varied from 80% (Saccharomycotina) to 14% (basal lineages). About 80% of respondents reported no problems with PCR amplification of ITS, 90% scored it as easy to obtain a high-quality PCR product, and 80% reported no significant sequencing concerns. In comparison, >70% reported PCR amplification problems for RPB1; 40–50% reported primer failure as the biggest problem.
Species Identification.
We performed several analyses to allow direct comparison of the barcoding use of the four main markers under consideration (i.e., ITS, LSU, SSU, and RPB1) (Figs. 2 and 3). To assess the PCI, data were divided into subsets according to taxonomic affinity. The combined four-marker PCI comparisons (Fig. 2) included 742 samples, with 142 species represented by more than one sample and 84 species represented with one sample. With all taxa considered, the PCI of ITS (0.73) was marginally lower than RPB1 (0.76). RPB1 consistently yielded high levels of species discrimination in all of the fungal groups except the early diverging lineages, which is comparable with multigene combinations (Fig. 2). Within Dikarya, ITS had the most resolving power for species discrimination in Basidiomycota (0.77 vs. 0.67 for RPB1). For Pezizomycotina, the PCI of RPB1 (0.80) outperformed ITS (0.71). ITS had lower discriminatory power than SSU and LSU in early diverging lineages, but margins of error were high. LSU had variable levels of PCI (0.66–0.75) among all groups but was generally lower than RPB1 or ITS (Fig. 2). In Saccharomycotina, LSU had the lowest PCI (0.67), but all four markers performed similarly. SSU was consistently the worst performing marker, with the lowest species discrimination in Pezizomycotina (Fig. 2) and Basidiomycota (Fig. 2). In the early diverging lineages (Fig. 2), SSU had a better PCI, on par with LSU and better than both ITS and RPB1.
Fig. 2.
Barcode gap probability of identification for the four-marker datasets of ITS, LSU, SSU, and RPB1. The plots show the combinations of barcode markers investigated on the y axis. I, ITS; L, LSU; S, SSU; R, RPB1. The x axis shows the barcode gap PCI estimate for Ascomycota, Pezizomycotina (142 species), Basidiomycota (43 species), Ascomycota, Saccharomycotina (13 species), early diverging lineages (8 species), and combined groups (206 species). The error bars indicate 95% confidence intervals for the PCI estimate.
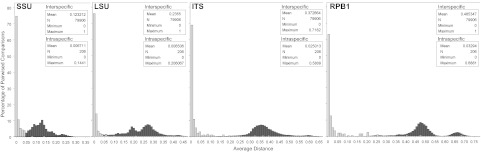
Fig. 3.
Barcode gap analyses using distance histograms for each marker. Histograms display intraspecific variation in light gray and interspecific variation in dark gray. Inserts summarize distance data.
In the multigene combinations, the most effective two genes in the combined analysis were either ITS and RPB1 or LSU and RPB1, both yielding a PCI of 0.78. This finding represented an increase of 0.02 from the highest-ranked single gene. The highest-ranked three- and four-gene combinations gave comparable increases.
Two supplementary three-marker comparisons expanded diversity for some major clades underrepresented in the four-gene analysis. For lichen-forming fungi, SSU was often absent, because the protocols favored amplicons from the photobiont rather than the fungus. Eliminating the requirement for SSU allowed more intensive sampling, yielding 683 sequences that included 179 species represented by more than one sample and 117 species represented by one sample (Fig. S5_A_). There was no apparent difference in ranking of the four candidate barcodes for the Pezizomycotina compared with the four-gene comparison in this analysis. Similarly, early diverging lineages yielded only 43 RPB1 sequences, and a comparison of ribosomal markers (ITS, SSU, and LSU) allowed inclusion of a larger set of 152 samples, with 34 species represented by more than one sample and 50 species by one sample. In this dataset, all sequences were unique to their species (Fig. S5_B_), and there was again no difference from the original four-gene comparison.
The barcode gap analyses (Fig. 3) largely confirmed the trends seen in the PCI analysis. The clearest indication of a barcode gap is seen for RPB1 followed by ITS. LSU and SSU performed poorly, each lacking a significant barcode gap.
To test whether other single-copy protein-coding markers might have a similar barcoding performance to RPB1, RPB2 and MCM7 sequences were tested for a subset of taxa. Neither yielded data from the early diverging lineages, but a combination of remaining groups yielded 207 strains, including 55 species with more than one sample and 23 species with one sample (Fig. S6). For both markers, all sequences were unique to their species. The two supplementary genes had a similar barcoding performance to RPB1, with RPB2 yielding slightly superior results followed by RPB1 and MCM7.
Discussion
Overall, ribosomal markers had fewer problems with PCR amplification than protein-coding markers (Fig. 1 and Fig. S4). Based on overall performance in species discrimination, SSU had almost no barcode gap (46) and the worst combined PCI, and it can be eliminated as a candidate locus (Figs. 2 and 3). LSU, a favored phylogenetic marker among many mycologists, had virtually no amplification, sequencing, alignment, or editing problems, and the barcode gap was superior to the SSU. However, across the fungal kingdom, ITS was generally superior to LSU in species discrimination and had a more clearly defined barcode gap (Fig. 3). The overall probability of correct species identification using ITS is comparable with the success reported for the two-marker plant barcode system (0.73 vs. 0.70) (7). Higher species identification success can be expected in the major macrofungal groups in Basidiomycota (0.79), and slightly lower success can be expected in the economically important microfungal groups in filamentous Ascomycota (0.75). ITS performed as a close second to the most heavily sampled of our protein-coding markers, RPB1. However, the much higher PCR amplification success rate for ITS is a critical difference in its performance as a barcode (Fig. 1). ITS primers used in this study were applied to a range of fungal lineages, and several primers function as almost universal primers. However, all primer sets have a range of biases, and an appropriate solution will be to use more than one primer combination (47).
Taking all these arguments into account, we propose ITS as the standard barcode for fungi. The proposal will satisfy most fungal biologists but not all. Given the fungal kingdom's age and genetic diversity, it is unlikely that a single-marker barcode system will be capable of identifying every specimen or culture to species level. Furthermore, the limitations of ITS sequences for identifying species in some groups and the failure of the universal ITS primers to work in a minority of other groups will have to be carefully documented (14, 43, 48). ITS sequences shared among different species have already been documented in species-rich Pezizomycotina genera with shorter amplicons, such as the economically important genera Cladosporium (49), Penicillium (50), and Fusarium (51). In Aspergillus, ITS sequences are identical in several complexes of critical mycotoxigenic, industrial, and medically important species, and additional markers are necessary (52). Although the ITS region is a potentially effective DNA barcode in several lichenized lineages (53), genetic drift may prevent lineage sorting of ancestral polymorphisms in some slowly evolving groups.
Intragenomic variation, such as the existence of multiple paralogous or nonorthologous copies within single fruiting bodies of basidiomycetes (54, 55) and ascomycetes (56) or within axenic cultures (51), may lead to higher estimates of infraspecific variability (57, 58) or generation of barcodes that act only as representative sequences of multiple variable repeats (59, 60). Highly variable lengths and high evolutionary rates for the nuclear ribosomal cistron in species of Cantharellus, Tulasnella (Cantherellales, Basidiomycota) (61–63), and some lichens (53) may provide challenges for sequencing and analysis. The upper range of this ITS region variation is likely found in the Glomeromycota, with up to 20% divergence within a single multinucleate spore (64, 65).
We acknowledge that species delimitations vary from one fungal group to another and are often influenced by scarcity of sampling and lack of detailed biological knowledge (43, 45). This influence is reflected when ITS distances are compared between phyla, subphyla, and species (Figs. S7 and S8). In an expanded dataset of ITS sequences from our fungal DNA barcoding database, the highest variation was most often found in the early diverging lineages. This finding confirms the fact that fungal diversity remains poorly sampled with DNA sequences for these lineages (43, 48). It is, therefore, very likely that high divergence reflects the presence of multiple cryptic species, indicating important focal points for additional study. Despite these challenges, ITS combines the highest resolving power for discriminating closely related species with a high PCR and sequencing success rate across a broad range of Fungi.
In addition to Fungi, ITS may also be applicable as a barcode for other organisms. Its use has already been shown in Chlorophyta and plants (66, 67) as well as in Oomycota (6). The possibility of multikingdom analyses of complex ecosystems like soil using the species-informative, stable, high copy number ITS mirrors the original vision of DNA barcoding, and it already seems feasible, for example, to amplify Fungi and other eukaryotes from soil (23).
Protein-coding genes are popular phylogenetic markers in mycology, and they are used as de facto barcodes of limited taxonomic scope in several groups of fungi. We chose RPB1 as a representative marker to include in our broad comparisons, with RPB2 and MCM7 analyzed for a smaller sampling. In general, such protein markers had more species resolving power, but PCR and sequencing failures eliminate them as potential universal barcodes for the broad phylogenetic scope of the kingdom Fungi. Reliable kingdom-wide PCR amplification needs to be tested for other widely used protein-coding markers, such as translation elongation factor 1-α, β-tubulin, or actin.
The possibility of a two-marker barcoding system for fungi, as adopted for plants, is often discussed among mycologists, particularly researchers working on ascomycetous yeasts (19–21) and Glomeromycota (68) who prefer a system combining ITS and LSU. Data from this study (Fig. S5) indicate that ITS and LSU perform very similarly as barcodes and that differences in these sequences correlate well with current species concepts. Combinations of both ITS and LSU sequences are also applied in environmental sampling (69), where tandem amplification can allow simultaneous species identification with ITS and phylogenetic analysis with LSU. Our analyses with two-, three-, or four-marker barcode systems (Fig. 2) reveal only a modest increase in the PCI over a single-marker ITS barcode. The need for a second marker depends on the intended purpose of an investigation (i.e., whether a broad and general survey is intended or whether particular critical species are being monitored). If these taxa are taxa with low ITS interspecific variability, secondary markers must be used to accurately report genetic diversity (70). Genome mining efforts have identified a few single-copy genes that might be amenable for broad-range priming, and these efforts should continue (71, 72).
Although the genome diversity of fungal species is studied with increasing intensity, the vast majority of fungal species remains unknown. The recent discovery of a ubiquitous fungal class from soil (73) and a diverse early diverging phylum, Cryptomycota, tied to Rozella (74–76) from riverine and marine sites illuminates this fact. More than 90% of Fungi may be awaiting discovery, posing a tremendous pressure to increase the pace of fungal species discovery (1, 2). In addition to this, the Melbourne Botanical Congress has recently approved large-scale changes to the process of naming fungi (77), and sequence data from type specimens will increasingly be essential to the stability of fungal nomenclature. Continuing discovery of novel biodiversity while classifying knowledge already available will demand well-coordinated initiatives, and DNA barcoding has a crucial role to play.
Materials and Methods
DNA Isolation, Amplification, and Sequencing.
DNA was isolated and purified from cultures or specimens using the methods routinely used by the participating laboratories. Similarly, PCR protocols (Table S1) and thermocyclers varied from laboratory to laboratory. PCR primers were those primers from the AFToL project (Table S1). Several samples were sent by contributors for PCR amplification and sequencing at LifeTech. For PCR at LifeTech, 1–2 μL fungal DNA were amplified in a final volume of 30 μL with 15 μL AmpliTaq Gold 360 Mastermix, PCR primers, and water. All forward primers contained the M13F-20 sequencing primer, and reverse primers included the M13R-27 sequencing primer. PCR products (3 μL) were enzymatically cleaned before cycle sequencing with 1 μL ExoSap-IT and 1 μL Tris EDTA and incubated at 37 °C for 20 min followed by 80 °C for 15 min. Cycle sequencing reactions contained 5 μL cleaned PCR product, 2 μL BigDye Terminator v3.1 Ready Reaction Mix, 1 μL 5× Sequencing Buffer, 1.6 pmol M13F or M13R sequencing primer, and water in a final volume of 10 μL. The standard cycle sequencing protocol was 27 cycles of 10 s at 96 °C, 5 s at 50 °C, 4 min at 60 °C, and hold at 4 °C. Sequencing cleaning was performed with the BigDye XTerminator Purification Kit as recommended by the manufacturer for 10-μL volumes. Sequencing reactions were analyzed on a 3730xl Genetic Analyzer.
Sampling.
Closely related but separately named asexual and sexual species were coded with one genus name and then divided into subsets to allow taxonomically targeted assessment of markers for each major clade (Fig. 1). From the barcoding database of 2,920 samples, we selected a subset of 742 strains with sequences for all four markers (ITS, LSU, SSU, and RPB1). This subset was divided into four taxonomically delimited datasets: 416 strains in Pezizomycotina (filamentous ascomycetes), 81 strains in Saccharomycotina (ascomycete yeasts), 202 strains in Basidiomycota, and 43 strains from the combined polyphyletic early diverging lineages. Two additional analyses were performed for samples with three markers to enhance evaluation of certain undersampled lineages: the first analysis for 683 strains of Pezizomycotina with ITS, LSU, and RPB1 sequences and the second analysis for 152 representatives of basal lineages with ITS, LSU, and SSU sequences. Finally, a six-marker comparison was made for a selection of 207 strains of Pezizomycotina, Basidiomycota, and Saccharomycotina, with the first four markers supplemented with the two optional markers, MCM7 and RPB2. The species and strains used in the analysis are shown in Dataset S1.
PCR success.
Participants recorded their experience on the success of PCR amplification and sequencing for the genes and taxa that they contributed to this study. They also documented specific problems with PCR, quality of PCR amplification, primer problems (PCR and sequencing), and whether cloning was required. The genes were ranked for their ability to discriminate species and their overall taxonomic and phylogenetic use in specialized taxonomic groups. Comments were parsed to identify taxon-specific problems and are summarized in Fig. S4.
Data Analyses.
Database.
A query-based BioloMICS database (78) was established for 2,920 strains (1,022 species including subspecies) provided by >70 members of the consortium (www.fungalbarcoding.org). The complete database sets consist of 213 different genera and 915 unique species; there was an average of four species per genus and three strains per species. Approximately one-third (1,029) of the strains were scored as sibling species of other species in the sample, with 156 unique sibling species groups. All data are based on deposited voucher specimens or cultures identified by taxonomic specialists. The database allowed pairwise sequence alignments or polyphasic identifications using one or any combination of the six genes used in this study. The taxon sampling covered 15 of 17 major lineages attributed to the Fungi (Fig. 1) that were weighted to species-rich higher taxa such as the Pezizomycotina (the largest group of Ascomycota) and the Agaricomycotina (mushrooms and other macrobasidiomycetes).
PCI.
For each dataset, we calculated the barcode gap PCI. All alignments used the BLAST default DNA scoring system (79, 80). Two kinds of sequence alignment were calculated between every sample pair, namely (i) a global alignment using the Needleman–Wunsch algorithm, which aligns the entire sequence length with penalties for gaps at the alignment ends (81), and (ii) a semiglobal alignment using a variant Needleman–Wunsch algorithm that includes both ends of one sequence and finds the alignment with the highest score without penalizing end gaps in the other sequence. The latter algorithm does the same for the other sequence, returning the alignment with the higher of the two scores. Thus, the global alignment matches the whole length of two sequences, and the semiglobal alignment matches one sequence to a subset of the other and then vice versa. Semiglobal alignment checks whether disparate sequence lengths degrade species identification; if they do not, global and semiglobal alignment should result in similar identifications. For the two types of alignment, the _p_-distance (the proportion of aligned nucleotide pairs consisting of differing nucleotides) was calculated. The sequence diameter of a species is defined as the greatest _p_-distance between any two samples from within a species. Based on the sequence diameter, correct identification of a species occurs if, for every sample in the species, no sample from another species lies within the sequence diameter. The corresponding barcode gap PCI is the fraction of species correctly identified (7). The Wilson score interval yielded 95% confidence intervals for each PCI estimate (82). PCI was also calculated for all possible combinations of two, three, or four genes to evaluate the potential payoff of a multigene barcoding system.
Sequence divergence and DNA gap analyses.
Using the same dataset as for the PCI analysis, a DNA barcode gap analysis was performed using matrix algebra and Statistic Analysis Software (SAS Institute) as described previously (6) except that the lower triangular uncorrected distance matrix was calculated using mothur (83). The results are indicated in Fig. 3. Additional comparisons were done and are described in Figs. S2, S3, and S7–S9.
Supplementary Material
Supporting Information
Acknowledgments
We thank David L. Hawksworth, Martin Bidartondo, and numerous other colleagues for critical comments. This work was organized under the Fungal Working Group of the Consortium for the Barcode of Life (CBOL), which provided support from its funding from the Alfred P. Sloan Foundation. Support was also provided by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health, Life Technologies Corporation, and the individual funders to authors who provided sequences for our analysis. Publication charges were provided by the International Barcode of Life Network from Genome Canada through the Ontario Genomics Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in GenBank. Sequences are listed in Dataset S1.
2A complete list of the Fungal Barcoding Consortium can be found in the SI Appendix.
References
- 1.Blackwell M. The Fungi: 1, 2, 3 … 5.1 million species? Am J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. How many species are there on Earth and in the ocean? PLoS Biol. 2011;9:e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003;270(Suppl 1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindel DE, Miller SE. DNA barcoding a useful tool for taxonomists. Nature. 2005;435:17. doi: 10.1038/435017b. [DOI] [PubMed] [Google Scholar]
- 6.Robideau GP, et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour. 2011;11:1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollingsworth PM, et al. CBOL Plant Working Group A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kress WJ, et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc Natl Acad Sci USA. 2009;106:18621–18626. doi: 10.1073/pnas.0909820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seifert KA, et al. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc Natl Acad Sci USA. 2007;104:3901–3906. doi: 10.1073/pnas.0611691104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dentinger BTM, Didukh MY, Moncalvo JM. Comparing COI and ITS as DNA barcode markers for mushrooms and allies (Agaricomycotina) PLoS One. 2011;6:e25081. doi: 10.1371/journal.pone.0025081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmore SR, Gräfenhan T, Louis-Seize G, Seifert KA. Multiple copies of cytochrome oxidase 1 in species of the fungal genus Fusarium. Mol Ecol Resour. 2009;9(Suppl S1):90–98. doi: 10.1111/j.1755-0998.2009.02636.x. [DOI] [PubMed] [Google Scholar]
- 12.Vialle A, et al. Evaluation of mitochondrial genes as DNA barcode for Basidiomycota. Mol Ecol Resour. 2009;9:99–113. doi: 10.1111/j.1755-0998.2009.02637.x. [DOI] [PubMed] [Google Scholar]
- 13.Rossman AY. Report of the planning workshop for all fungi DNA Barcoding. Inoculum. 2007;58:1–5. [Google Scholar]
- 14.Seifert KA. The all-fungi barcoding campaign (FunBOL) Persoonia. 2008;20:106. [Google Scholar]
- 15.Bullerwell CE, Lang BF. Fungal evolution: The case of the vanishing mitochondrion. Curr Opin Microbiol. 2005;8:362–369. doi: 10.1016/j.mib.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Begerow D, Nilsson H, Unterseher M, Maier W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl Microbiol Biotechnol. 2010;87:99–108. doi: 10.1007/s00253-010-2585-4. [DOI] [PubMed] [Google Scholar]
- 17.Eberhardt U. A constructive step towards selecting a DNA barcode for fungi. New Phytol. 2010;187:265–268. doi: 10.1111/j.1469-8137.2010.03329.x. [DOI] [PubMed] [Google Scholar]
- 18.Stackebrandt E, Goebel BM. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44:846–849. [Google Scholar]
- 19.Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- 20.Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 21.Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. Systematics of basidiomycetous yeasts: A comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 22.Buée M, et al. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 2009;184:449–456. doi: 10.1111/j.1469-8137.2009.03003.x. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 2005;71:5544–5550. doi: 10.1128/AEM.71.9.5544-5550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geml J, et al. Molecular phylogenetic biodiversity assessment of arctic and boreal ectomycorrhizal Lactarius Pers. (Russulales; Basidiomycota) in Alaska, based on soil and sporocarp DNA. Mol Ecol. 2009;18:2213–2227. doi: 10.1111/j.1365-294X.2009.04192.x. [DOI] [PubMed] [Google Scholar]
- 25.Taylor DL, et al. Increasing ecological inference from high throughput sequencing of fungi in the environment through a tagging approach. Mol Ecol Resour. 2008;8:742–752. doi: 10.1111/j.1755-0998.2008.02094.x. [DOI] [PubMed] [Google Scholar]
- 26.Del-Prado R, et al. Genetic distances within and among species in monophyletic lineages of Parmeliaceae (Ascomycota) as a tool for taxon delimitation. Mol Phylogenet Evol. 2010;56:125–133. doi: 10.1016/j.ympev.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Geml J, Laursen GA, Taylor DL. Molecular diversity assessment of arctic and boreal Agaricus taxa. Mycologia. 2008;100:577–589. doi: 10.3852/07-042r1. [DOI] [PubMed] [Google Scholar]
- 28.Porter TM, Golding GB. Are similarity- or phylogeny-based methods more appropriate for classifying internal transcribed spacer (ITS) metagenomic amplicons? New Phytol. 2011;192:775–782. doi: 10.1111/j.1469-8137.2011.03838.x. [DOI] [PubMed] [Google Scholar]
- 29.Schoch CL, et al. The Ascomycota tree of life: A phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- 30.O'Donnell K, et al. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J Clin Microbiol. 2010;48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frisvad JC, Samson RA. Polyphasic taxonomy of Penicillium subgenus Penicillium—a guide to identification of food and air-borne terverticillate penicillia and their mycotoxins. Stud Mycol. 2004;49:1–173. [Google Scholar]
- 32.Tanabe Y, Watanabe , Sugiyama Are Microsporidia really related to Fungi?: A reappraisal based on additional gene sequences from basal fungi. Mycol Res. 2002;106:1380–1391. [Google Scholar]
- 33.Cheney SA, Lafranchi-Tristem NJ, Bourges D, Canning EU. Relationships of microsporidian genera, with emphasis on the polysporous genera, revealed by sequences of the largest subunit of RNA polymerase II (RPB1) J Eukaryot Microbiol. 2001;48:111–117. doi: 10.1111/j.1550-7408.2001.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 34.Garnica S, Weiß M, Oertel B, Ammirati J, Oberwinkler F. Phylogenetic relationships in Cortinarius, section Calochroi, inferred from nuclear DNA sequences. BMC Evol Biol. 2009;9:1. doi: 10.1186/1471-2148-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matheny PB, Liu YJJ, Ammirati JF, Hall BD. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales) Am J Bot. 2002;89:688–698. doi: 10.3732/ajb.89.4.688. [DOI] [PubMed] [Google Scholar]
- 36.Tanabe Y, Saikawa M, Watanabe MM, Sugiyama J. Molecular phylogeny of Zygomycota based on EF-1alpha and RPB1 sequences: Limitations and utility of alternative markers to rDNA. Mol Phylogenet Evol. 2004;30:438–449. doi: 10.1016/s1055-7903(03)00185-4. [DOI] [PubMed] [Google Scholar]
- 37.Longet D, Pawlowski J. Higher-level phylogeny of Foraminifera inferred from the RNA polymerase II (RPB1) gene. Eur J Protistol. 2007;43:171–177. doi: 10.1016/j.ejop.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin DJ, Hibbett DS, Lutzoni F, Spatafora JW, Vilgalys R. The search for the fungal tree of life. Trends Microbiol. 2009;17:488–497. doi: 10.1016/j.tim.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 39.James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 40.Aguileta G, et al. Assessing the performance of single-copy genes for recovering robust phylogenies. Syst Biol. 2008;57:613–627. doi: 10.1080/10635150802306527. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt I, et al. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia. 2009;23:35–40. doi: 10.3767/003158509X470602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raja HA, Schoch CL, Hustad VP, Shearer CA, Miller AN. Testing the phylogenetic utility of MCM7 in the Ascomycota. MycoKeys. 2011;1:63–94. [Google Scholar]
- 43.Voigt K, Kirk PM. Recent developments in the taxonomic affiliation and phylogenetic positioning of fungi: Impact in applied microbiology and environmental biotechnology. Appl Microbiol Biotechnol. 2011;90:41–57. doi: 10.1007/s00253-011-3143-4. [DOI] [PubMed] [Google Scholar]
- 44.Lee SC, et al. Evolution of the sex-related locus and genomic features shared in Microsporidia and Fungi. PLoS One. 2010;5:e10539. doi: 10.1371/journal.pone.0010539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor JW, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Anonymous Guidelines for CBOL Approval of Non-COI Barcode Regions. Available at http://www.barcoding.si.edu/pdf/guidelines%20for%20non-co1%20selection%20final.pdf. Accessed March 8, 2012.
- 47.Bellemain E, et al. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinform Online. 2008;4:193–201. doi: 10.4137/ebo.s653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert K, et al. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol. 2007;58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skouboe P, et al. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycol Res. 1999;103:873–881. [Google Scholar]
- 51.O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 52.Geiser DM, et al. The current status of species recognition and identification in Aspergillus. Stud Mycol. 2007;59:1–10. doi: 10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly LJ, et al. DNA barcoding of lichenized fungi demonstrates high identification success in a floristic context. New Phytol. 2011;191:288–300. doi: 10.1111/j.1469-8137.2011.03677.x. [DOI] [PubMed] [Google Scholar]
- 54.Lindner DL, Banik MT. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia. 2011;103:731–740. doi: 10.3852/10-331. [DOI] [PubMed] [Google Scholar]
- 55.Smith ME, Douhan GW, Rizzo DM. Intra-specific and intra-sporocarp ITS variation of ectomycorrhizal fungi as assessed by rDNA sequencing of sporocarps and pooled ectomycorrhizal roots from a Quercus woodland. Mycorrhiza. 2007;18:15–22. doi: 10.1007/s00572-007-0148-z. [DOI] [PubMed] [Google Scholar]
- 56.Kovács GM, Balázs TK, Calonge FD, Martín MP. The diversity of Terfezia desert truffles: New species and a highly variable species complex with intrasporocarpic nrDNA ITS heterogeneity. Mycologia. 2011;103:841–853. doi: 10.3852/10-312. [DOI] [PubMed] [Google Scholar]
- 57.Gomes EA, Kasuya MCM, de Barros EG, Borges AC, Araujo EF. Polymorphism in the internal transcribed spacer (ITS) of the ribosomal DNA of 26 isolates of ectomycorrhizal fungi. Genet Mol Biol. 2002;25:477–483. [Google Scholar]
- 58.Simon UK, Weiß M. Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol Biol Evol. 2008;25:2251–2254. doi: 10.1093/molbev/msn188. [DOI] [PubMed] [Google Scholar]
- 59.Ganley ARD, Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007;17:184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.James SA, et al. Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res. 2009;19:626–635. doi: 10.1101/gr.084517.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor DL, McCormick MK. Internal transcribed spacer primers and sequences for improved characterization of basidiomycetous orchid mycorrhizas. New Phytol. 2008;177:1020–1033. doi: 10.1111/j.1469-8137.2007.02320.x. [DOI] [PubMed] [Google Scholar]
- 62.Moncalvo JM, et al. The cantharelloid clade: Dealing with incongruent gene trees and phylogenetic reconstruction methods. Mycologia. 2006;98:937–948. doi: 10.3852/mycologia.98.6.937. [DOI] [PubMed] [Google Scholar]
- 63.Buyck B, Cruaud C, Couloux A, Hofstetter V. Cantharellus texensis sp. nov. from Texas, a southern lookalike of C. cinnabarinus revealed by tef-1 sequence data. Mycologia. 2011;103:1037–1046. doi: 10.3852/10-261. [DOI] [PubMed] [Google Scholar]
- 64.Stockinger H, Walker C, Schüssler A. ‘Glomus intraradices DAOM197198’, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 2009;183:1176–1187. doi: 10.1111/j.1469-8137.2009.02874.x. [DOI] [PubMed] [Google Scholar]
- 65.Krüger M, Krüger C, Walker C, Stockinger H, Schüßler A. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol. 2012;193:970–984. doi: 10.1111/j.1469-8137.2011.03962.x. [DOI] [PubMed] [Google Scholar]
- 66.Buchheim MA, et al. Internal transcribed spacer 2 (nu ITS2 rRNA) sequence-structure phylogenetics: Towards an automated reconstruction of the green algal tree of life. PLoS One. 2011;6:e16931. doi: 10.1371/journal.pone.0016931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D-Z, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA. 2011;108:19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stockinger H, Krüger M, Schüßler A. DNA barcoding of arbuscular mycorrhizal fungi. New Phytol. 2010;187:461–474. doi: 10.1111/j.1469-8137.2010.03262.x. [DOI] [PubMed] [Google Scholar]
- 69.Gorfer M, et al. Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fungal Divers. 2010;44:65–75. doi: 10.1007/s13225-010-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gazis R, Rehner S, Chaverri P. Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Mol Ecol. 2011;20:3001–3013. doi: 10.1111/j.1365-294X.2011.05110.x. [DOI] [PubMed] [Google Scholar]
- 71.Lewis CA, et al. Identification of fungal DNA barcode targets and PCR primers based on Pfam protein families and taxonomic hierarchy. Open Appl Inform J. 2011;5:30–44. [Google Scholar]
- 72.Robert V, et al. The quest for a general and reliable fungal DNA barcode. Open Appl Inform J. 2011;5:45–61. [Google Scholar]
- 73.Rosling A, et al. Archaeorhizomycetes: Unearthing an ancient class of ubiquitous soil fungi. Science. 2011;333:876–879. doi: 10.1126/science.1206958. [DOI] [PubMed] [Google Scholar]
- 74.Jones MDM, et al. Discovery of novel intermediate forms redefines the fungal tree of life. Nature. 2011;474:200–203. doi: 10.1038/nature09984. [DOI] [PubMed] [Google Scholar]
- 75.Lara E, Moreira D, López-García P. The environmental clade LKM11 and Rozella form the deepest branching clade of fungi. Protist. 2010;161:116–121. doi: 10.1016/j.protis.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 76.Jones MDM, Richards TA, Hawksworth DL, Bass D. Validation and justification of the phylum name Cryptomycota phyl. nov. IMA Fungus. 2011;2:173–175. doi: 10.5598/imafungus.2011.02.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hawksworth D. A new dawn for the naming of fungi: Impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys. 2011;1:7–20. doi: 10.5598/imafungus.2011.02.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robert VA, et al. BioloMICS software: Biological data management, identification, classification and statistics. Open Appl Inform J. 2011;5:87–98. [Google Scholar]
- 79.Altschul SF. Hot papers in Bioinformatics—Gapped BLAST and PSI-BLAST: A new generation of protein database search programs by S.F. Altschul, T.L. Madden, A.A. Schaffer, J.H. Zhang, Z. Zhang, W. Miller, D.J. Lipman—Comments. Scientist. 1999;13:15. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 82.Wilson EB. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc. 1927;22:209–212. [Google Scholar]
- 83.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information